A Deep Learning Approach for Featureless Robust Quality Assessment of Intermittent Atrial Fibrillation Recordings from Portable and Wearable Devices
Abstract
1. Introduction
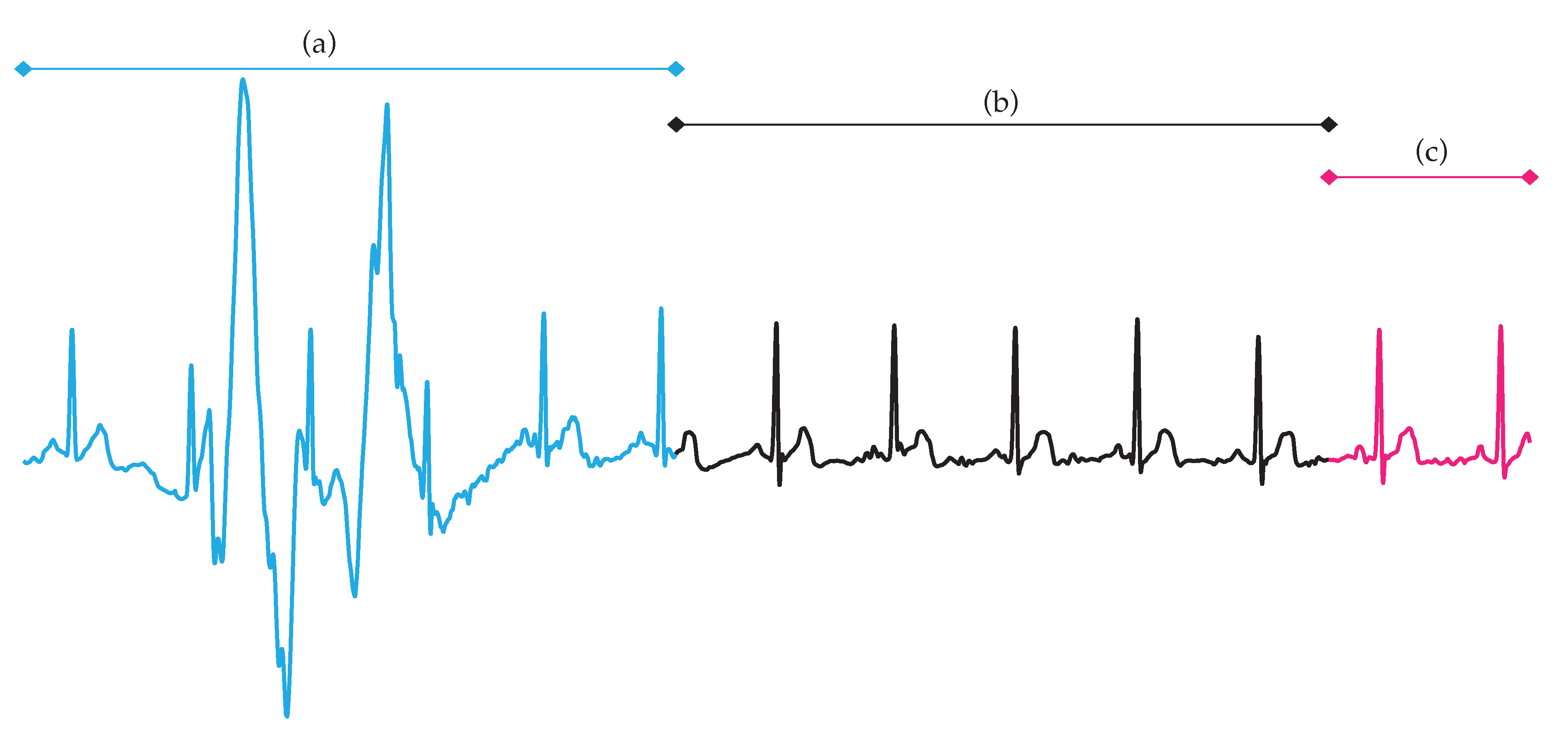
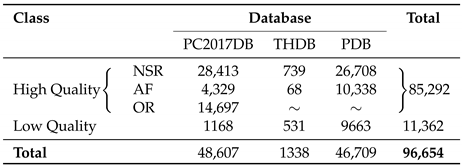
2. Databases
3. Methodology
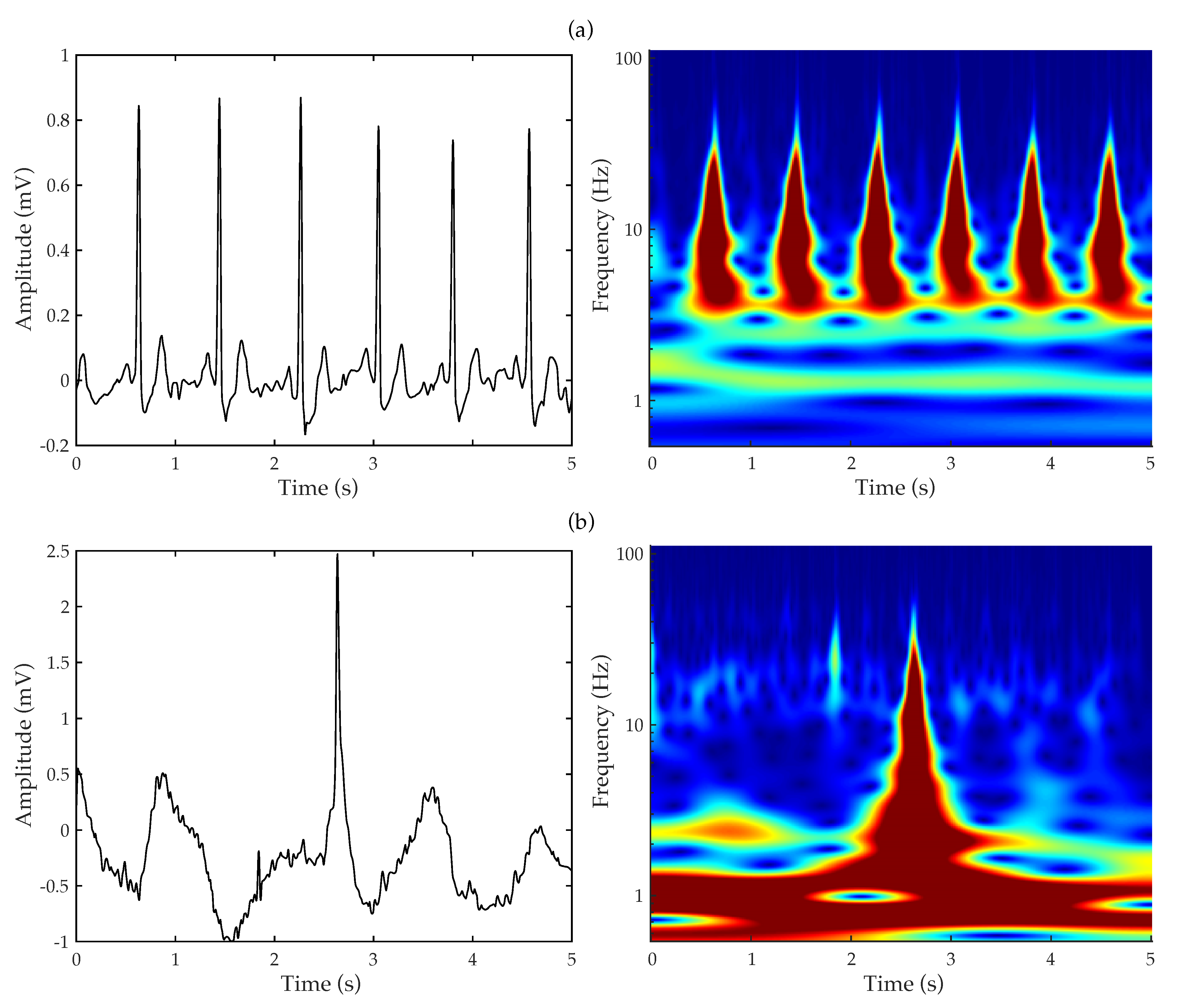
3.1. Continuous Wavelet Transform
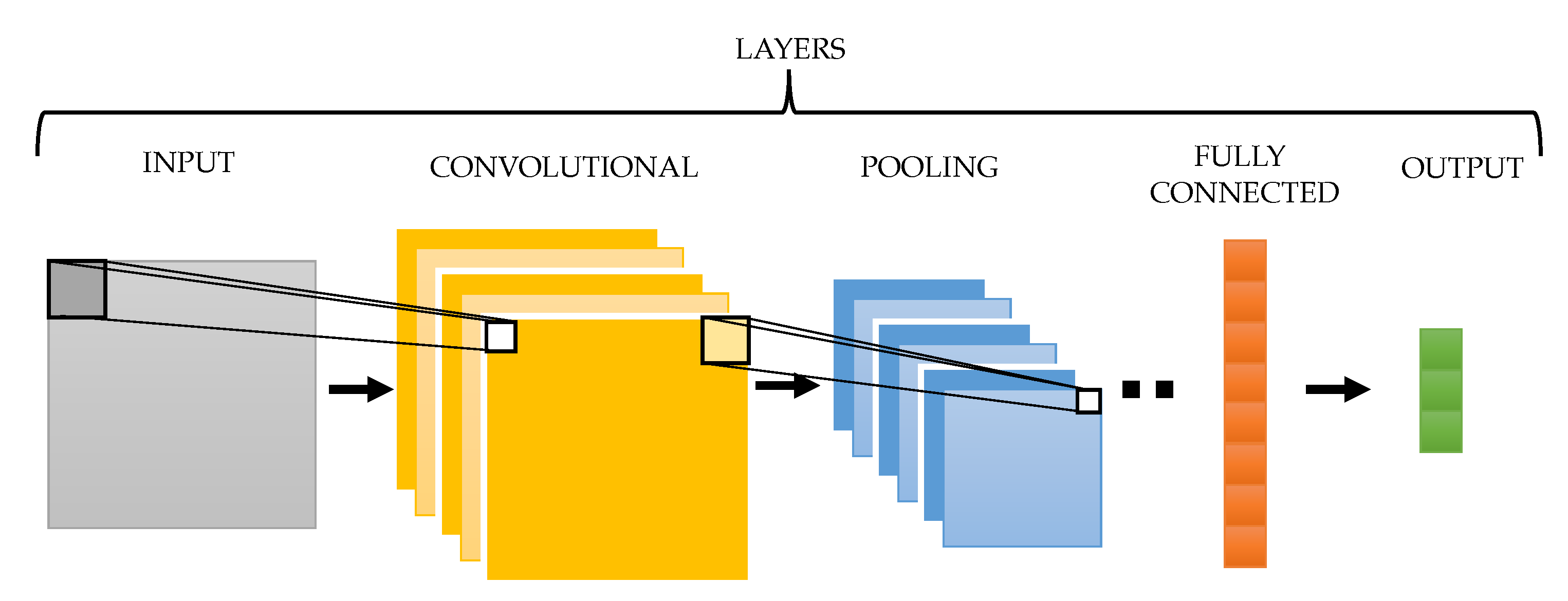
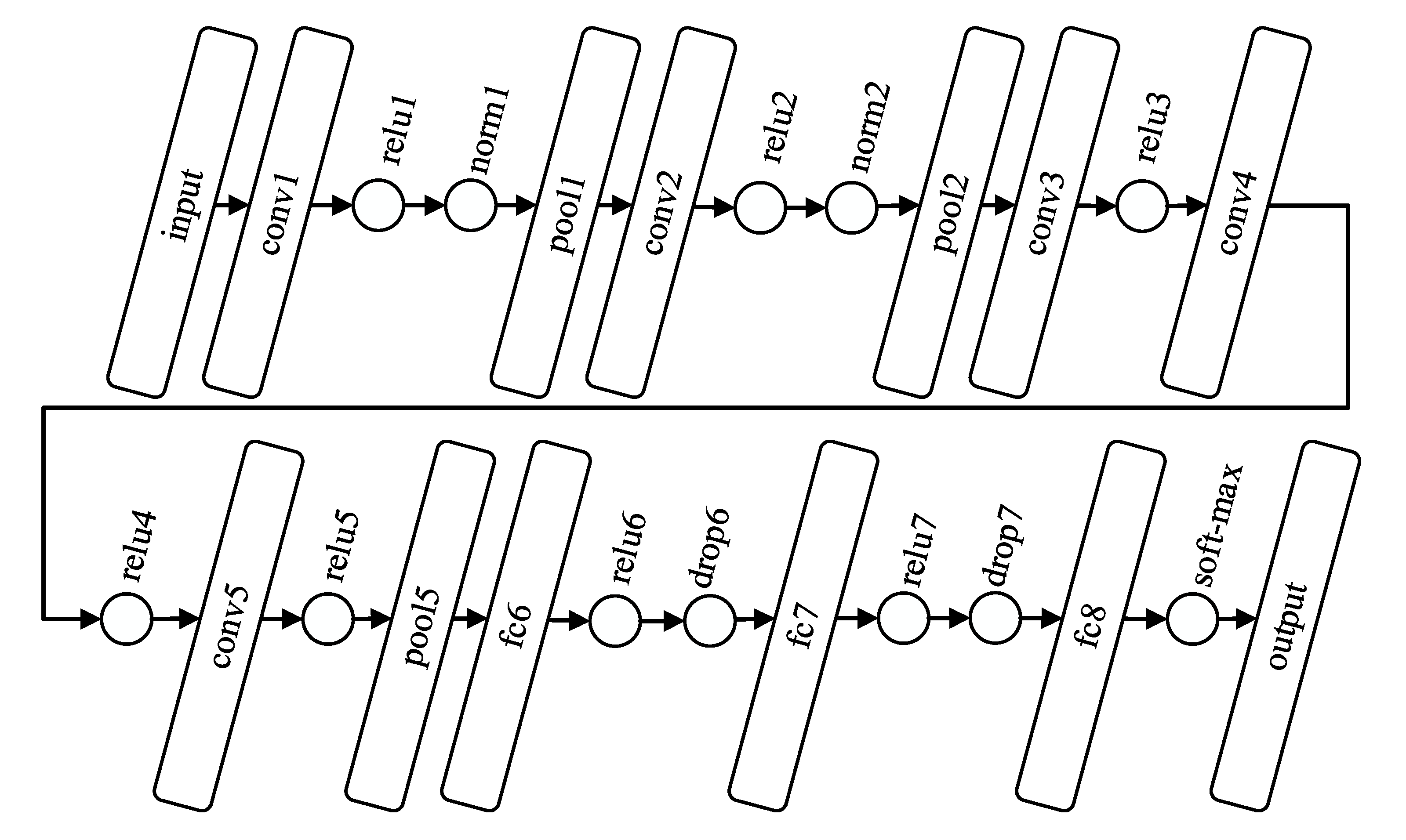
3.2. Convolutional Neural Network
3.3. Experimental Setup and Performance Assessment
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lippi, G.; Sanchis-Gomar, F.; Cervellin, G. Global epidemiology of atrial fibrillation: An increasing epidemic and public health challenge. Int. J. Stroke 2020, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Krijthe, B.P.; Kunst, A.; Benjamin, E.J.; Lip, G.Y.H.; Franco, O.H.; Hofman, A.; Witteman, J.C.M.; Stricker, B.H.; Heeringa, J. Projections on the number of individuals with atrial fibrillation in the European Union, from 2000 to 2060. Eur. Heart J. 2013, 34, 2746–2751. [Google Scholar] [CrossRef]
- Colilla, S.; Crow, A.; Petkun, W.; Singer, D.E.; Simon, T.; Liu, X. Estimates of current and future incidence and prevalence of atrial fibrillation in the U.S. adult population. Am. J. Cardiol. 2013, 112, 1142–1147. [Google Scholar] [CrossRef] [PubMed]
- Khoo, C.W.; Krishnamoorthy, S.; Lim, H.S.; Lip, G.Y.H. Atrial fibrillation, arrhythmia burden and thrombogenesis. Int. J. Cardiol. 2012, 157, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Warmus, P.; Niedziela, N.; Huć, M.; Wierzbicki, K.; Adamczyk-Sowa, M. Assessment of the manifestations of atrial fibrillation in patients with acute cerebral stroke—A single-center study based on 998 patients. Neurol. Res. 2020, 42, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Sposato, L.A.; Cipriano, L.E.; Saposnik, G.; Ruíz Vargas, E.; Riccio, P.M.; Hachinski, V. Diagnosis of atrial fibrillation after stroke and transient ischaemic attack: A systematic review and meta-analysis. Lancet Neurol. 2015, 14, 377–387. [Google Scholar] [CrossRef]
- Schotten, U.; Dobrev, D.; Platonov, P.G.; Kottkamp, H.; Hindricks, G. Current controversies in determining the main mechanisms of atrial fibrillation. J. Intern. Med. 2016, 279, 428–438. [Google Scholar] [CrossRef]
- Ferrari, R.; Bertini, M.; Blomstrom-Lundqvist, C.; Dobrev, D.; Kirchhof, P.; Pappone, C.; Ravens, U.; Tamargo, J.; Tavazzi, L.; Vicedomini, G.G. An update on atrial fibrillation in 2014: From pathophysiology to treatment. Int. J. Cardiol. 2016, 203, 22–29. [Google Scholar] [CrossRef]
- Meyre, P.; Blum, S.; Berger, S.; Aeschbacher, S.; Schoepfer, H.; Briel, M.; Osswald, S.; Conen, D. Risk of Hospital Admissions in Patients With Atrial Fibrillation: A Systematic Review and Meta-analysis. Can. J. Cardiol. 2019, 35, 1332–1343. [Google Scholar] [CrossRef]
- Van Wagoner, D.R.; Piccini, J.P.; Albert, C.M.; Anderson, M.E.; Benjamin, E.J.; Brundel, B.; Califf, R.M.; Calkins, H.; Chen, P.S.; Chiamvimonvat, N.; et al. Progress toward the prevention and treatment of atrial fibrillation: A summary of the Heart Rhythm Society Research Forum on the Treatment and Prevention of Atrial Fibrillation, Washington, DC, December 9-10, 2013. Heart Rhythm 2015, 12, e5–e29. [Google Scholar] [CrossRef] [PubMed]
- Amara, W.; Montagnier, C.; Cheggour, S.; Boursier, M.; Gully, C.; Barnay, C.; Georger, F.; Deplagne, A.; Fromentin, S.; Mlotek, M.; et al. Early Detection and Treatment of Atrial Arrhythmias Alleviates the Arrhythmic Burden in Paced Patients: The SETAM Study. Pacing Clin. Electrophysiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Stachon, P.; Ahrens, I.; Faber, T.; Bode, C.; Zirlik, A. Asymptomatic atrial fibrillation and risk of stroke. Panminerva Med. 2015, 57, 211–215. [Google Scholar]
- de Vos, C.B.; Pisters, R.; Nieuwlaat, R.; Prins, M.H.; Tieleman, R.G.; Coelen, R.J.S.; van den Heijkant, A.C.; Allessie, M.A.; Crijns, H.J.G.M. Progression from paroxysmal to persistent atrial fibrillation clinical correlates and prognosis. J. Am. Coll. Cardiol. 2010, 55, 725–731. [Google Scholar] [CrossRef]
- Schuchert, A.; Behrens, G.; Meinertz, T. Impact of long-term ECG recording on the detection of paroxysmal atrial fibrillation in patients after an acute ischemic stroke. Pacing Clin. Electrophysiol. 1999, 22, 1082–1084. [Google Scholar] [CrossRef] [PubMed]
- Pagola, J.; Juega, J.; Francisco-Pascual, J.; Moya, A.; Sanchis, M.; Bustamante, A.; Penalba, A.; Usero, M.; Cortijo, E.; Arenillas, J.F.; et al. Yield of atrial fibrillation detection with Textile Wearable Holter from the acute phase of stroke: Pilot study of Crypto-AF registry. Int. J. Cardiol. 2018, 251, 45–50. [Google Scholar] [CrossRef]
- Luong, D.T.; Ha, N.T.; Thuan, N.D. Android Smart Phones Application in Tele-monitoring Electrocardiogram (ECG). Am. J. Biomed. Sci. 2019, 11. [Google Scholar] [CrossRef]
- Haverkamp, H.T.; Fosse, S.O.; Schuster, P. Accuracy and usability of single-lead ECG from smartphones—A clinical study. Indian Pacing Electrophysiol. J. 2019, 19, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Rahman, T.; Ghani, N.H.; Hossain, S.; Uddin, J. IoT based patient monitoring system using ECG sensor. In Proceedings of the 2019 International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST), Dhaka, Bangladesh, 10–12 January 2019; pp. 378–382. [Google Scholar]
- Nagai, S.; Anzai, D.; Wang, J. Motion artefact removals for wearable ECG using stationary wavelet transform. Healthc. Technol. Lett. 2017, 4, 138–141. [Google Scholar] [CrossRef]
- Satija, U.; Ramkumar, B.; Manikandan, M.S. A Review of Signal Processing Techniques for Electrocardiogram Signal Quality Assessment. IEEE Rev. Biomed. Eng. 2018, 11, 36–52. [Google Scholar] [CrossRef]
- Aboukhalil, A.; Nielsen, L.; Saeed, M.; Mark, R.G.; Clifford, G.D. Reducing false alarm rates for critical arrhythmias using the arterial blood pressure waveform. J. Biomed. Inf. 2008, 41, 442–451. [Google Scholar] [CrossRef]
- Bashar, S.K.; Ding, E.; Walkey, A.J.; McManus, D.D.; Chon, K.H. Noise Detection in Electrocardiogram Signals for Intensive Care Unit Patients. IEEE Access 2019, 7, 88357–88368. [Google Scholar] [CrossRef]
- Yoon, D.; Lim, H.S.; Jung, K.; Kim, T.Y.; Lee, S. Deep Learning-Based Electrocardiogram Signal Noise Detection and Screening Model. Healthc. Inform. Res. 2019, 25, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Oster, J.; Behar, J.; Sayadi, O.; Nemati, S.; Johnson, A.E.; Clifford, G.D. Semisupervised ECG ventricular beat classification with novelty detection based on switching Kalman filters. IEEE Trans. Biomed. Eng. 2015, 62, 2125–2134. [Google Scholar] [CrossRef]
- Levkov, C.; Mihov, G.; Ivanov, R.; Daskalov, I.; Christov, I.; Dotsinsky, I. Removal of power-line interference from the ECG: A review of the subtraction procedure. Biomed. Eng. Online 2005, 4, 50. [Google Scholar] [CrossRef]
- Luo, S.; Johnston, P. A review of electrocardiogram filtering. J. Electrocardiol. 2010, 43, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A.; Alcaraz, R.; Rieta, J.J. Application of the phasor transform for automatic delineation of single-lead ECG fiducial points. Physiol. Meas. 2010, 31, 1467–1485. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, M.S.; Ramkumar, B. Straightforward and robust QRS detection algorithm for wearable cardiac monitor. Healthc. Technol. Lett. 2014, 1, 40–44. [Google Scholar] [CrossRef]
- Aizawa, Y.; Watanabe, H.; Okumura, K. Electrocardiogram for the Prediction of Incident Atrial Fibrillation: An Overview. J. Atr. Fibrillation 2017, 10, 1724. [Google Scholar] [CrossRef]
- Satija, U.; Ramkumar, B.; Manikandan, M.S. An automated ECG signal quality assessment method for unsupervised diagnostic systems. Biocybern. Biomed. Eng. 2018, 38, 54–70. [Google Scholar] [CrossRef]
- Behar, J.; Oster, J.; Li, Q.; Clifford, G.D. ECG signal quality during arrhythmia and its application to false alarm reduction. IEEE Trans. Biomed. Eng. 2013, 60, 1660–1666. [Google Scholar] [CrossRef]
- Satija, U.; Ramkumar, B.; Manikandan, M.S. Automated ECG Noise Detection and Classification System for Unsupervised Healthcare Monitoring. IEEE J. Biomed. Health Inf. 2018, 22, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Fu, L.; Gu, L. A Cascaded Convolutional Neural Network for Assessing Signal Quality of Dynamic ECG. Comput. Math. Methods Med. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wei, S.; Ma, C.; Luo, K.; Zhang, L.; Liu, C. Atrial Fibrillation Beat Identification Using the Combination of Modified Frequency Slice Wavelet Transform and Convolutional Neural Networks. J. Healthc. Eng. 2018, 2018, 2102918. [Google Scholar] [CrossRef]
- Al Rahhal, M.M.; Bazi, Y.; Al Zuair, M.; Othman, E.; BenJdira, B. Convolutional neural networks for electrocardiogram classification. J. Med. Biol. Eng. 2018, 38, 1014–1025. [Google Scholar] [CrossRef]
- Mashrur, F.R.; Roy, A.D.; Saha, D.K. Automatic Identification of Arrhythmia from ECG Using AlexNet Convolutional Neural Network. In Proceedings of the 2019 4th International Conference on Electrical Information and Communication Technology (EICT), Khulna, Bangladesh, 20–22 December 2019; pp. 1–5. [Google Scholar]
- He, R.; Wang, K.; Zhao, N.; Liu, Y.; Yuan, Y.; Li, Q.; Zhang, H. Automatic Detection of Atrial Fibrillation Based on Continuous Wavelet Transform and 2D Convolutional Neural Networks. Front. Physiol. 2018, 9, 1206. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Feng, X.; Yang, C. A Deep Learning Method to Detect Atrial Fibrillation Based on Continuous Wavelet Transform. In Proceedings of the 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 1908–1912. [Google Scholar]
- Yildirim, O.; Talo, M.; Ay, B.; Baloglu, U.B.; Aydin, G.; Acharya, U.R. Automated detection of diabetic subject using pre-trained 2D-CNN models with frequency spectrum images extracted from heart rate signals. Comput. Biol. Med. 2019, 113, 103387. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.A.; Majumder, S. A novel approach for OSA detection using single-lead ECG scalogram based on deep neural network. J. Mech. Med. Biol. 2019, 19, 1950026. [Google Scholar] [CrossRef]
- Rastegar, S.; Gholamhosseini, H.; Lowe, A.; Mehdipour, F.; Lindén, M. Estimating Systolic Blood Pressure Using Convolutional Neural Networks. Stud. Health Technol. Inf. 2019, 261, 143–149. [Google Scholar]
- Byeon, Y.H.; Pan, S.B.; Kwak, K.C. Intelligent Deep Models Based on Scalograms of Electrocardiogram Signals for Biometrics. Sensors 2019, 19, 935. [Google Scholar] [CrossRef]
- Clifford, G.D.; Liu, C.; Moody, B.; Lehman, L.W.H.; Silva, I.; Li, Q.; Johnson, A.E.; Mark, R.G. AF Classification from a Short Single Lead ECG Recording: The PhysioNet/Computing in Cardiology Challenge 2017. Comput. Cardiol. (2010) 2017, 44. [Google Scholar] [CrossRef]
- Redmond, S.J.; Xie, Y.; Chang, D.; Basilakis, J.; Lovell, N.H. Electrocardiogram signal quality measures for unsupervised telehealth environments. Physiol. Meas. 2012, 33, 1517. [Google Scholar] [CrossRef]
- Ródenas, J.; García, M.; Alcaraz, R.; Rieta, J.J. Combined nonlinear analysis of atrial and ventricular series for automated screening of atrial fibrillation. Complexity 2017, 2017, 2163610. [Google Scholar]
- Shoeb, A.; Cliford, G. Chapter 16—Wavelets; multiscale activity in physiological signals. In Biomedical Signal Image Process; Springer: New York, NY, USA, 2005. [Google Scholar]
- Li, T.; Zhou, M. ECG classification using wavelet packet entropy and random forests. Entropy 2016, 18, 285. [Google Scholar] [CrossRef]
- Khorrami, H.; Moavenian, M. A comparative study of DWT, CWT and DCT transformations in ECG arrhythmias classification. Expert Syst. Appl. 2010, 37, 5751–5757. [Google Scholar] [CrossRef]
- Addison, P.S. The Illustrated Wavelet Transform Handbook: Introductory Theory and Applications in Science, Engineering, Medicine and Finance; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Bolós, V.J.; Benítez, R. The wavelet scalogram in the study of time series. In Advances in Differential Equations and Applications; Springer: Berlin, Germany, 2014; pp. 147–154. [Google Scholar]
- Lee, T.S. Image representation using 2D Gabor wavelets. IEEE Trans. Pattern Anal. Mach. Intell. 1996, 18, 959–971. [Google Scholar]
- Lyon, A.; Mincholé, A.; Martínez, J.P.; Laguna, P.; Rodriguez, B. Computational techniques for ECG analysis and interpretation in light of their contribution to medical advances. J. R. Soc. Interface 2018, 15. [Google Scholar] [CrossRef]
- Mincholé, A.; Rodriguez, B. Artificial intelligence for the electrocardiogram. Nat. Med. 2019, 25, 22–23. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, Y.; Oerlemans, A.; Lao, S.; Wu, S.; Lew, M.S. Deep learning for visual understanding: A review. Neurocomputing 2016, 187, 27–48. [Google Scholar] [CrossRef]
- Chaib, S.; Yao, H.; Gu, Y.; Amrani, M. Deep feature extraction and combination for remote sensing image classification based on pre-trained CNN models. In Proceedings of the Ninth International Conference on Digital Image Processing (ICDIP 2017), Hong Kong, China, 19–22 May 2017; Volume 10420, p. 104203D. [Google Scholar]
- Gogul, I.; Kumar, V.S. Flower species recognition system using convolution neural networks and transfer learning. In Proceedings of the 2017 Fourth International Conference on Signal Processing, Communication and Networking (ICSCN), Chennai, India, 16–18 March 2017; pp. 1–6. [Google Scholar]
- Jadhav, P.; Rajguru, G.; Datta, D.; Mukhopadhyay, S. Automatic sleep stage classification using time–frequency images of CWT and transfer learning using convolution neural network. Biocybern. Biomed. Eng. 2020, 40, 494–504. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 1097–1105. [Google Scholar] [CrossRef]
- Aloysius, N.; Geetha, M. A review on deep convolutional neural networks. In Proceedings of the 2017 International Conference on Communication and Signal Processing (ICCSP), Tamilnadu, India, 6–8 April 2017; pp. 0588–0592. [Google Scholar]
- Pak, M.; Kim, S. A review of deep learning in image recognition. In Proceedings of the 2017 4th International Conference on Computer Applications and Information Processing Technology (CAIPT), Kuta Bali, Indonesia, 8–10 August 2017; pp. 1–3. [Google Scholar]
- Al-Saffar, A.A.M.; Tao, H.; Talab, M.A. Review of deep convolution neural network in image classification. In Proceedings of the 2017 International Conference on Radar, Antenna, Microwave, Electronics, and Telecommunications (ICRAMET), Jakarta Selatan, Indonesia, 23–24 October 2017; pp. 26–31. [Google Scholar]
- Li, Q.; Rajagopalan, C.; Clifford, G.D. A machine learning approach to multi-level ECG signal quality classification. Comput. Methods Progr. Biomed. 2014, 117, 435–447. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, Y. SQI Quality Evaluation Mechanism of Single-Lead ECG Signal Based on Simple Heuristic Fusion and Fuzzy Comprehensive Evaluation. Front. Physiol. 2018, 9, 727. [Google Scholar] [CrossRef] [PubMed]
- Moeyersons, J.; Smets, E.; Morales, J.; Villa, A.; De Raedt, W.; Testelmans, D.; Buyse, B.; Van Hoof, C.; Willems, R.; Van Huffel, S.; et al. Artefact detection and quality assessment of ambulatory ECG signals. Comput. Methods Progr. Biomed. 2019, 182, 105050. [Google Scholar] [CrossRef]
- Clifford, G.; Behar, J.; Li, Q.; Rezek, I. Signal quality indices and data fusion for determining clinical acceptability of electrocardiograms. Physiol. Meas. 2012, 33, 1419. [Google Scholar] [CrossRef]
- Loong, T.W. Understanding sensitivity and specificity with the right side of the brain. BMJ 2003, 327, 716–719. [Google Scholar] [CrossRef] [PubMed]
- Silva, I.; Moody, G.B.; Celi, L. Improving the quality of ECGs collected using mobile phones: The Physionet/Computing in Cardiology Challenge 2011. Comput. Cardiol. 2011, 38, 273–276. [Google Scholar]
- Orphanidou, C.; Bonnici, T.; Charlton, P.; Clifton, D.; Vallance, D.; Tarassenko, L. Signal-quality indices for the electrocardiogram and photoplethysmogram: Derivation and applications to wireless monitoring. IEEE J. Biomed. Health Inf. 2015, 19, 832–838. [Google Scholar] [CrossRef] [PubMed]
- Hayn, D.; Jammerbund, B.; Schreier, G. QRS detection based ECG quality assessment. Physiol. Meas. 2012, 33, 1449–1461. [Google Scholar] [CrossRef] [PubMed]
- Casey, S.; Avalos, G.; Dowling, M. Critical care nurses’ knowledge of alarm fatigue and practices towards alarms: A multicentre study. Intensive Crit. Care Nurs. 2018, 48, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Nattel, S.; Guasch, E.; Savelieva, I.; Cosio, F.G.; Valverde, I.; Halperin, J.L.; Conroy, J.M.; Al-Khatib, S.M.; Hess, P.L.; Kirchhof, P.; et al. Early management of atrial fibrillation to prevent cardiovascular complications. Eur. Heart J. 2014, 35, 1448–1456. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, X.; Nakamura, K.; Mahito, N. ECG Quality Assessment Using 1D-Convolutional Neural Network. In Proceedings of the 2018 14th IEEE International Conference on Signal Processing (ICSP), Beijing, China, 12–16 August 2018; pp. 780–784. [Google Scholar]
- Zhao, Z.; Liu, C.; Li, Y.; Li, Y.; Wang, J.; Lin, B.S.; Li, J. Noise rejection for wearable ECGs using modified frequency slice wavelet transform and convolutional neural networks. IEEE Access 2019, 7, 34060–34067. [Google Scholar] [CrossRef]
- Petrėnas, A.; Marozas, V.; Sörnmo, L. Low-complexity detection of atrial fibrillation in continuous long-term monitoring. Comput. Biol. Med. 2015, 65, 184–191. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: The convolutional neural network AlexNet is well-known and its implementation is freely available for several software platforms (such as Matlab and Tensorflow). The databases PC2017DB and THDB are public and freely available. The PDB dataset is proprietary and cannot be made publicly available. However, access to the data for scientific purposes will be granted upon reasonable request. The trained CNN model applied in this work is also available from the corresponding author upon request. |
| Database | Value | Se (%) | Sp (%) | Acc (%) | (%) | (%) | (%) |
|---|---|---|---|---|---|---|---|
| PC2017DB | Mean | 86.91 | 91.00 | 88.95 | 89.28 | 85.92 | 85.69 |
| Std | 2.64 | 2.66 | 1.03 | 2.80 | 3.18 | 3.58 | |
| Maximum | 93.10 | 96.40 | 91.80 | 95.55 | 91.61 | 93.84 | |
| Minimum | 81.70 | 85.00 | 87.05 | 82.33 | 77.91 | 79.28 | |
| THDB | Mean | 95.49 | 85.00 | 91.42 | 96.50 | 94.04 | — |
| Std | 2.71 | 4.74 | 1.08 | 5.59 | 2.85 | — | |
| Maximum | 98.78 | 88.46 | 92.91 | 100 | 99.31 | — | |
| Minimum | 92.68 | 76.92 | 90.30 | 87.50 | 92.92 | — | |
| PDB | Mean | 97.17 | 92.42 | 94.79 | 99.58 | 93.86 | — |
| Std | 1.57 | 3.14 | 1.41 | 0.94 | 2.75 | — | |
| Maximum | 98.75 | 96.25 | 96.88 | 100 | 97.09 | — | |
| Minimum | 94.58 | 88.75 | 93.33 | 97.90 | 89.69 | — | |
| Samples from previous datasets | Mean | 94.42 | 90.61 | 92.51 | 92.87 | 92.05 | — |
| Std | 3.44 | 3.45 | 1.08 | 1.35 | 2.23 | — | |
| Maximum | 96.65 | 92.29 | 91.43 | 97.51 | 94.59 | — | |
| Minimum | 89.08 | 86.08 | 89.40 | 94.21 | 88.56 | — |
| Database | Value | Se (%) | Sp (%) | Acc (%) | (%) | (%) |
|---|---|---|---|---|---|---|
| PC2017DB | Mean | 80.84 | 81.17 | 81.01 | 79.97 | 81.73 |
| Std | 1.72 | 1.61 | 1.08 | 2.10 | 2.29 | |
| Maximum | 84.10 | 86.20 | 83.70 | 83.80 | 86.30 | |
| Minimum | 77.10 | 78.50 | 78.80 | 75.82 | 76.48 | |
| THDB | Mean | 84.09 | 66.03 | 76.92 | 83.68 | 89.10 |
| Std | 4.20 | 5.25 | 3.06 | 4.46 | 8.85 | |
| Maximum | 90.68 | 72.64 | 82.02 | 90.54 | 100 | |
| Minimum | 79.50 | 59.43 | 73.78 | 78.37 | 76.92 | |
| PDB | Mean | 87.97 | 86.94 | 87.45 | 94.51 | 81.08 |
| Std | 1.08 | 1.61 | 0.82 | 0.85 | 1.81 | |
| Maximum | 89.90 | 90.40 | 89.50 | 96.20 | 84.11 | |
| Minimum | 85.60 | 83.90 | 85.55 | 92.41 | 76.93 | |
| Samples from previous datasets | Mean | 84.49 | 85.43 | 84.43 | 90.24 | 80.11 |
| Std | 0.79 | 1.50 | 0.96 | 1.14 | 1.26 | |
| Maximum | 85.22 | 86.50 | 85.54 | 91.80 | 81.85 | |
| Minimum | 83.29 | 82.86 | 83.08 | 88.70 | 78.92 |
| Work | Methodology | Classes | Main Results |
|---|---|---|---|
| Behar et al. [31] | Seven ECG-based indices | High- and | = 98.4% |
| combined with a SVM classifier | low-quality ECGs | ||
| Moeyersons et al. [64] | Descriptive features from autocorrelation | High- and | = 97.7% |
| function combined with a RUSBoost classifier | low-quality ECGs | = 94.7% | |
| Clifford et al. [65] | Four ECG-based indices | High- and | = 95.8% |
| combined with a | low-quality | = 97.2% | |
| SVM classifier | ECGs | = 96.5% | |
| Orphanidou et al. [68] | Analysis detected R-peaks and | High- and | = 97% |
| correlation QRS complexes with a template | low-quality ECGs | = 94% | |
| Hayn et al. [69] | Multiple QRS-based parameters | High- and | = 91.3% |
| combined with rules | low-quality ECGs | ||
| Zhao & Zhang [63] | Multiple R-peak-based parameters | High- and | = 97.33% |
| combined with rules and | low-quality | = 88.67% | |
| Fuzzy synthesis | ECGs | = 92.57% | |
| Satija et al. [30] | Parameters extracted from wavelet | High- and | = 99.53% |
| decomposition of the ECG and | low-quality | = 98.95% | |
| combined with rules | ECGs | = 99.16% | |
| Satija et al. [32] | Parameters extracted from empirical mode | High- and | = 98.56% |
| decomposition of the ECG and | low-quality | = 99.12% | |
| combined with rules | ECGs | = 98.90% |
| Work | Methodology | Classes | Main Results |
|---|---|---|---|
| Zhou et al. [72] | A 1-D CNN fed with the ECG | High- and | = 95.5% |
| low-quality | = 91.3% | ||
| ECGs | = 94.3% | ||
| Yoon et al. [23] | Two 1-D CNNs working | High- | = 89% |
| in parallel with | and | = 88% | |
| ECG and its | low-quality | = 88% | |
| spectral distribution | ECGs | = 50% | |
| Zhang et al. [33] | Two stages with two CNNs | ECGs with three | = 91.8% |
| (1D and 2D) working in parallel | levels of noise | = 75–83% | |
| Zhao et al. [73] | A 2-D CNN fed with wavelet | ECGs with three | = 86.3% |
| scalogram of the ECG | levels of noise |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herraiz, Á.H.; Martínez-Rodrigo, A.; Bertomeu-González, V.; Quesada, A.; Rieta, J.J.; Alcaraz, R. A Deep Learning Approach for Featureless Robust Quality Assessment of Intermittent Atrial Fibrillation Recordings from Portable and Wearable Devices. Entropy 2020, 22, 733. https://doi.org/10.3390/e22070733
Herraiz ÁH, Martínez-Rodrigo A, Bertomeu-González V, Quesada A, Rieta JJ, Alcaraz R. A Deep Learning Approach for Featureless Robust Quality Assessment of Intermittent Atrial Fibrillation Recordings from Portable and Wearable Devices. Entropy. 2020; 22(7):733. https://doi.org/10.3390/e22070733
Chicago/Turabian StyleHerraiz, Álvaro Huerta, Arturo Martínez-Rodrigo, Vicente Bertomeu-González, Aurelio Quesada, José J. Rieta, and Raúl Alcaraz. 2020. "A Deep Learning Approach for Featureless Robust Quality Assessment of Intermittent Atrial Fibrillation Recordings from Portable and Wearable Devices" Entropy 22, no. 7: 733. https://doi.org/10.3390/e22070733
APA StyleHerraiz, Á. H., Martínez-Rodrigo, A., Bertomeu-González, V., Quesada, A., Rieta, J. J., & Alcaraz, R. (2020). A Deep Learning Approach for Featureless Robust Quality Assessment of Intermittent Atrial Fibrillation Recordings from Portable and Wearable Devices. Entropy, 22(7), 733. https://doi.org/10.3390/e22070733






