Information Transfer in Linear Multivariate Processes Assessed through Penalized Regression Techniques: Validation and Application to Physiological Networks
Abstract
1. Introduction
2. Materials and Methods
2.1. Vector Autoregressive Model Identification
2.2. Measures of Information Transfer
2.3. Computation of the Measures of Information Transfer for Multivariate Gaussian Processes
Formulation of State–Space Models
2.4. Testing the Significance of the Conditional Transfer Entropy
3. Simulation Experiments
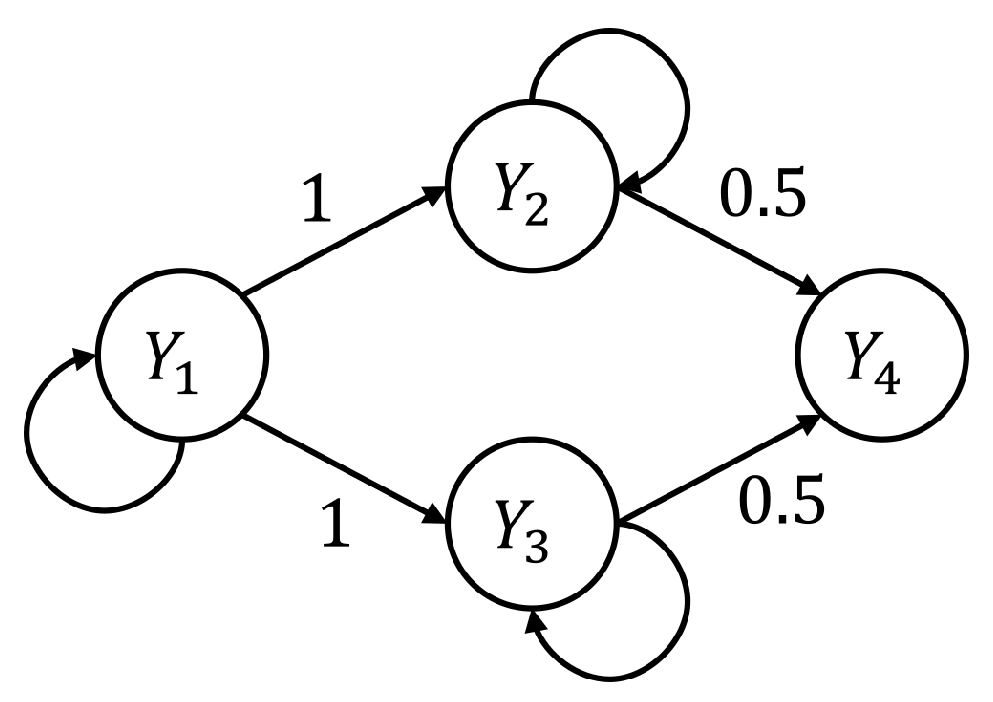
3.1. Simulation Study I
3.1.1. Simulation Design and Realization
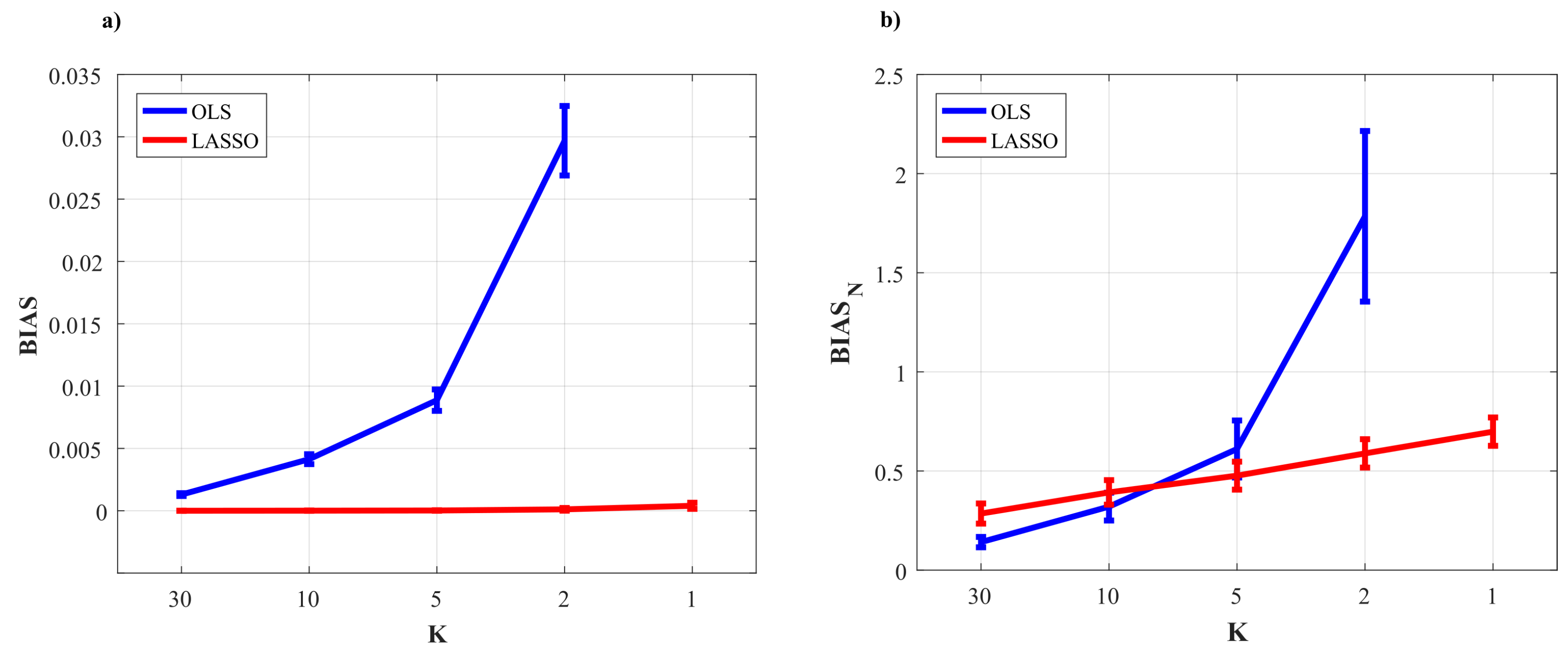
3.1.2. Simulation Results
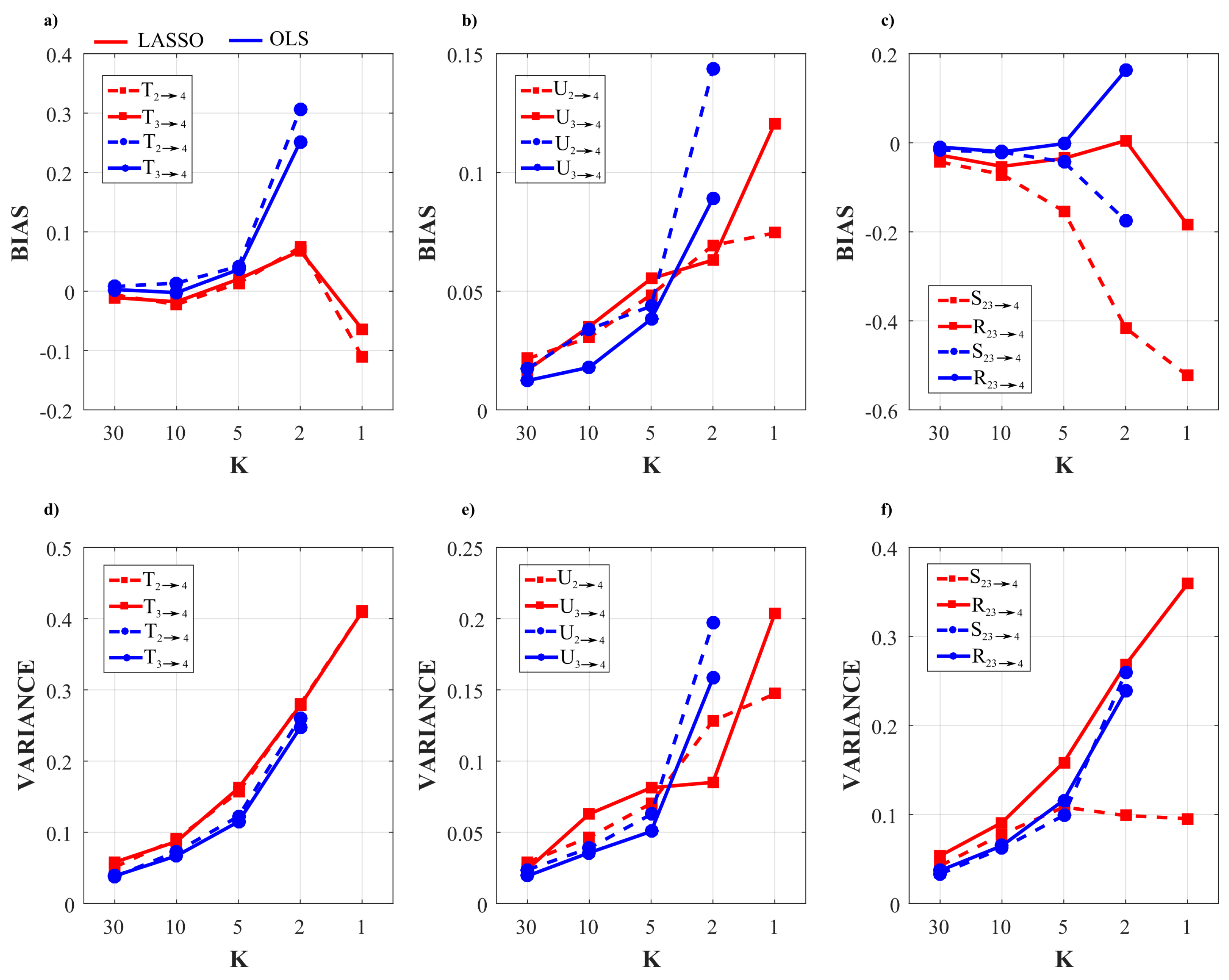
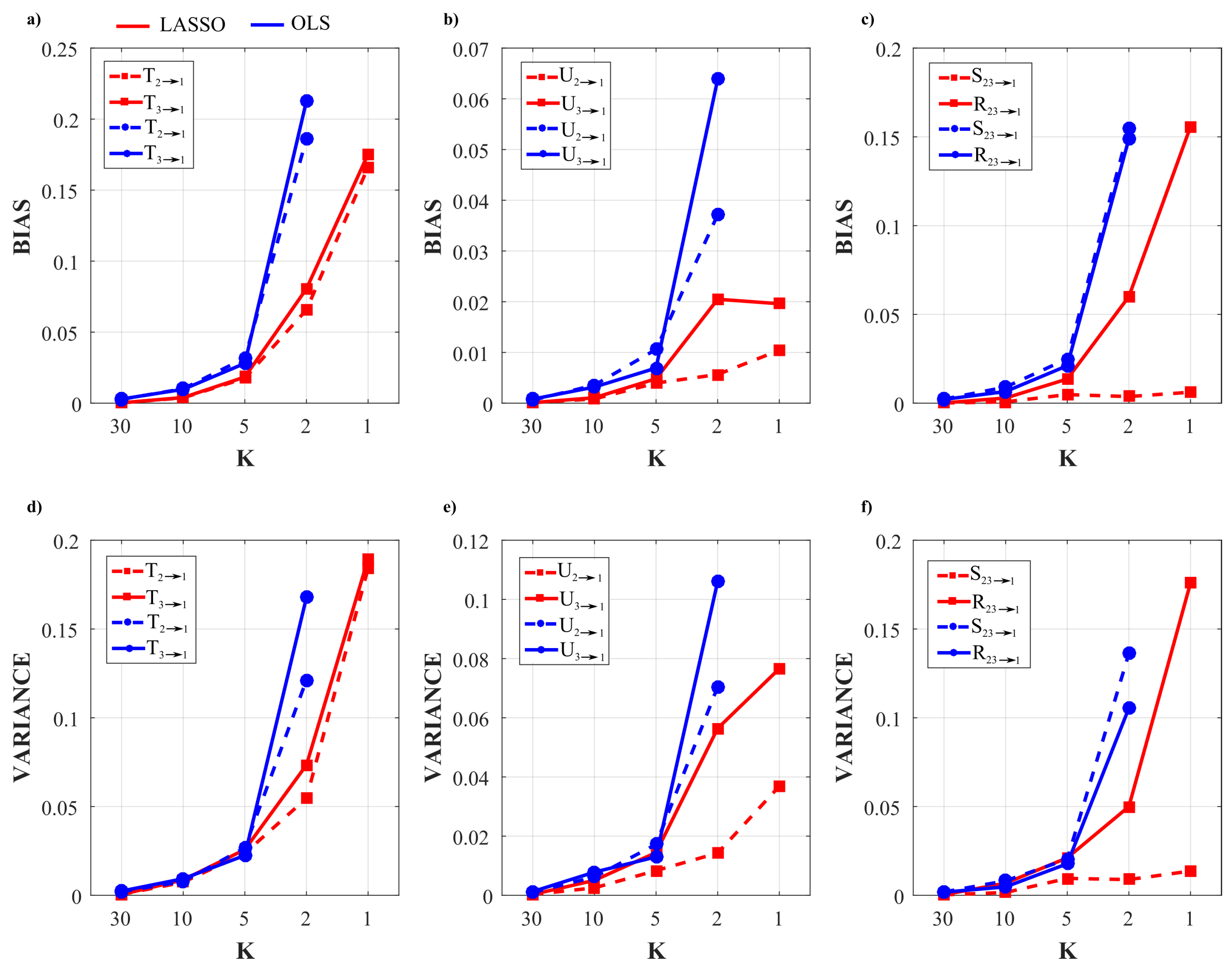
3.2. Simulation Study II
3.2.1. Simulation Design and Realization
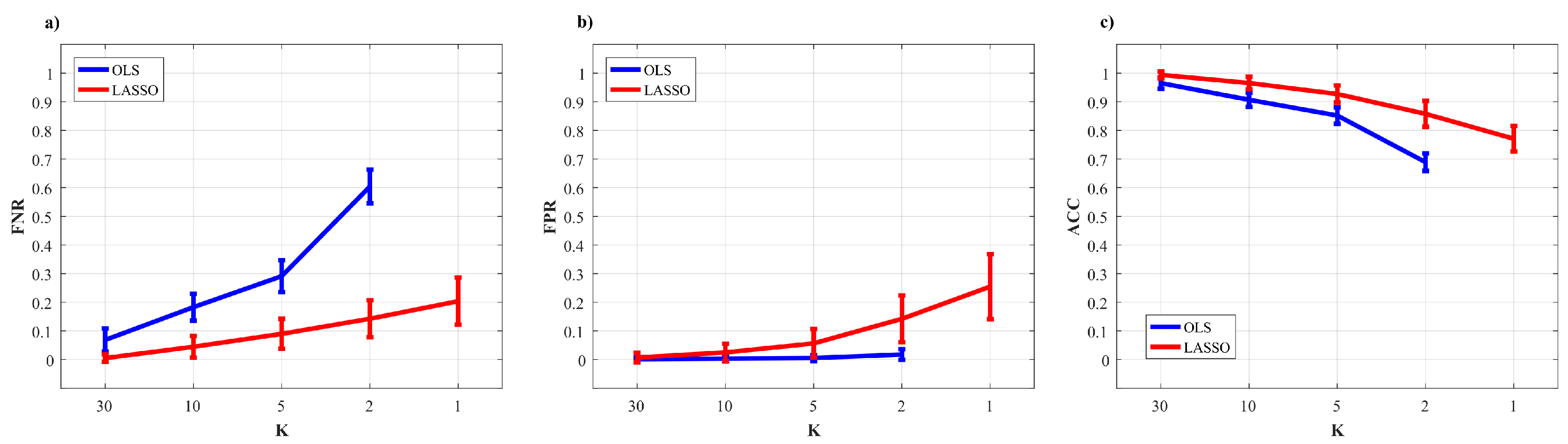
3.2.2. Performance Evaluation
3.2.3. Statistical Analysis
3.2.4. Results of the Simulation Study
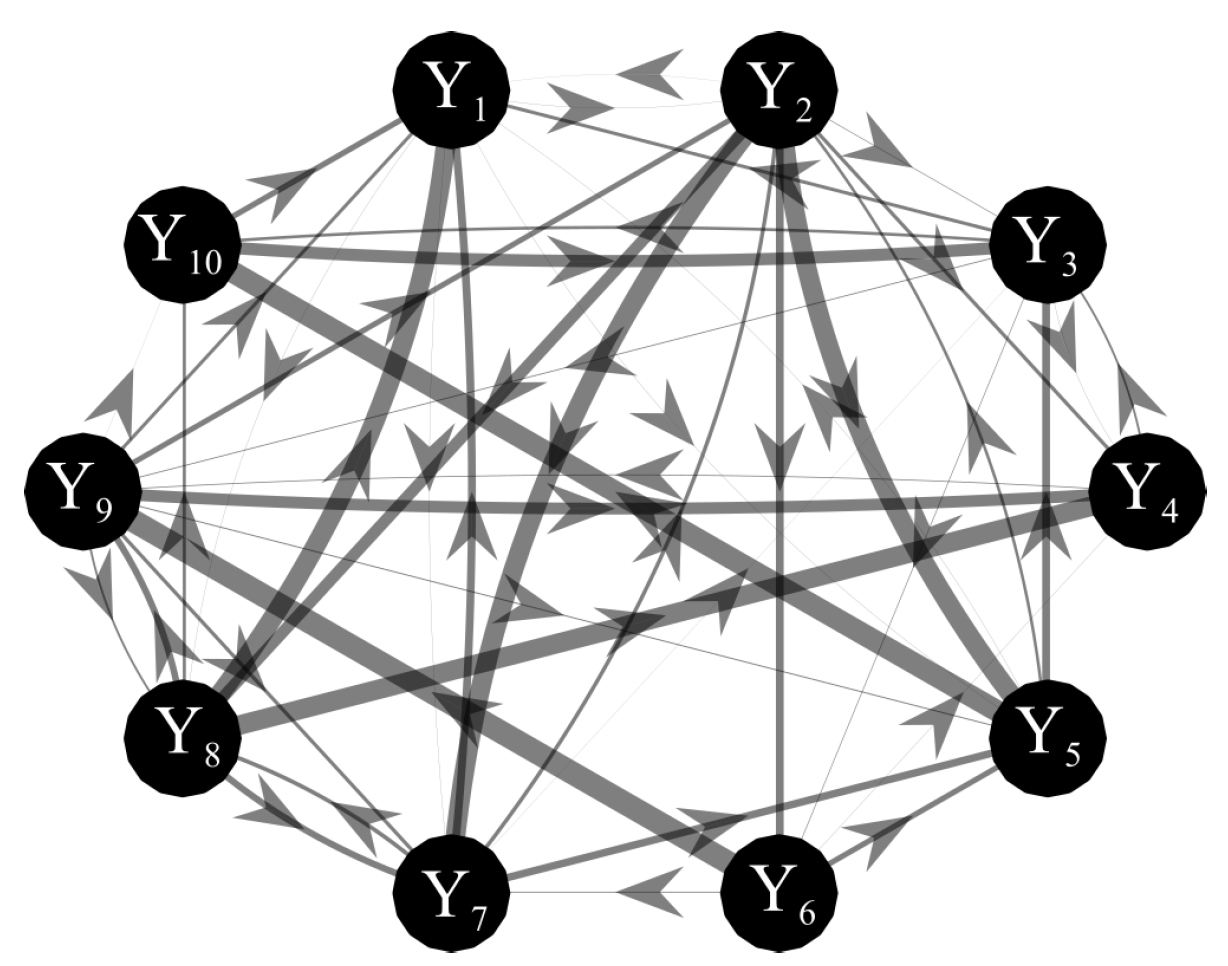
4. Application to Physiological Time Series
4.1. Data Acquisition and Pre-Processing
4.2. Information Transfer Analysis
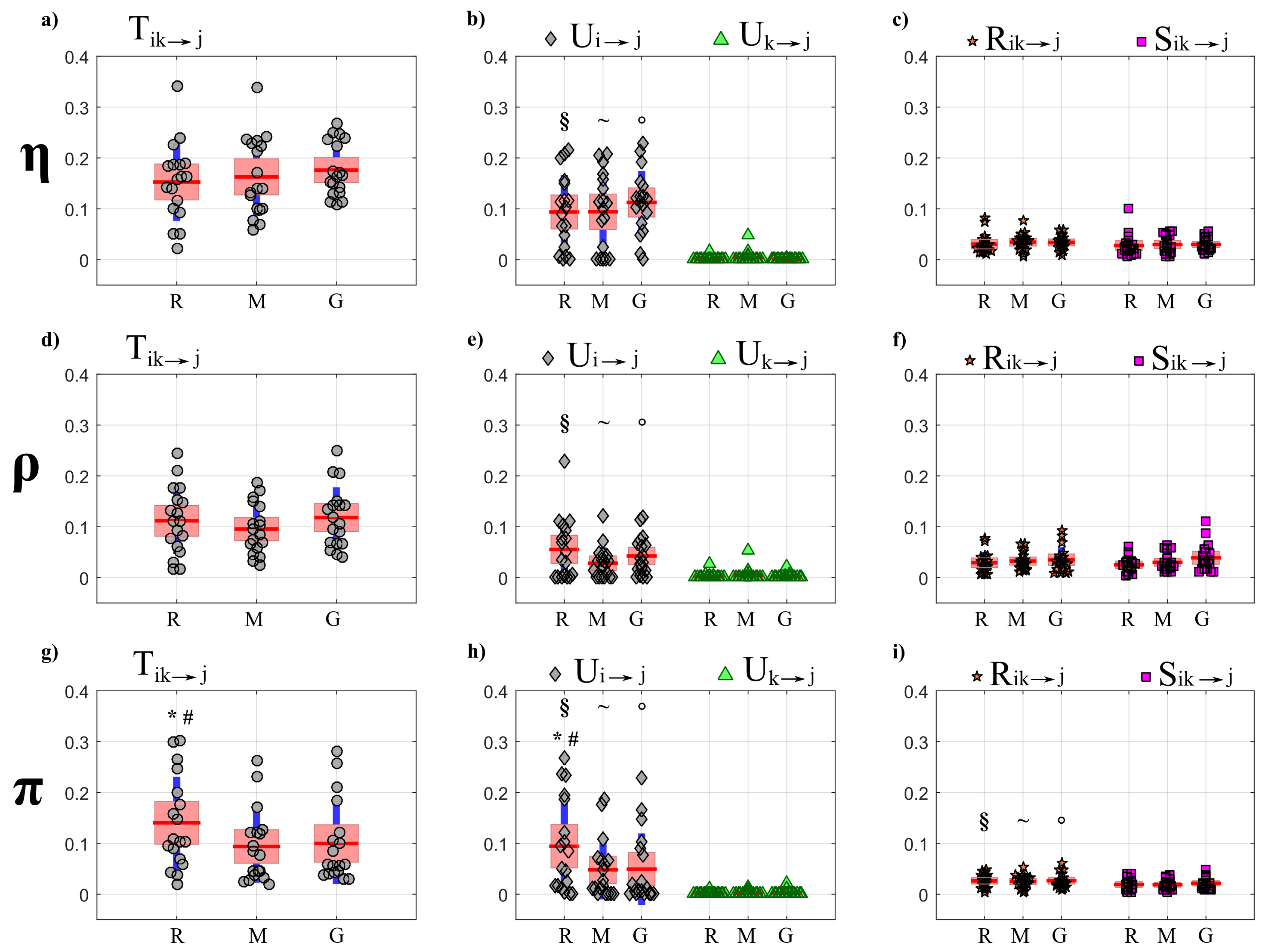
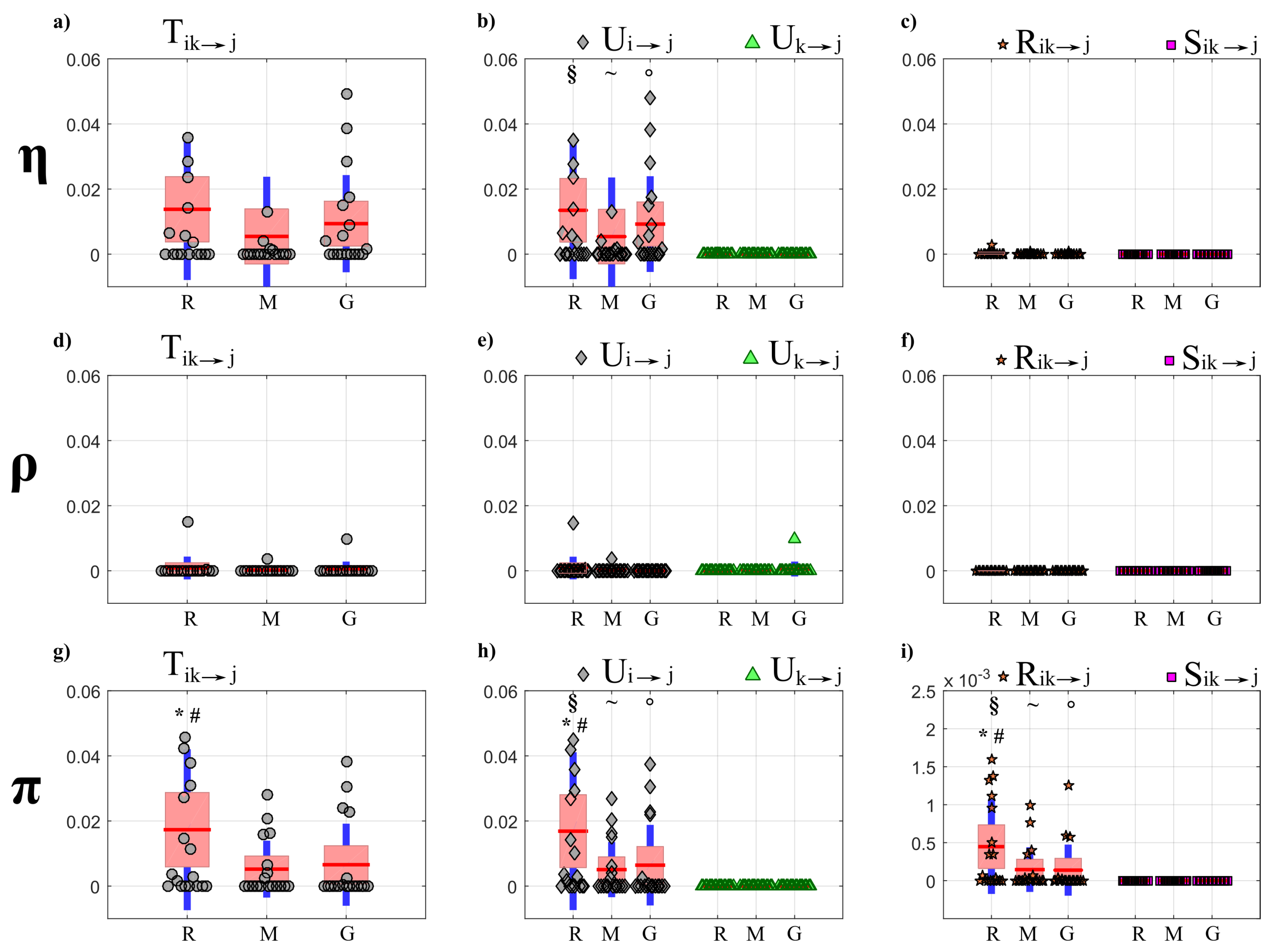
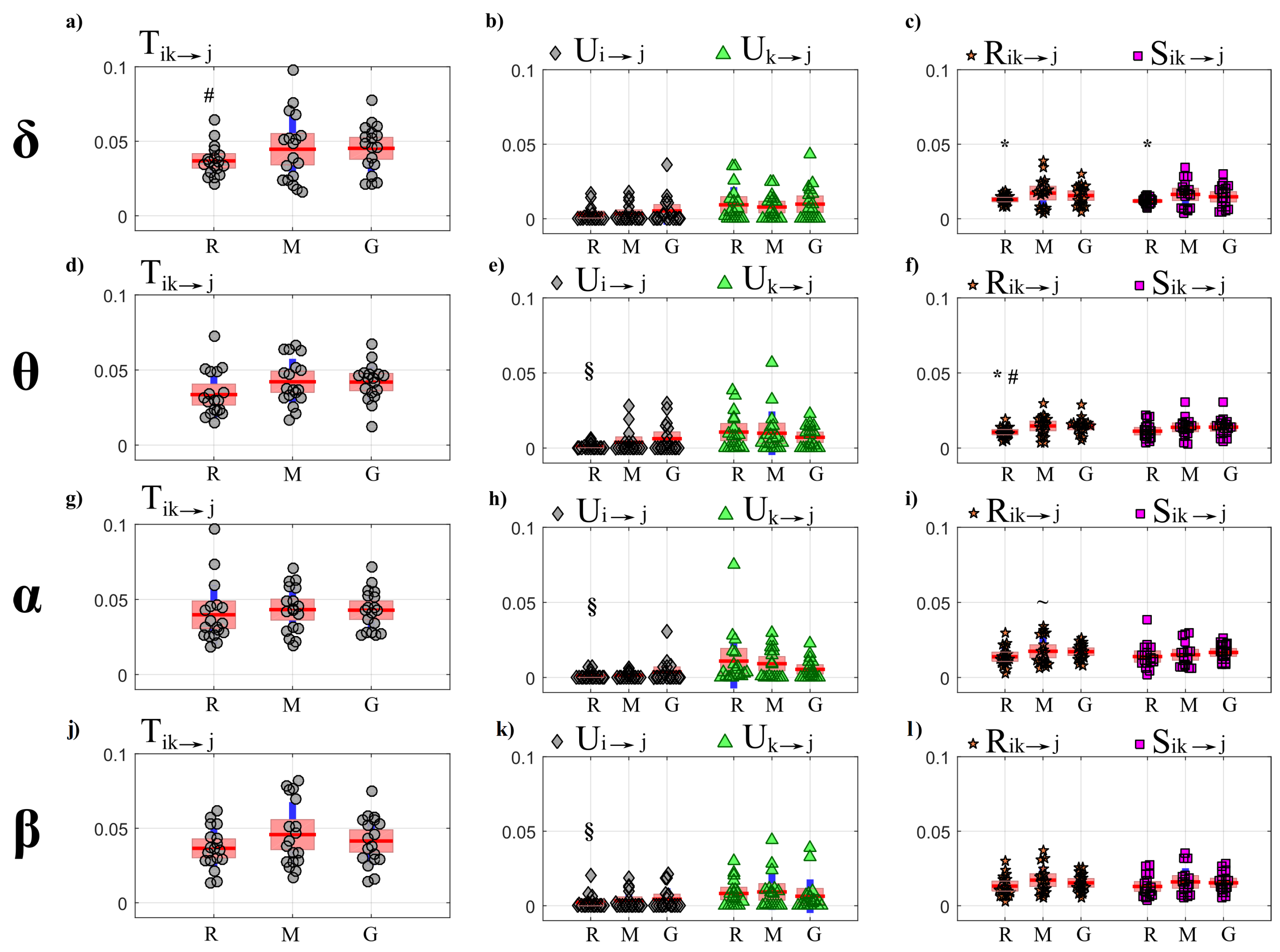
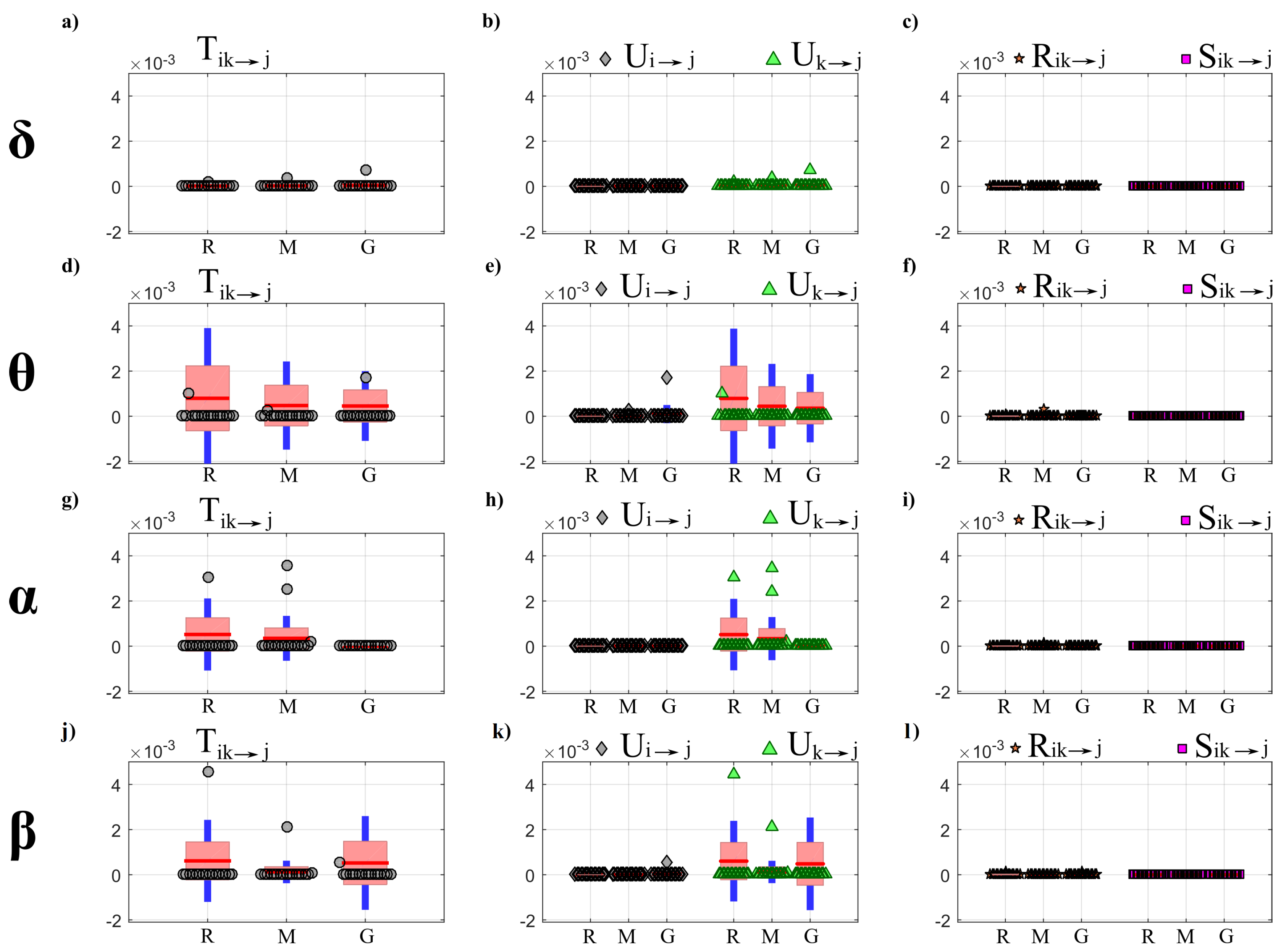
- First, a PID analysis was performed for OLS and LASSO through the computation of the joint information transfer and the terms of its decomposition , , , . The analysis was performed collecting in the first source (index i) the processes forming the so-called “body” sub-network that accounts for cardiac, cardiovascular and respiratory dynamics, and in the second source (index k) the processes forming the “brain” sub-network that accounts for the different brain wave amplitudes; the analysis was repeated considering each one of the seven processes as the target process and excluding it from the set of sources.
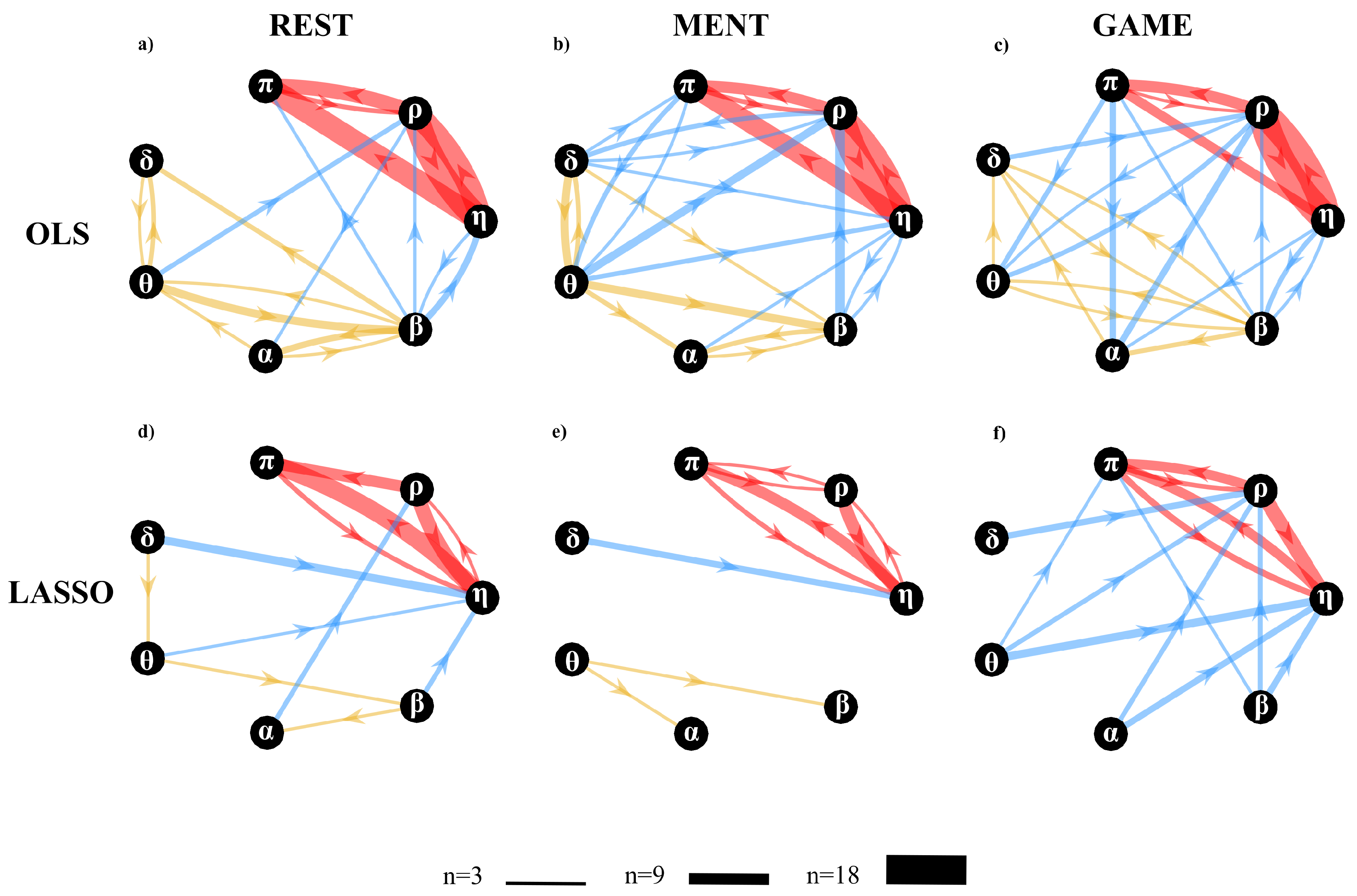
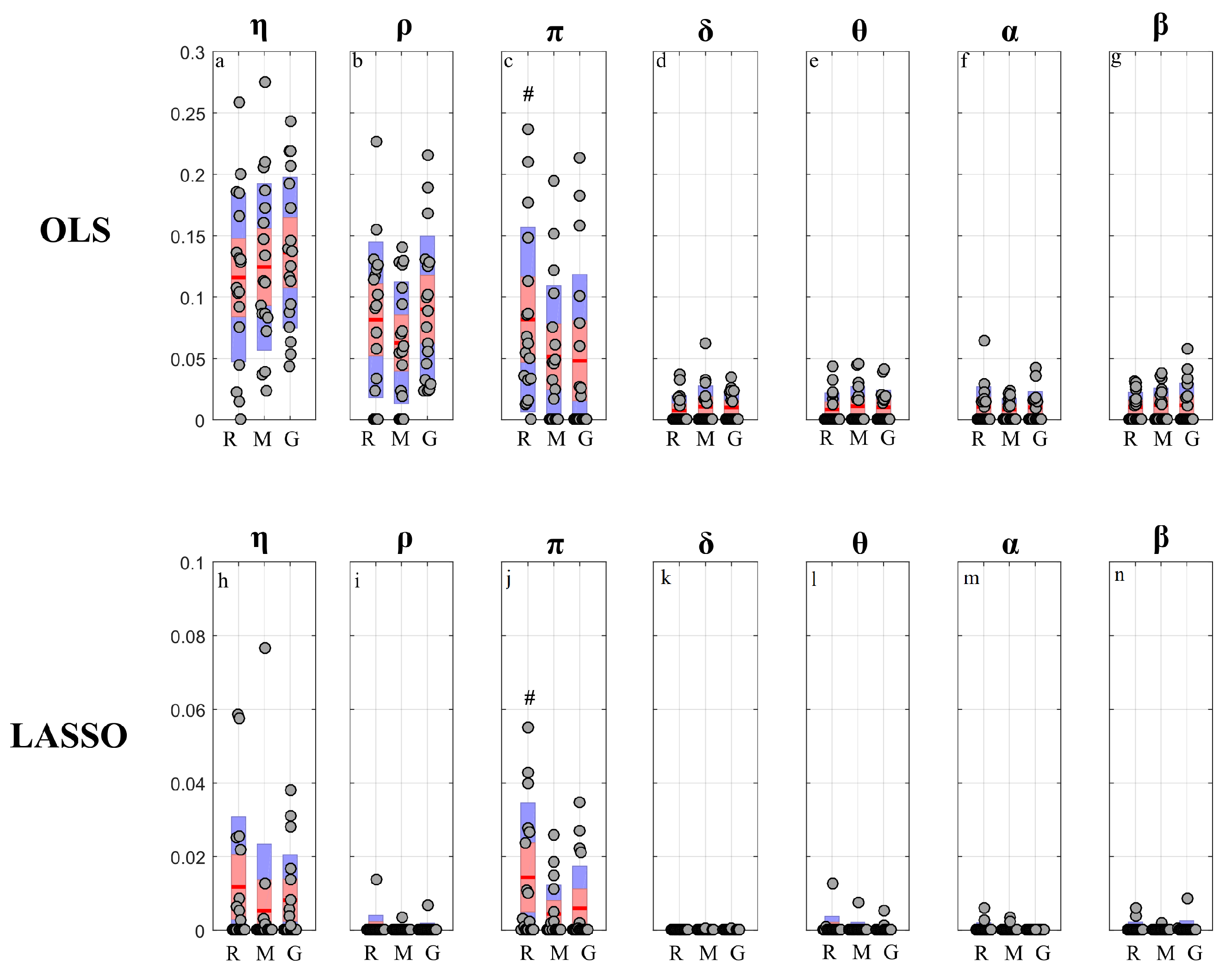
- Second, the topological structure of the network of physiological interactions was detected computing the conditional transfer entropy based on the two VAR identification methods combined with their method for assessing the statistical significance of cTE (i.e., using surrogate data for OLS and exploiting the intrinsic sparseness for LASSO). The analysis was performed between each pair of processes as driver and target and collecting the remaining five processes in the conditioning vector with index s. As a quantitative descriptor of the network was used the in-strength, defined as the sum of all weighted inward links connected to one node [63]. Moreover, to describe the overall brain–body interactions the in-strength of the body sub-network due to brain sub-network (and vice-versa) was computed considering as link weights the percentage of subjects showing at least one statistically significant brain-to-body connection (and vice-versa). To study the involvement of each specific node in the network, the in-strength of each node was computed considering as link weights the cTE values of all network links pointing into the considered node.
4.3. Statistical Analysis
4.4. Results of Real Data Application
4.4.1. Partial Information Decomposition
4.4.2. Conditional Information Transfer
5. Discussion
5.1. Simulation Study I
5.2. Simulation Study II
5.3. Real Data Application
5.3.1. Partial Information Decomposition Analysis
5.3.2. Conditional Information Transfer Analysis
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bashan, A.; Bartsch, R.P.; Kantelhardt, J.W.; Havlin, S.; Ivanov, P.C. Network physiology reveals relations between network topology and physiological function. Nat. Commun. 2012, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Faes, L.; Porta, A.; Nollo, G. Information decomposition in bivariate systems: Theory and application to cardiorespiratory dynamics. Entropy 2015, 17, 277–303. [Google Scholar] [CrossRef]
- Zanetti, M.; Faes, L.; Nollo, G.; De Cecco, M.; Pernice, R.; Maule, L.; Pertile, M.; Fornaser, A. Information dynamics of the brain, cardiovascular and respiratory network during different levels of mental stress. Entropy 2019, 21, 275. [Google Scholar] [CrossRef]
- Malik, M. Heart rate variability: Standards of measurement, physiological interpretation, and clinical use: Task force of the European Society of Cardiology and the North American Society for Pacing and Electrophysiology. Ann. Noninvasive Electrocardiol. 1996, 1, 151–181. [Google Scholar] [CrossRef]
- Pereda, E.; Quiroga, R.Q.; Bhattacharya, J. Nonlinear multivariate analysis of neurophysiological signals. Prog. Neurobiol. 2005, 77, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Adochiei, F.C.; Edu, I.R.; Schroeder, R.; Costin, H.; Bär, K.J.; Voss, A. Cardiovascular and cardiorespiratory coupling analyses: A review. Philos. Trans. R. Soc. A 2013, 371, 20120191. [Google Scholar] [CrossRef]
- Faes, L.; Nollo, G.; Jurysta, F.; Marinazzo, D. Information dynamics of brain–heart physiological networks during sleep. New J. Phys. 2014, 16, 105005. [Google Scholar] [CrossRef]
- Bartsch, R.P.; Liu, K.K.; Bashan, A.; Ivanov, P.C. Network physiology: How organ systems dynamically interact. PLoS ONE 2015, 10, e0142143. [Google Scholar] [CrossRef]
- Lizier, J.T.; Prokopenko, M.; Zomaya, A.Y. Local measures of information storage in complex distributed computation. Inf. Sci. 2012, 208, 39–54. [Google Scholar] [CrossRef]
- Faes, L.; Javorka, M.; Nollo, G. Information-Theoretic Assessment of Cardiovascular Variability During Postural and Mental Stress. In Proceedings of the XIV Mediterranean Conference on Medical and Biological Engineering and Computing 2016, Paphos, Cyprus, 31 March–2 April 2016; pp. 67–70. [Google Scholar]
- Schreiber, T. Measuring information transfer. Phys. Rev. Lett. 2000, 85, 461. [Google Scholar] [CrossRef]
- Lizier, J.T.; Prokopenko, M.; Zomaya, A.Y. Information modification and particle collisions in distributed computation. Chaos Interdiscip. J. Nonlinear Sci. 2010, 20, 037109. [Google Scholar] [CrossRef] [PubMed]
- Schneidman, E.; Bialek, W.; Berry, M.J. Synergy, redundancy, and independence in population codes. J. Neurosci. 2003, 23, 11539–11553. [Google Scholar] [CrossRef] [PubMed]
- Barnett, L.; Barrett, A.B.; Seth, A.K. Granger causality and transfer entropy are equivalent for Gaussian variables. Phys. Rev. Lett. 2009, 103, 238701. [Google Scholar] [CrossRef]
- Barrett, A.B. Exploration of synergistic and redundant information sharing in static and dynamical Gaussian systems. Phys. Rev. E 2015, 91, 052802. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, F.; Wang, J.W.; Zhang, X.; Ivanov, P.C. Power-law correlations and coupling of active and quiet states underlie a class of complex systems with self-organization at criticality. EPJ Web Conf. 2020, 230, 00005. [Google Scholar] [CrossRef]
- Lombardi, F.; Gómez-Extremera, M.; Bernaola-Galván, P.; Vetrivelan, R.; Saper, C.B.; Scammell, T.E.; Ivanov, P.C. Critical dynamics and coupling in bursts of cortical rhythms indicate non-homeostatic mechanism for sleep-stage transitions and dual role of VLPO neurons in both sleep and wake. J. Neurosci. 2020, 40, 171–190. [Google Scholar] [CrossRef]
- Porta, A.; Bari, V.; De Maria, B.; Baumert, M. A network physiology approach to the assessment of the link between sinoatrial and ventricular cardiac controls. Physiol. Meas. 2017, 38, 1472. [Google Scholar] [CrossRef]
- Krohova, J.; Faes, L.; Czippelova, B.; Turianikova, Z.; Mazgutova, N.; Pernice, R.; Busacca, A.; Marinazzo, D.; Stramaglia, S.; Javorka, M. Multiscale information decomposition dissects control mechanisms of heart rate variability at rest and during physiological stress. Entropy 2019, 21, 526. [Google Scholar] [CrossRef]
- Widjaja, D.; Montalto, A.; Vlemincx, E.; Marinazzo, D.; Van Huffel, S.; Faes, L. Cardiorespiratory information dynamics during mental arithmetic and sustained attention. PLoS ONE 2015, 10, e0129112. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M.; Mizumoto, T.; Faes, L.; Fornaser, A.; De Cecco, M.; Maule, L.; Valente, M.; Nollo, G. Multilevel assessment of mental stress via network physiology paradigm using consumer wearable devices. J. Ambient Intell. Hum. Comput. 2019. [Google Scholar] [CrossRef]
- Valenza, G.; Greco, A.; Gentili, C.; Lanata, A.; Sebastiani, L.; Menicucci, D.; Gemignani, A.; Scilingo, E. Combining electroencephalographic activity and instantaneous heart rate for assessing brain–heart dynamics during visual emotional elicitation in healthy subjects. Philos. Trans. R. Soc. A 2016, 374, 20150176. [Google Scholar] [CrossRef] [PubMed]
- Greco, A.; Faes, L.; Catrambone, V.; Barbieri, R.; Scilingo, E.P.; Valenza, G. Lateralization of directional brain-heart information transfer during visual emotional elicitation. Am. J. Physiol.-Regul. Integr. and Comp. Physiol. 2019, 317, R25–R38. [Google Scholar] [CrossRef] [PubMed]
- Wibral, M.; Lizier, J.; Vögler, S.; Priesemann, V.; Galuske, R. Local active information storage as a tool to understand distributed neural information processing. Front. Neuroinf. 2014, 8, 1. [Google Scholar] [CrossRef]
- Barnett, L.; Lizier, J.T.; Harré, M.; Seth, A.K.; Bossomaier, T. Information flow in a kinetic Ising model peaks in the disordered phase. Phys. Rev. Lett. 2013, 111, 177203. [Google Scholar] [CrossRef]
- Dimpfl, T.; Peter, F.J. Using transfer entropy to measure information flows between financial markets. Stud. Nonlinear Dyn. Econom. 2013, 17, 85–102. [Google Scholar] [CrossRef]
- Stramaglia, S.; Cortes, J.M.; Marinazzo, D. Synergy and redundancy in the Granger causal analysis of dynamical networks. New J. Phys. 2014, 16, 105003. [Google Scholar] [CrossRef]
- Barnett, L.; Seth, A.K. Granger causality for state-space models. Phys. Rev. E 2015, 91, 040101. [Google Scholar] [CrossRef]
- Solo, V. State-space analysis of Granger-Geweke causality measures with application to fMRI. Neural Comput. 2016, 28, 914–949. [Google Scholar] [CrossRef]
- Faes, L.; Marinazzo, D.; Stramaglia, S. Multiscale information decomposition: Exact computation for multivariate Gaussian processes. Entropy 2017, 19, 408. [Google Scholar] [CrossRef]
- Schlögl, A. A comparison of multivariate autoregressive estimators. Signal Process. 2006, 86, 2426–2429. [Google Scholar] [CrossRef]
- Hoerl, A.E.; Kennard, R.W. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics 1970, 12, 55–67. [Google Scholar] [CrossRef]
- Antonacci, Y.; Toppi, J.; Caschera, S.; Anzolin, A.; Mattia, D.; Astolfi, L. Estimating brain connectivity when few data points are available: Perspectives and limitations. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Seogwipo, Korea, 11–15 July 2017; pp. 4351–4354. [Google Scholar]
- Antonacci, Y.; Toppi, J.; Mattia, D.; Pietrabissa, A.; Astolfi, L. Single-trial Connectivity Estimation through the Least Absolute Shrinkage and Selection Operator. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 6422–6425. [Google Scholar]
- Marinazzo, D.; Pellicoro, M.; Stramaglia, S. Causal information approach to partial conditioning in multivariate data sets. Comput. Math. Methods Med. 2012, 2012, 303601. [Google Scholar] [CrossRef]
- Siggiridou, E.; Kugiumtzis, D. Granger causality in multivariate time series using a time-ordered restricted vector autoregressive model. IEEE Trans. Signal Process. 2015, 64, 1759–1773. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Wainwright, M. Statistical Learning with Sparsity: The Lasso and Generalizations; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Haufe, S.; Müller, K.R.; Nolte, G.; Krämer, N. Sparse causal discovery in multivariate time series. arXiv 2009, arXiv:0901.2234. [Google Scholar]
- Antonacci, Y.; Toppi, J.; Mattia, D.; Pietrabissa, A.; Astolfi, L. Estimation of brain connectivity through Artificial Neural Networks. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 636–639. [Google Scholar]
- Billinger, M.; Brunner, C.; Müller-Putz, G.R. Single-trial connectivity estimation for classification of motor imagery data. J. Neural Eng. 2013, 10, 046006. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Sosa, P.A.; Sánchez-Bornot, J.M.; Lage-Castellanos, A.; Vega-Hernández, M.; Bosch-Bayard, J.; Melie-García, L.; Canales-Rodríguez, E. Estimating brain functional connectivity with sparse multivariate autoregression. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 969–981. [Google Scholar] [CrossRef]
- Smeekes, S.; Wijler, E. Macroeconomic forecasting using penalized regression methods. Int. J. Forecasting 2018, 34, 408–430. [Google Scholar] [CrossRef]
- Pernice, R.; Zanetti, M.; Nollo, G.; De Cecco, M.; Busacca, A.; Faes, L. Mutual Information Analysis of Brain-Body Interactions during different Levels of Mental stress. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 6176–6179. [Google Scholar]
- Lütkepohl, H. Introduction to Multiple Time Series Analysis; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Zou, H.; Hastie, T.; Tibshirani, R. On the “degrees of freedom” of the lasso. Ann. Stat. 2007, 35, 2173–2192. [Google Scholar] [CrossRef]
- Sun, X. The Lasso and Its Implementation for Neural Networks. Ph.D. Thesis, National Library of Canada = Bibliothèque nationale du Canada, University of Toronto, Toronto, ON, Canada, February 1999. [Google Scholar]
- Tibshirani, R.J.; Taylor, J. Degrees of freedom in lasso problems. Ann. Stat. 2012, 40, 1198–1232. [Google Scholar] [CrossRef]
- Faes, L.; Porta, A.; Nollo, G.; Javorka, M. Information decomposition in multivariate systems: Definitions, implementation and application to cardiovascular networks. Entropy 2017, 19, 5. [Google Scholar] [CrossRef]
- Bossomaier, T.; Barnett, L.; Harré, M.; Lizier, J. An Introduction to Transfer Entropy: Information Flow in Complex Systems; Springer International Publishing: Berlin, Germany; Cham, Switzerland, 2016. [Google Scholar]
- Williams, P.L.; Beer, R.D. Nonnegative decomposition of multivariate information. arXiv 2010, arXiv:1004.2515. [Google Scholar]
- Barrett, A.B.; Barnett, L.; Seth, A.K. Multivariate Granger causality and generalized variance. Phys. Rev. E 2010, 81, 041907. [Google Scholar] [CrossRef]
- Faes, L.; Nollo, G.; Porta, A. Information decomposition: A tool to dissect cardiovascular and cardiorespiratory complexity. In Complexity Nonlinearity Cardiovasc. Signals; Springer: Cham, Switzerland, 2017; pp. 87–113. [Google Scholar]
- Faes, L.; Nollo, G.; Stramaglia, S.; Marinazzo, D. Multiscale granger causality. Phys. Rev. E 2017, 96, 042150. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, T.; Schmitz, A. Improved surrogate data for nonlinearity tests. Phys. Rev. Lett. 1996, 77, 635. [Google Scholar] [CrossRef]
- Toppi, J.; Mattia, D.; Risetti, M.; Formisano, R.; Babiloni, F.; Astolfi, L. Testing the significance of connectivity networks: Comparison of different assessing procedures. IEEE Trans. Biomed. Eng. 2016, 63, 2461–2473. [Google Scholar] [PubMed]
- Porta, A.; Faes, L.; Nollo, G.; Bari, V.; Marchi, A.; De Maria, B.; Takahashi, A.C.; Catai, A.M. Conditional self-entropy and conditional joint transfer entropy in heart period variability during graded postural challenge. PLoS ONE 2015, 10, e0132851. [Google Scholar] [CrossRef] [PubMed]
- Anzolin, A.; Astolfi, L. Statistical Causality in the EEG for the Study of Cognitive Functions in Healthy and Pathological Brains; Sapienza University of Rome: Rome, Italy, 2018; Available online: https://iris.uniroma1.it (accessed on 19 February 2018).
- Kim, S.; Kim, H. A new metric of absolute percentage error for intermittent demand forecasts. Int. J. Forecast. 2016, 32, 669–679. [Google Scholar] [CrossRef]
- Toppi, J.; Sciaraffa, N.; Antonacci, Y.; Anzolin, A.; Caschera, S.; Petti, M.; Mattia, D.; Astolfi, L. Measuring the agreement between brain connectivity networks. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 68–71. [Google Scholar]
- Porta, A.; D’addio, G.; Guzzetti, S.; Lucini, D.; Pagani, M. Testing the presence of non stationarities in short heart rate variability series. In Proceedings of the Computers in Cardiology, Chicago, IL, USA, 19–22 September 2004; pp. 645–648. [Google Scholar]
- Schwarz, G. Estimating the dimension of a model. Annu. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Rubinov, M.; Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 2010, 52, 1059–1069. [Google Scholar] [CrossRef]
- Lizier, J.T.; Bertschinger, N.; Jost, J.; Wibral, M. Information decomposition of target effects from multi-source interactions: Perspectives on previous, current and future work. Entropy 2018, 220, 307. [Google Scholar] [CrossRef]
- Silvey, S. Multicollinearity and imprecise estimation. J. R. Stat. Soc. Ser. B (Methodol.) 1969, 31, 539–552. [Google Scholar] [CrossRef]
- Rish, I.; Grabarnik, G. Sparse Modeling: Theory, Algorithms, and Applications; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Zou, H. The adaptive lasso and its oracle properties. J. Am. Stat. Assoc. 2006, 101, 1418–1429. [Google Scholar] [CrossRef]
- Fan, J.; Li, R. Variable selection via nonconcave penalized likelihood and its oracle properties. J. Am. Stat. Assoc. 2001, 96, 1348–1360. [Google Scholar] [CrossRef]
- Irfan, M.; Javed, M.; Raza, M.A. Comparison of shrinkage regression methods for remedy of multicollinearity problem. Middle-East J. Sci. Res. 2013, 14, 570–579. [Google Scholar]
- Abd Elrahman, S.M.; Abraham, A. A review of class imbalance problem. J. Netw. Innov. Comput. 2013, 1, 332–340. [Google Scholar]
- Chetverikov, D.; Liao, Z.; Chernozhukov, V. On Cross-Validated LASSO in High Dimensions; Technical Report, Working Paper; UCLA: Los Angeles, CA, USA, February 2020. [Google Scholar]
- Schulz, S.; Haueisen, J.; Bär, K.J.; Voss, A. Multivariate assessment of the central-cardiorespiratory network structure in neuropathological disease. Physiol. Meas. 2018, 39, 074004. [Google Scholar] [CrossRef]
- Lin, A.; Liu, K.K.; Bartsch, R.P.; Ivanov, P.C. Dynamic network interactions among distinct brain rhythms as a hallmark of physiologic state and function. Commun. Biol. 2020, 3, 1–11. [Google Scholar]
- Kuipers, N.T.; Sauder, C.L.; Carter, J.R.; Ray, C.A. Neurovascular responses to mental stress in the supine and upright postures. J. Appl. Physiol. 2008, 104, 1129–1136. [Google Scholar] [CrossRef]
- Berntson, G.G.; Cacioppo, J.T.; Quigley, K.S. Respiratory sinus arrhythmia: Autonomic origins, physiological mechanisms, and psychophysiological implications. Psychophysiology 1993, 30, 183–196. [Google Scholar] [CrossRef]
- Schäfer, C.; Rosenblum, M.G.; Kurths, J.; Abel, H.H. Heartbeat synchronized with ventilation. Nature 1998, 392, 239–240. [Google Scholar] [CrossRef]
- Drinnan, M.J.; Allen, J.; Murray, A. Relation between heart rate and pulse transit time during paced respiration. Physiol. Meas. 2001, 22, 425. [Google Scholar] [CrossRef]
- Vrieze, S.I. Model selection and psychological theory: A discussion of the differences between the Akaike information criterion (AIC) and the Bayesian information criterion (BIC). Psychol. Methods 2012, 17, 228. [Google Scholar] [CrossRef] [PubMed]
- Faes, L.; Stramaglia, S.; Marinazzo, D. On the interpretability and computational reliability of frequency-domain Granger causality. F1000Research 2017, 6, 1710. [Google Scholar] [CrossRef]
- Kuo, T.B.; Chen, C.Y.; Hsu, Y.C.; Yang, C.C. EEG beta power and heart rate variability describe the association between cortical and autonomic arousals across sleep. Auton. Neurosci. 2016, 194, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Sato, W.; Toichi, M.; Murai, T.; Okada, T.; Hayashi, A.; Sengoku, A. Frontal midline theta rhythm is correlated with cardiac autonomic activities during the performance of an attention demanding meditation procedure. Cognit. Brain Res. 2001, 11, 281–287. [Google Scholar] [CrossRef]
- Behzadnia, A.; Ghoshuni, M.; Chermahini, S. EEG Activities and the Sustained Attention Performance. Neurophysiology 2017, 49, 226–233. [Google Scholar] [CrossRef]
- Tort, A.B.; Ponsel, S.; Jessberger, J.; Yanovsky, Y.; Brankačk, J.; Draguhn, A. Parallel detection of theta and respiration-coupled oscillations throughout the mouse brain. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pernice, R.; Javorka, M.; Krohova, J.; Czippelova, B.; Turianikova, Z.; Busacca, A.; Faes, L. Comparison of short-term heart rate variability indexes evaluated through electrocardiographic and continuous blood pressure monitoring. Med. Biol. Eng. Comput. 2019, 57, 1247–1263. [Google Scholar] [CrossRef]
- Silvani, A.; Calandra-Buonaura, G.; Dampney, R.A.; Cortelli, P. Brain–heart interactions: Physiology and clinical implications. Philos. Trans. R. Soc. A 2016, 374, 20150181. [Google Scholar] [CrossRef]
- Jurysta, F.; Lanquart, J.P.; Sputaels, V.; Dumont, M.; Migeotte, P.F.; Leistedt, S.; Linkowski, P.; Van De Borne, P. The impact of chronic primary insomnia on the heart rate–EEG variability link. Clin. Neurophysiol. 2009, 120, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Jurysta, F.; Lanquart, J.P.; Van De Borne, P.; Migeotte, P.F.; Dumont, M.; Degaute, J.P.; Linkowski, P. The link between cardiac autonomic activity and sleep delta power is altered in men with sleep apnea-hypopnea syndrome. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2006, 291, R1165–R1171. [Google Scholar] [CrossRef][Green Version]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Seri. B (Stat. Method.) 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Milde, T.; Leistritz, L.; Astolfi, L.; Miltner, W.H.; Weiss, T.; Babiloni, F.; Witte, H. A new Kalman filter approach for the estimation of high-dimensional time-variant multivariate AR models and its application in analysis of laser-evoked brain potentials. Neuroimage 2010, 50, 960–969. [Google Scholar] [CrossRef] [PubMed]

| Factor | |||||
|---|---|---|---|---|---|
| K | 8582 ** | 1694 ** | 2204 ** | 197.2 ** | 2492 ** |
| TYPE | 1640 ** | 377 ** | 3538 ** | 223.4 ** | 1575 ** |
| K × TYPE | 8633 ** | 848 ** | 1055 ** | 114.5 ** | 339 ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonacci, Y.; Astolfi, L.; Nollo, G.; Faes, L. Information Transfer in Linear Multivariate Processes Assessed through Penalized Regression Techniques: Validation and Application to Physiological Networks. Entropy 2020, 22, 732. https://doi.org/10.3390/e22070732
Antonacci Y, Astolfi L, Nollo G, Faes L. Information Transfer in Linear Multivariate Processes Assessed through Penalized Regression Techniques: Validation and Application to Physiological Networks. Entropy. 2020; 22(7):732. https://doi.org/10.3390/e22070732
Chicago/Turabian StyleAntonacci, Yuri, Laura Astolfi, Giandomenico Nollo, and Luca Faes. 2020. "Information Transfer in Linear Multivariate Processes Assessed through Penalized Regression Techniques: Validation and Application to Physiological Networks" Entropy 22, no. 7: 732. https://doi.org/10.3390/e22070732
APA StyleAntonacci, Y., Astolfi, L., Nollo, G., & Faes, L. (2020). Information Transfer in Linear Multivariate Processes Assessed through Penalized Regression Techniques: Validation and Application to Physiological Networks. Entropy, 22(7), 732. https://doi.org/10.3390/e22070732






