Causal Discovery in High-Dimensional Point Process Networks with Hidden Nodes
Abstract
:1. Introduction
2. The Hawkes Processes with Unobserved Components
2.1. The Hawkes Process
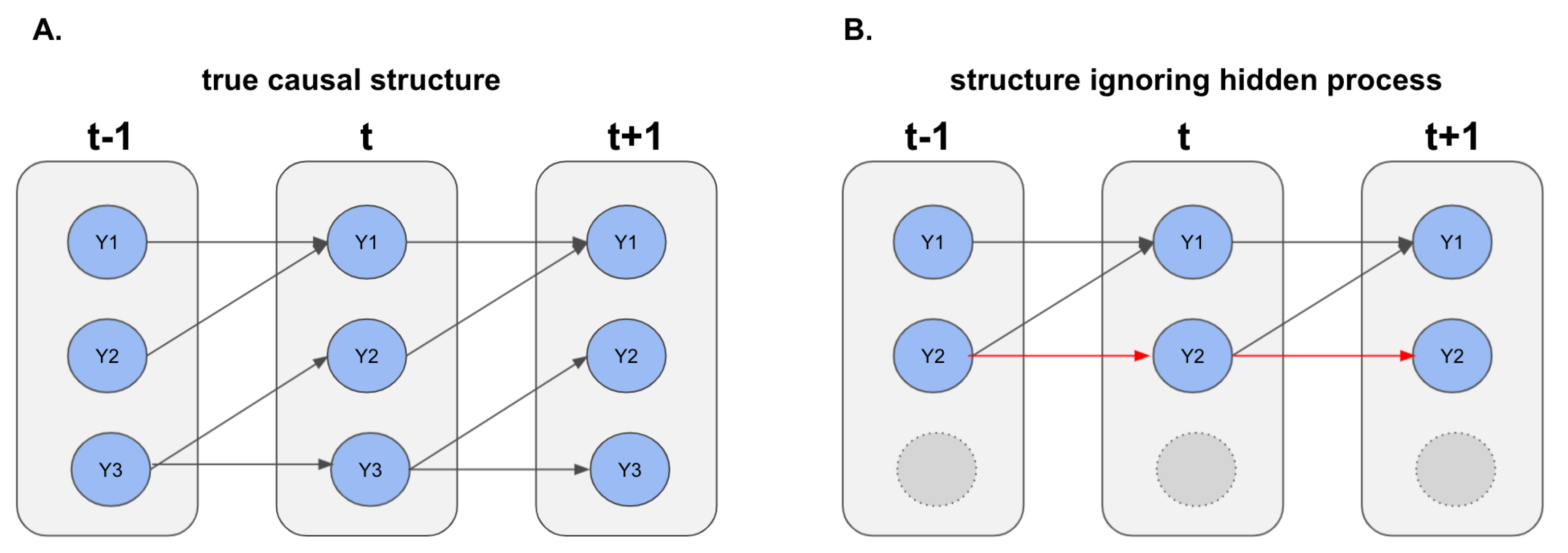
2.2. The Confounded Hawkes Process
3. Estimating Causal Effects in Confounded Hawkes Processes
3.1. Extending Trim Regression to Hawkes Processes
3.2. An Alternative Approach
4. Theoretical Properties
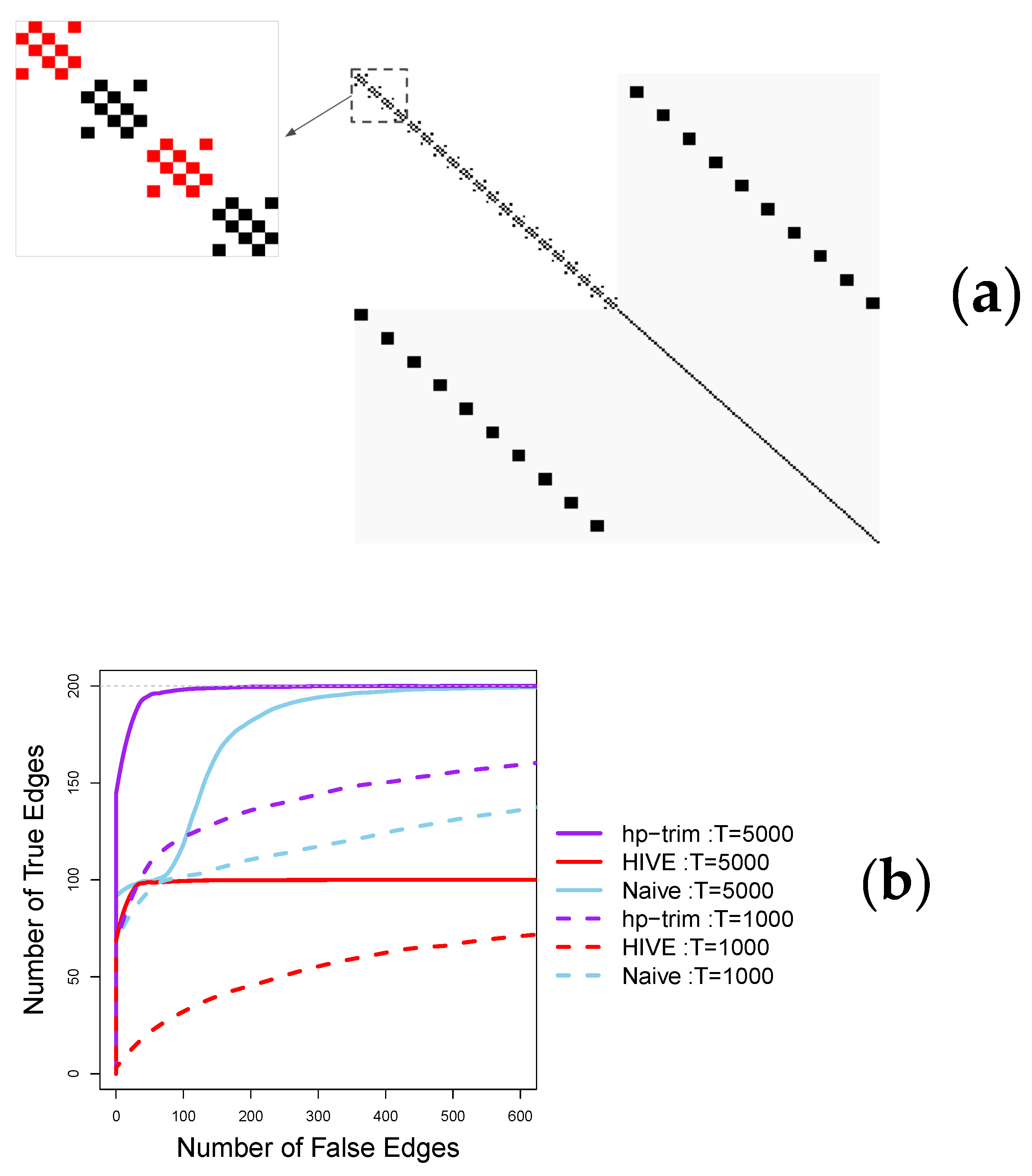
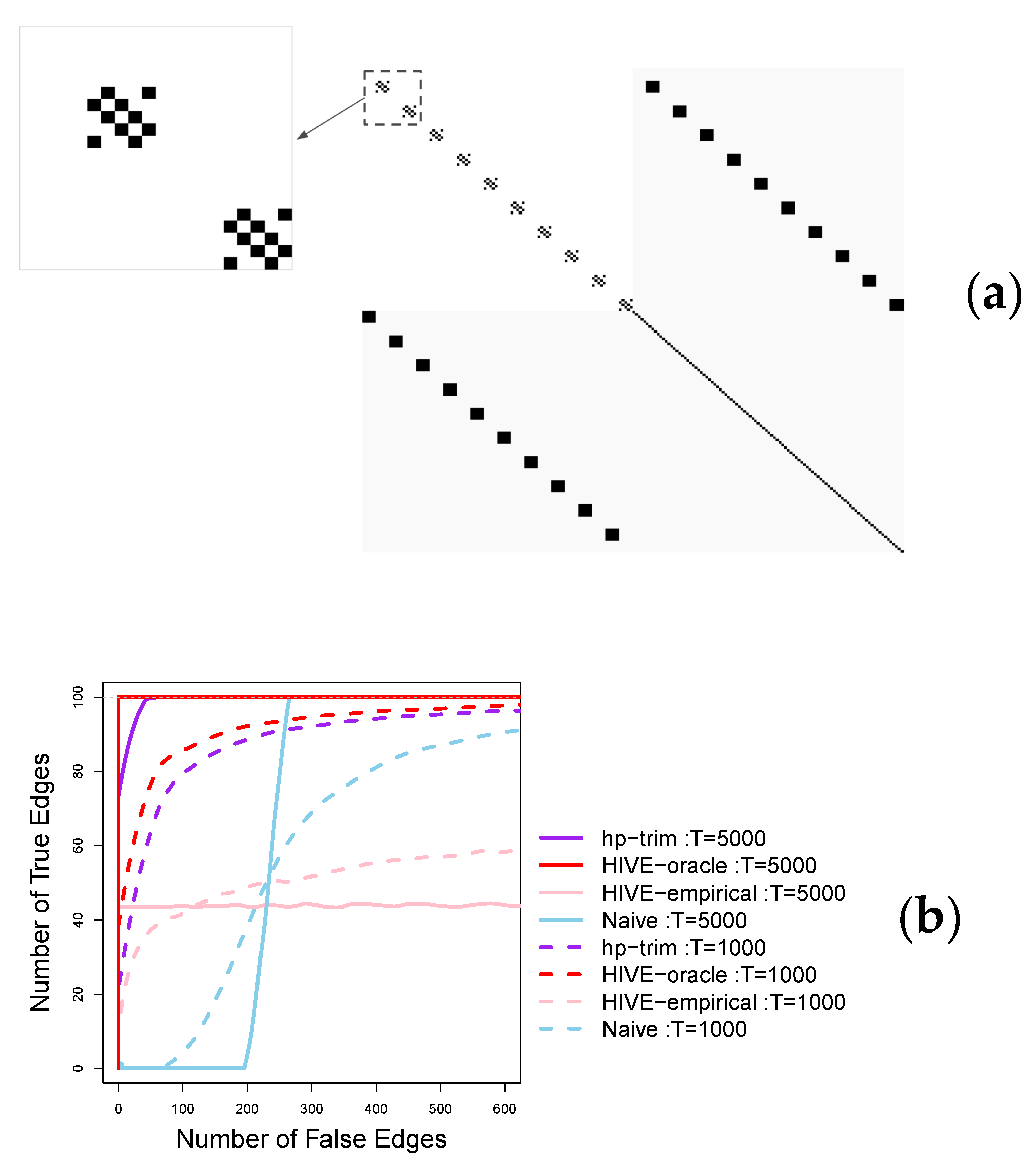
5. Simulation Studies
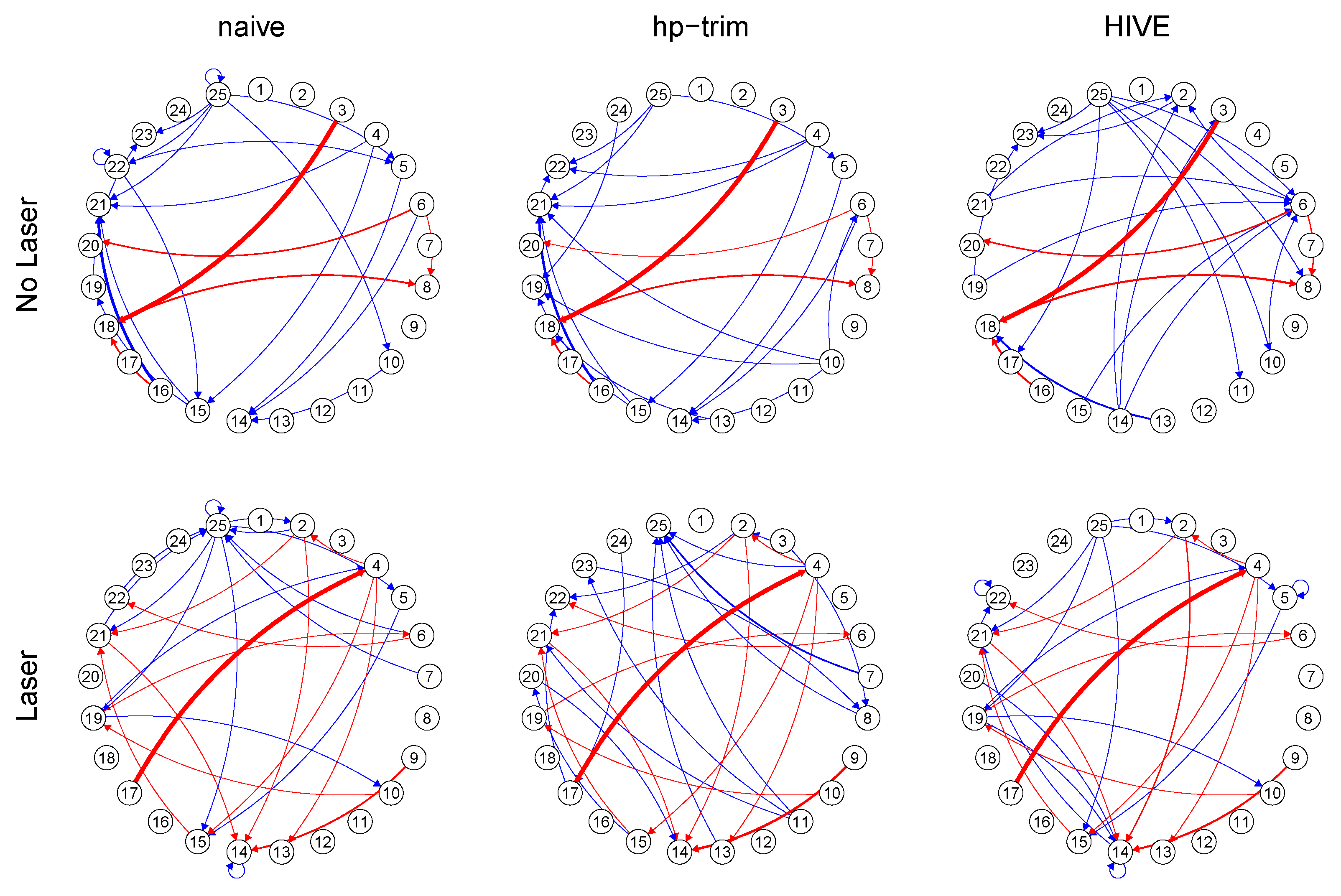
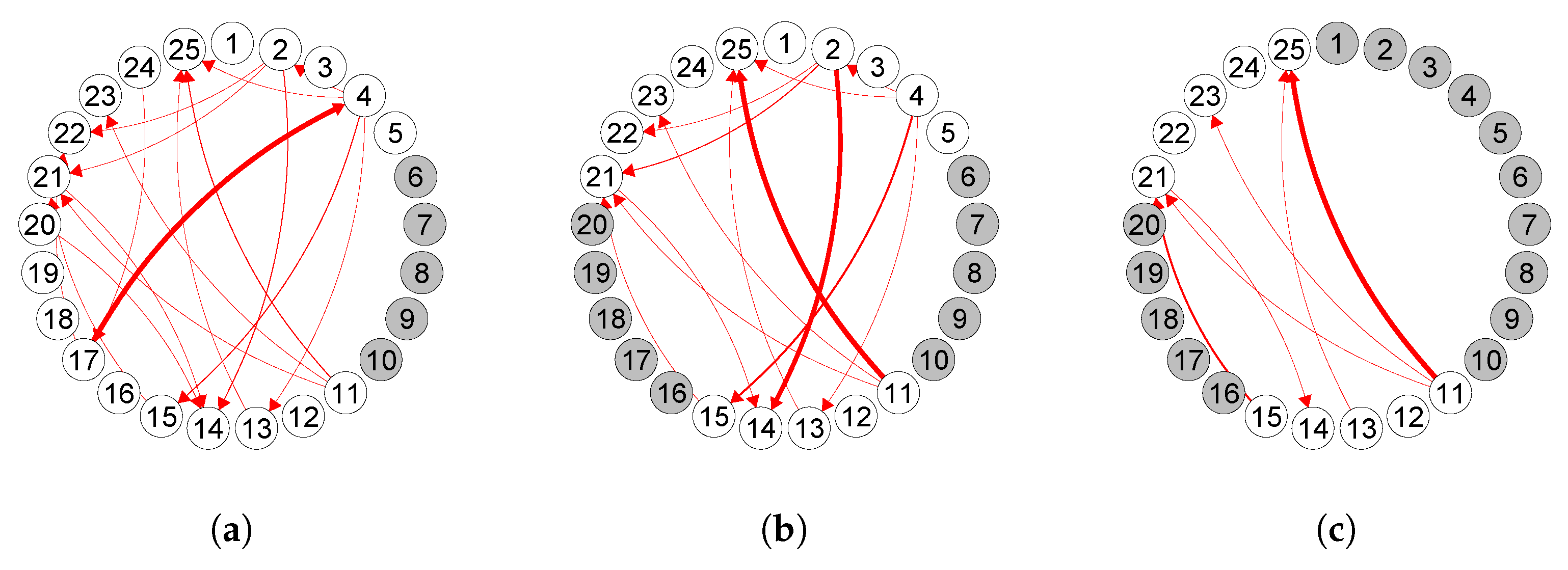
6. Analysis of Mouse Spike Train Data
7. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Additional Details on HIVE
Appendix B. Proof of Main Results
Appendix C. Parameter Estimation Performance

References
- Glymour, C.; Zhang, K.; Spirtes, P. Review of causal discovery methods based on graphical models. Front. Genet. 2019, 10, 524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shojaie, A.; Fox, E.B. Granger causality: A review and recent advances. arXiv 2021, arXiv:2105.02675. [Google Scholar] [CrossRef]
- Reid, A.T.; Headley, D.B.; Mill, R.D.; Sanchez-Romero, R.; Uddin, L.Q.; Marinazzo, D.; Lurie, D.J.; Valdés-Sosa, P.A.; Hanson, S.J.; Biswal, B.B.; et al. Advancing functional connectivity research from association to causation. Nat. Neurosci. 2019, 22, 1751–1760. [Google Scholar] [CrossRef]
- Breitung, J.; Swanson, N.R. Temporal aggregation and spurious instantaneous causality in multiple time series models. J. Time Ser. Anal. 2002, 23, 651–665. [Google Scholar] [CrossRef]
- Silvestrini, A.; Veredas, D. Temporal aggregation of univariate and multivaraite time series models: A survey. J. Econ. Surv. 2008, 22, 458–497. [Google Scholar] [CrossRef]
- Tank, A.; Fox, E.B.; Shojaie, A. Identifiability and estimation of structural vector autoregressive models for subsampled and mixed-frequency time series. Biometrika 2019, 106, 433–452. [Google Scholar] [CrossRef]
- Soudry, D.; Keshri, S.; Stinson, P.; hwan Oh, M.; Iyengar, G.; Paninski, L. A shotgun sampling solution for the common input problem in neural connectivity inference. arXiv 2014, arXiv:1309.3724. [Google Scholar]
- Yang, Y.; Qiao, S.; Sani, O.G.; Sedillo, J.I.; Ferrentino, B.; Pesaran, B.; Shanechi, M.M. Modelling and prediction of the dynamic responses of large-scale brain networks during direct electrical stimulation. Nat. Biomed. Eng. 2021, 5, 324–345. [Google Scholar] [CrossRef] [PubMed]
- Bloch, J.; Greaves-Tunnell, A.; Shea-Brown, E.; Harchaoui, Z.; Shojaie, A.; Yazdan-Shahmorad, A. Cortical network structure mediates response to stimulation: An optogenetic study in non-human primates. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lin, F.H.; Ahveninen, J.; Raij, T.; Witzel, T.; Chu, Y.H.; Jääskeläinen, I.P.; Tsai, K.W.K.; Kuo, W.J.; Belliveau, J.W. Increasing fMRI sampling rate improves Granger causality estimates. PLoS ONE 2014, 9, e100319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Zhang, Y.; Xiao, Y.; Cai, D. Analysis of sampling artifacts on the Granger causality analysis for topology extraction of neuronal dynamics. Front. Comput. Neurosci. 2014, 8, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prevedel, R.; Yoon, Y.G.; Hoffmann, M.; Pak, N.; Wetzstein, G.; Kato, S.; Schrödel, T.; Raskar, R.; Zimmer, M.; Boyden, E.S.; et al. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat. Methods 2014, 11, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Okatan, M.; Wilson, M.A.; Brown, E.N. Analyzing functional connectivity using a network likelihood model of ensemble neural spiking activity. Neural Comput. 2005, 17, 1927–1961. [Google Scholar] [CrossRef] [PubMed]
- Bolding, K.A.; Franks, K.M. Recurrent cortical circuits implement concentration-invariant odor coding. Science 2018, 361, 6407. [Google Scholar] [CrossRef] [PubMed]
- Berényi, A.; Somogyvári, Z.; Nagy, A.J.; Roux, L.; Long, J.D.; Fujisawa, S.; Stark, E.; Leonardo, A.; Harris, T.D.; Buzsáki, G. Large-scale, high-density (up to 512 channels) recording of local circuits in behaving animals. J. Neurophysiol. 2014, 111, 1132–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trong, P.K.; Rieke, F. Origin of correlated activity between parasol retinal ganglion cells. Nat. Neurosci. 2008, 11, 1343–1351. [Google Scholar] [CrossRef] [Green Version]
- Tchumatchenko, T.; Geisel, T.; Volgushev, M.; Wolf, F. Spike correlations—What can they tell about synchrony? Front. Neurosci. 2011, 5, 68. [Google Scholar] [CrossRef] [Green Version]
- Huang, H. Effects of hidden nodes on network structure inference. J. Phys. A Math. Theor. 2015, 48, 355002. [Google Scholar] [CrossRef] [Green Version]
- Spirtes, P.; Glymour, C.; Scheines, R. Causation, Prediction, and Search, 2nd ed.; MIT Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Malinsky, D.; Spirtes, P. Causal structure learning from multivariate time series in settings with unmeasured confounding. Proceedings of 2018 ACM SIGKDD Workshop on Causal Disocvery, London, UK, 20 August 2018; Le, T.D., Zhang, K., Kıcıman, E., Hyvärinen, A., Liu, L., Eds.; PMLR: London, UK, 2018; Volume 92, pp. 23–47. [Google Scholar]
- Chen, W.; Drton, M.; Shojaie, A. Causal structural learning via local graphs. arXiv 2021, arXiv:2107.03597. [Google Scholar]
- Shojaie, A.; Michailidis, G. Penalized likelihood methods for estimation of sparse high-dimensional directed acyclic graphs. Biometrika 2010, 97, 519–538. [Google Scholar] [CrossRef]
- Hawkes, A.G. Spectra of some self-exciting and mutually exciting point processes. Biometrika 1971, 58, 83–90. [Google Scholar] [CrossRef]
- Eichler, M.; Dahlhaus, R.; Dueck, J. Graphical modeling for multivariate Hawkes processes with nonparametric link functions. J. Time Ser. Anal. 2017, 38, 225–242. [Google Scholar] [CrossRef] [Green Version]
- Bacry, E.; Muzy, J. First- and second-order statistics characterization of Hawkes processes and non-parametric estimation. IEEE Trans. Inf. Theory 2016, 62, 2184–2202. [Google Scholar] [CrossRef]
- Brillinger, D.R. Maximum likelihood analysis of spike trains of interacting nerve cells. Biol. Cybern. 1988, 59, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.H. Point process models of single-neuron discharges. J. Comput. Neurosci. 1996, 3, 275–299. [Google Scholar] [CrossRef]
- Krumin, M.; Reutsky, I.; Shoham, S. Correlation-based analysis and generation of multiple spike trains using Hawkes models with an exogenous input. Front. Comput. Neurosci. 2010, 4, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pernice, V.; Staude, B.; Cardanobile, S.; Rotter, S. How structure determines correlations in neuronal networks. PLoS Comput. Biol. 2011, 7, e1002059. [Google Scholar] [CrossRef] [PubMed]
- Reynaud-Bouret, P.; Rivoirard, V.; Tuleau-Malot, C. Inference of functional connectivity in neurosciences via Hawkes processes. In Proceedings of the 2013 IEEE Global Conference on Signal and Information Processing, Austin, TX, USA, 3–5 December 2013; pp. 317–320. [Google Scholar]
- Truccolo, W. From point process observations to collective neural dynamics: Nonlinear Hawkes process GLMs, low-dimensional dynamics and coarse graining. J. Physiol.-Paris 2016, 110, 336–347. [Google Scholar] [CrossRef]
- Lambert, R.C.; Tuleau-Malot, C.; Bessaih, T.; Rivoirard, V.; Bouret, Y.; Leresche, N.; Reynaud-Bouret, P. Reconstructing the functional connectivity of multiple spike trains using Hawkes models. J. Neurosci. Methods 2018, 297, 9–21. [Google Scholar] [CrossRef]
- Hansen, N.R.; Reynaud-Bouret, P.; Rivoirard, V. Lasso and probabilistic inequalities for multivariate point processes. Bernoulli 2015, 21, 83–143. [Google Scholar] [CrossRef]
- Chen, S.; Shojaie, A.; Shea-Brown, E.; Witten, D. The multivariate Hawkes process in high dimensions: Beyond mutual excitation. arXiv 2019, arXiv:1707.04928. [Google Scholar]
- Wang, X.; Kolar, M.; Shojaie, A. Statistical inference for networks of high-dimensional point processes. arXiv 2020, arXiv:2007.07448. [Google Scholar]
- Bing, X.; Ning, Y.; Xu, Y. Adaptive estimation of multivariate regression with hidden variables. arXiv 2020, arXiv:2003.13844. [Google Scholar]
- Ćevid, D.; Bühlmann, P.; Meinshausen, N. Spectral deconfounding via perturbed sparse linear models. arXiv 2020, arXiv:1811.05352. [Google Scholar]
- Linderman, S.; Adams, R. Discovering latent network structure in point process data. In International Conference on Machine Learning; PMLR: Beijing, China, 2014; Volume 32. [Google Scholar]
- De Abril, I.M.; Yoshimoto, J.; Doya, K. Connectivity inference from neural recording data: Challenges, mathematical bases and research directions. Neural Netw. 2018, 102, 120–137. [Google Scholar]
- Bacry, E.; Mastromatteo, I.; Muzy, J. Hawkes processes in finance. Mark. Microstruct. Liq. 2015, 1, 1550005. [Google Scholar] [CrossRef]
- Etesami, J.; Kiyavash, N.; Zhang, K.; Singhal, K. Learning network of multivariate Hawkes processes: A time series approach. arXiv 2016, arXiv:1603.04319. [Google Scholar]
- Costa, M.; Graham, C.; Marsalle, L.; Tran, V.C. Renewal in Hawkes processes with self-excitation and inhibition. arXiv 2018, arXiv:1801.04645. [Google Scholar] [CrossRef]
- Babington, P. Neuroscience, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2001. [Google Scholar]
- Brémaud, P.; Massoulié, L. Stability of nonlinear Hawkes processes. Ann. Probab. 1996, 24, 1563–1588. [Google Scholar] [CrossRef]
- Daley, D.J.; Vere-Jones, D. An Introduction to the Theory of Point Processes: Volume I: Elementary Theory and Methods; Probability and its Applications; Springer: New York, NY, USA, 2003. [Google Scholar]
- Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. Ser. B (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Lee, S.; Sun, W.; Wright, F.A.; Zou, F. An improved and explicit surrogate variable analysis procedure by coefficient adjustment. Biometrika 2017, 104, 303–316. [Google Scholar] [CrossRef]
- Cai, B.; Zhang, J.; Guan, Y. Latent network structure learning from high dimensional multivariate point processes. arXiv 2020, arXiv:2004.03569. [Google Scholar]
- Basu, S.; Michailidis, G. Regularized estimation in sparse high-dimensional time series models. Ann. Stat. 2015, 43, 1535–1567. [Google Scholar] [CrossRef] [Green Version]
- Safikhani, A.; Shojaie, A. Joint structural break detection and parameter estimation in high-dimensional nonstationary VAR models. J. Am. Stat. Assoc. 2020, 1–14. [Google Scholar] [CrossRef]
- Shojaie, A.; Basu, S.; Michailidis, G. Adaptive thresholding for reconstructing regulatory networks from time-course gene expression data. Stat. Biosci. 2012, 4, 66–83. [Google Scholar] [CrossRef] [Green Version]
- Van de Geer, S.; Bühlmann, P.; Zhou, S. The adaptive and the thresholded Lasso for potentially misspecified models (and a lower bound for the Lasso). Electron. J. Stat. 2011, 5, 688–749. [Google Scholar] [CrossRef]
- Buhlmann, P. Statistical significance in high-dimensional linear models. Bernoulli 2013, 19, 1212–1242. [Google Scholar] [CrossRef] [Green Version]
- Paninski, L.; Pillow, J.; Lewi, J. Statistical models for neural encoding, decoding, and optimal stimulus design. In Computational Neuroscience: Theoretical Insights into Brain Function; Elsevier: Amsterdam, The Netherlands, 2007; Volume 165, pp. 493–507. [Google Scholar]
- Pillow, J.; Shlens, J.; Paninski, L.; Sher, A.; Litke, A.; Chichilnisky, E.; Simoncelli, E. Spatio-temporal correlations and visual signaling in a complete neuronal population. Nature 2008, 454, 995–999. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Cai, T.T.; Wu, Y. Heteroskedastic PCA: Algorithm, optimality, and applications. arXiv 2019, arXiv:1810.08316. [Google Scholar]
- van de Geer, S. Exponential inequalities for martingales, with application to maximum likelihood estimation for counting processes. Ann. Stat. 1995, 23, 1779–1801. [Google Scholar] [CrossRef]
- Negahban, S.; Wainwright, M. Restricted strong convexity and weighted matrix completion: Optimal bounds with noise. J. Mach. Learn. Res. 2010, 13, 1665–1697. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Shojaie, A. Causal Discovery in High-Dimensional Point Process Networks with Hidden Nodes. Entropy 2021, 23, 1622. https://doi.org/10.3390/e23121622
Wang X, Shojaie A. Causal Discovery in High-Dimensional Point Process Networks with Hidden Nodes. Entropy. 2021; 23(12):1622. https://doi.org/10.3390/e23121622
Chicago/Turabian StyleWang, Xu, and Ali Shojaie. 2021. "Causal Discovery in High-Dimensional Point Process Networks with Hidden Nodes" Entropy 23, no. 12: 1622. https://doi.org/10.3390/e23121622
APA StyleWang, X., & Shojaie, A. (2021). Causal Discovery in High-Dimensional Point Process Networks with Hidden Nodes. Entropy, 23(12), 1622. https://doi.org/10.3390/e23121622




