Binary Expression Enhances Reliability of Messaging in Gene Networks
Abstract
1. Introduction
2. Model and Methods
2.1. Qualitative Model and Noise Quantification
2.2. A Stochastic Model for a Constitutive Gene
2.3. A Stochastic Model for the Binary Gene
2.3.1. Interpretation of the Parameters , , and N.
2.3.2. The Mean Number and the Conditional Mean Number of the Stochastic Model for a Binary Gene
2.3.3. The Variance and Bursting Noise of N on the Stochastic Model for a Binary Gene
2.4. Analyzing the Information Content of the Message
2.4.1. Entropy for the Constitutive Source
2.4.2. Entropy, Conditional Entropy and Mutual Information for the Binary Source
3. Results
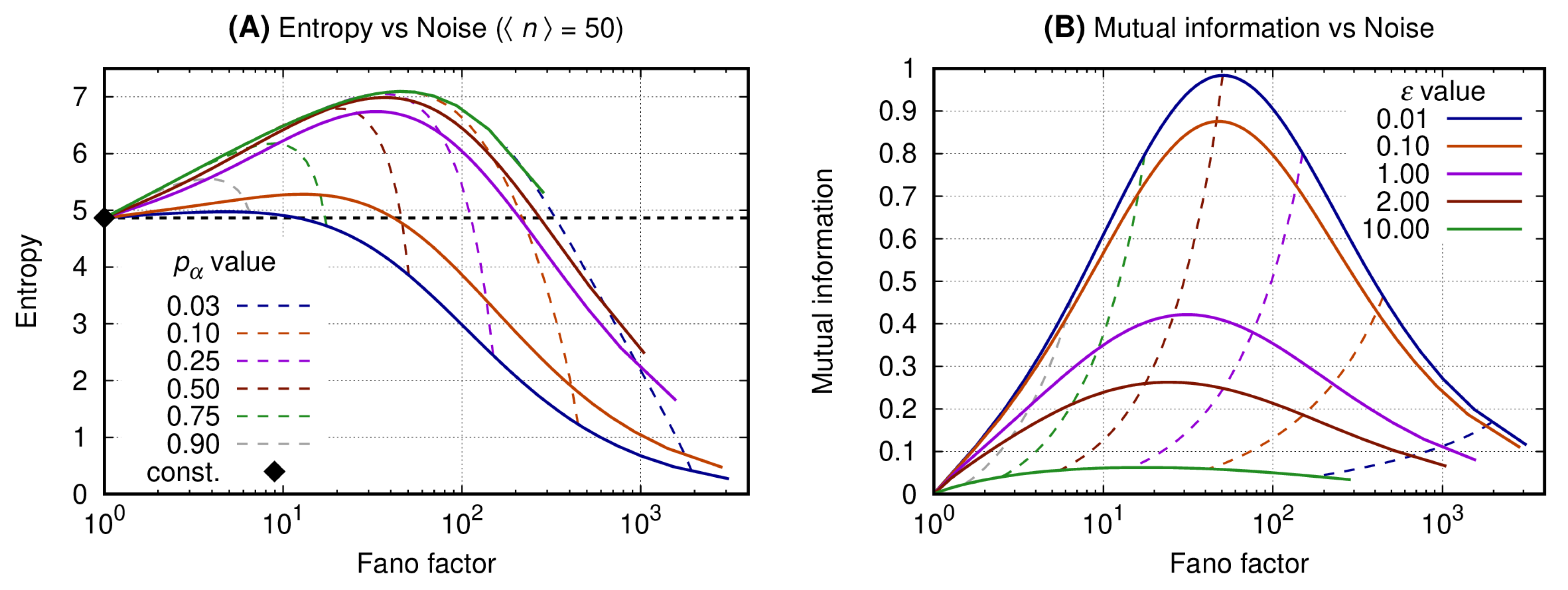
3.1. Binary Expression Enables Entropy Reduction and Mutual Information Increase
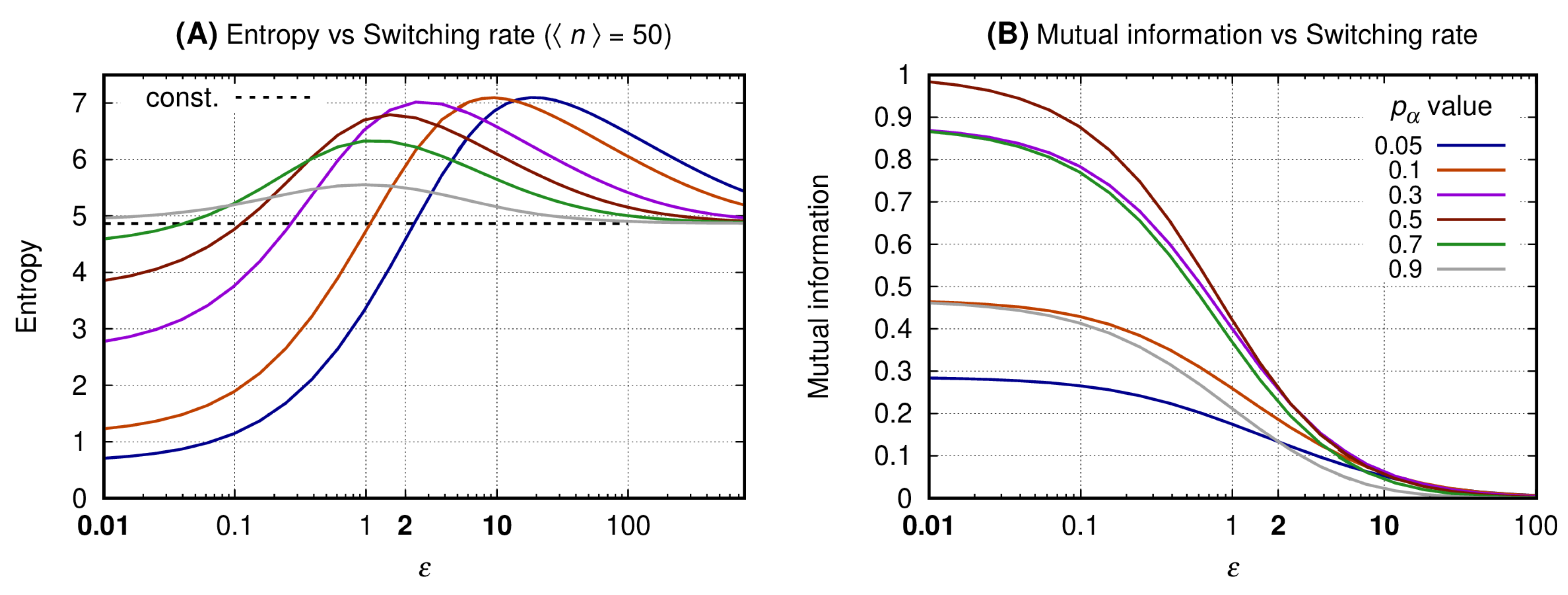
3.2. The Slow Switching Genes Generate Reduced Entropy and Increased Values for Mutual Information
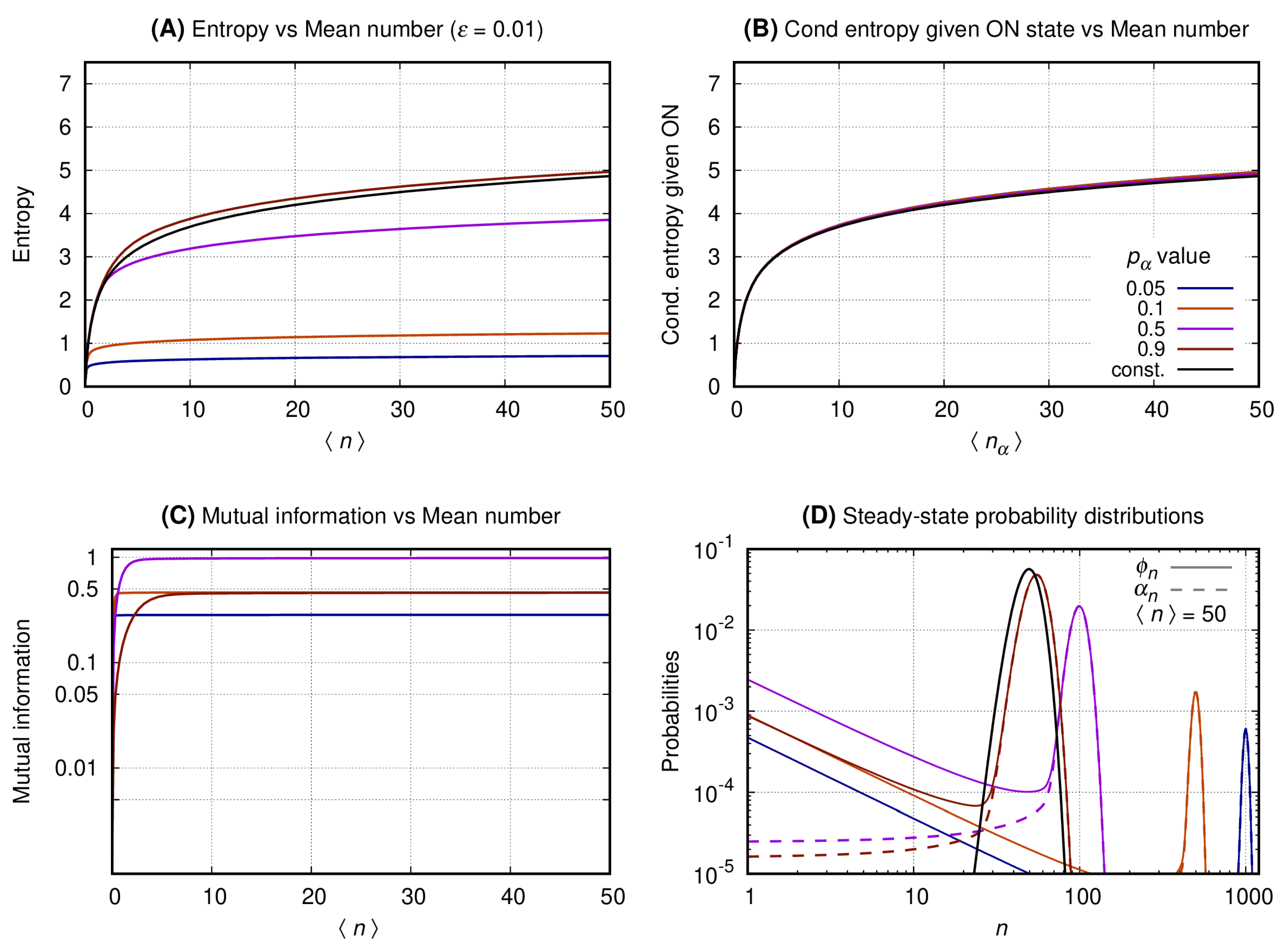
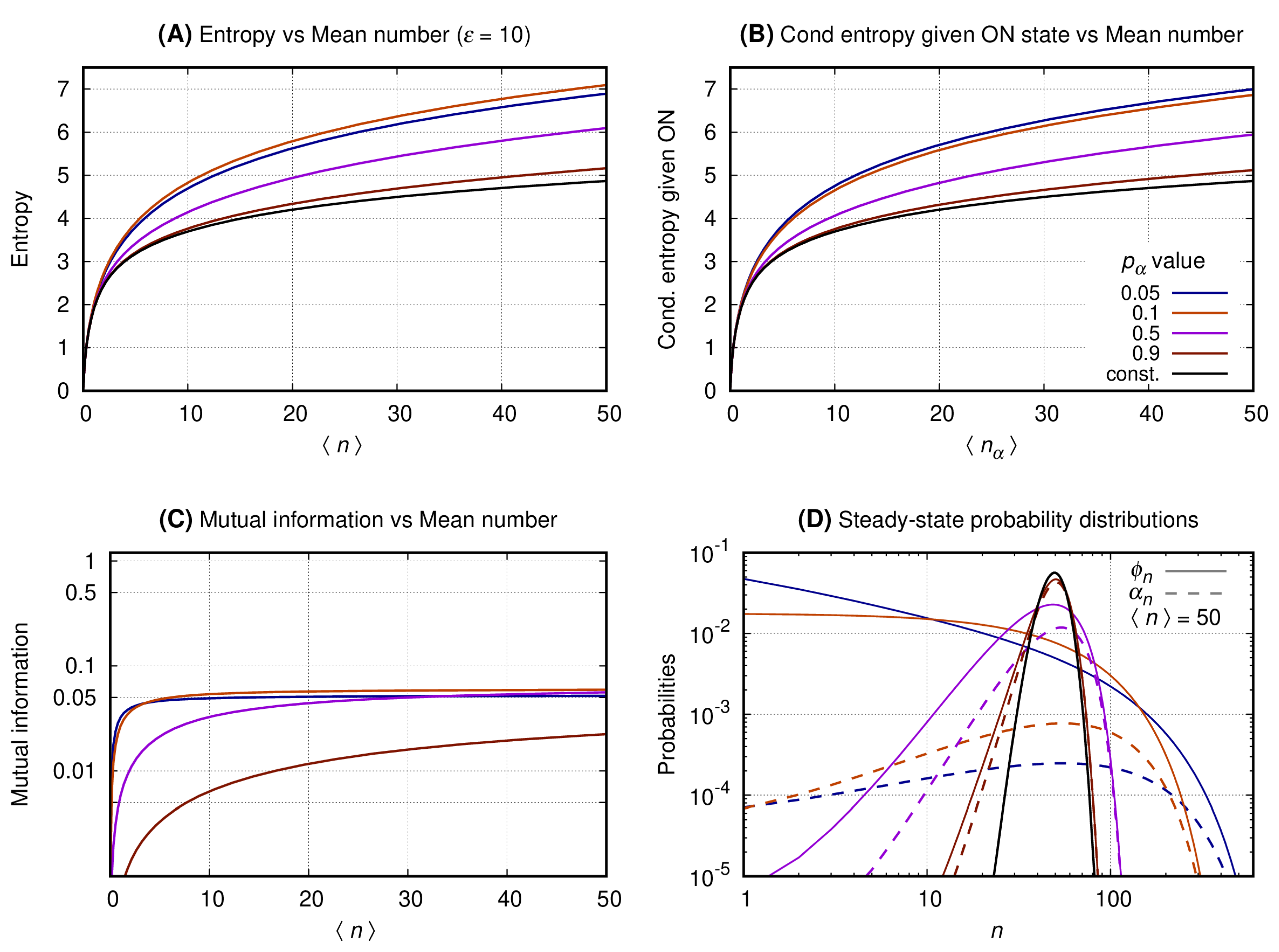
3.3. The Distributions of the Slow Switching Bursty Regime with Reduced Entropy and Maximal Mutual Information Are Bimodal
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Delbrück, M. Statistical fluctuations in autocatalytic reactions. J. Chem. Phys. 1940, 8, 120–124. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Swain, P.S. Stochastic gene expression in a single cell. Science 2002, 297, 1183–1186. [Google Scholar] [CrossRef]
- Blake, W.J.; KÆrn, M.; Cantor, C.R.; Collins, J.J. Noise in eukaryotic gene expression. Nature 2003, 422, 633–637. [Google Scholar] [CrossRef]
- Chubb, J.R.; Trcek, T.; Shenoy, S.M.; Singer, R.H. Transcriptional Pulsing of a Developmental Gene. Curr. Biol. 2006, 16, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- Boettiger, A.N.; Levine, M. Synchronous and Stochastic Patterns of Gene Activation in the Drosophila Embryo. Science 2009, 325, 471–473. [Google Scholar] [CrossRef] [PubMed]
- Paré, A.; Lemons, D.; Kosman, D.; Beaver, W.; Freund, Y.; McGinnis, W. Visualization of individual Scr mRNAs during Drosophila embryogenesis yields evidence for transcriptional bursting. Curr. Biol. 2009, 19, 2037–2042. [Google Scholar] [CrossRef]
- Little, S.C.; Tikhonov, M.; Gregor, T. Precise Developmental Gene Expression Arises from Globally Stochastic Transcriptional Activity. Cell 2013, 154, 789–800. [Google Scholar] [CrossRef]
- Suter, D.M.; Molina, N.; Gatfield, D.; Schneider, K.; Schibler, U.; Naef, F. Mammalian genes are transcribed with widely different bursting kinetics. Science 2011, 332, 472–474. [Google Scholar] [CrossRef]
- Raser, J.M.; O’Shea, E.K. Noise in gene expression: Origins, consequences, and control. Science 2005, 309, 2010–2013. [Google Scholar] [CrossRef]
- Hooshangi, S.; Weiss, R. The effect of negative feedback on noise propagation in transcriptional gene networks. Chaos 2006, 16, 026108. [Google Scholar] [CrossRef]
- Balázsi, G.; van Oudenaarden, A.; Collins, J.J. Cellular decision making and biological noise: From microbes to mammals. Cell 2011, 144, 910–925. [Google Scholar] [CrossRef] [PubMed]
- Chalancon, G.; Ravarani, C.N.; Balaji, S.; Martinez-Arias, A.; Aravind, L.; Jothi, R.; Babu, M.M. Interplay between gene expression noise and regulatory network architecture. Trends Genet. 2012, 28, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Bothma, J.P.; Garcia, H.G.; Esposito, E.; Schlissel, G.; Gregor, T.; Levine, M. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl. Acad. Sci. USA 2014, 111, 10598–10603. [Google Scholar] [CrossRef]
- Zoller, B.; Little, S.C.; Gregor, T. Diverse spatial expression patterns emerge from unified kinetics of transcriptional bursting. Cell 2018, 175, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Holloway, D.M.; Spirov, A.V. Transcriptional bursting in Drosophila development: Stochastic dynamics of eve stripe 2 expression. PLoS ONE 2017, 12, e0176228. [Google Scholar] [CrossRef] [PubMed]
- Falo-Sanjuan, J.; Bray, S.J. Decoding the Notch signal. Dev. Growth Differ. 2019, 62, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Lammers, N.C.; Galstyan, V.; Reimer, A.; Medin, S.A.; Wiggins, C.H.; Garcia, H.G. Multimodal transcriptional control of pattern formation in embryonic development. Proc. Natl. Acad. Sci. USA 2020, 117, 836–847. [Google Scholar] [CrossRef]
- Ziv, E.; Nemenman, I.; Wiggins, C.H. Optimal Signal Processing in Small Stochastic Biochemical Networks. PLoS ONE 2007, 2, e1077. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tkačik, G.; Callan, C.G., Jr.; Bialek, W. Information flow and optimization in transcriptional regulation. Proc. Natl. Acad. Sci. USA 2008, 105, 12265–12270. [Google Scholar] [CrossRef]
- Bowsher, C.G.; Swain, P.S. Identifying sources of variation and the flow of information in biochemical networks. Proc. Natl. Acad. Sci. USA 2012, 109, E1320–E1328. [Google Scholar] [CrossRef]
- Garcia, H.G.; Tikhonov, M.; Lin, A.; Gregor, T. Quantitative live imaging of transcription in Drosophila embryos links polymerase activity to macroscopic patterns. Curr. Biol. 2013, 23, 2140–2145. [Google Scholar] [CrossRef] [PubMed]
- Fukaya, T.; Lim, B.; Levine, M. Enhancer Control of Transcriptional Bursting. Cell 2016, 166, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Reinitz, J.; Hou, S.; Sharp, D.H. Transcriptional control in Drosophila. Complexus 2003, 1, 54–64. [Google Scholar] [CrossRef]
- Janssens, H.; Hou, S.; Jaeger, J.; Kim, A.; Myasnikova, E.; Sharp, D.; Reinitz, J. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even-skipped gene. Nat. Genet. 2006, 38, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.R.; Martinez, C.; Ionides, J.; Ramos, A.F.; Ludwig, M.Z.; Ogawa, N.; Sharp, D.H.; Reinitz, J. Rearrangements of 2.5 Kilobases of Noncoding DNA from the Drosophila even-skipped Locus Define Predictive Rules of Genomic cis-Regulatory Logic. PLoS Genet. 2013, 9, e1003243. [Google Scholar] [CrossRef]
- Barr, K.A.; Martinez, C.; Moran, J.R.; Kim, A.R.; Ramos, A.F.; Reinitz, J. Synthetic enhancer design by in silico compensatory evolution reveals flexibility and constraint in cis-regulation. BMC Syst. Biol. 2017, 11, 116:1–116:15. [Google Scholar] [CrossRef] [PubMed]
- Granados, A.A.; Pietsch, J.M.J.; Cepeda-Humerez, S.A.; Farquhar, I.L.; Tkačik, G.; Swain, P.S. Distributed and dynamic intracellular organization of extracellular information. Proc. Natl. Acad. Sci. USA 2018, 115, 6088–6093. [Google Scholar] [CrossRef]
- Liu, Y.; Barr, K.; Reinitz, J. Fully Interpretable Deep Learning Model of Transcriptional Control. bioRxiv 2019, 1–11. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Ashall, L.; Horton, C.A.; Nelson, D.E.; Paszek, P.; Harper, C.V.; Sillitoe, K.; Ryan, S.; Spiller, D.G.; Unitt, J.F.; Broomhead, D.S.; et al. Pulsatile Stimulation Determines Timing and Specificity of NF-κB-Dependent Transcription. Science 2009, 324, 242–246. [Google Scholar] [CrossRef]
- Cheong, R.; Rhee, A.; Wang, C.; Nemenman, I.; Levchenko, A. Information Transduction Capacity of Noisy Biochemical Signaling Networks. Science 2011, 334, 354–358. [Google Scholar] [CrossRef]
- Levchenko, A.; Nemenman, I. Cellular noise and information transmission. Curr. Opin. Biotechnol. 2014, 28, 156–164. [Google Scholar] [CrossRef]
- Ghorbani, M.; Jonckheere, E.A.; Bogdan, P. Gene Expression Is Not Random: Scaling, Long-Range Cross-Dependence, and Fractal Characteristics of Gene Regulatory Networks. Front. Physiol. 2018, 9, 1446:1–1446:12. [Google Scholar] [CrossRef] [PubMed]
- Reinitz, J.; Mjolsness, E.; Sharp, D.H. Cooperative control of positional information in Drosophila by bicoid and maternal hunchback. J. Exp. Zool. 1995, 271, 47–56. [Google Scholar] [CrossRef]
- Jaeger, J.; Surkova, S.; Blagov, M.; Janssens, H.; Kosman, D.; Kozlov, K.N.; Myasnikova, E.; Vanario-Alonso, C.E.; Samsonova, M.; Sharp, D.H.; et al. Dynamic control of positional information in the early Drosophila embryo. Nature 2004, 430, 368–371. [Google Scholar] [CrossRef]
- Gregor, T.; Tank, D.W.; Wieschaus, E.F.; Bialek, W. Probing the limits to positional information. Cell 2007, 130, 153–164. [Google Scholar] [CrossRef] [PubMed]
- West, J.; Bianconi, G.; Severini, S.; Teschendorff, A.E. Differential network entropy reveals cancer system hallmarks. Sci. Rep. 2012, 2, 802:1–802:8. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Frieden, B.R. The critical roles of information and nonequilibrium thermodynamics in evolution of living systems. Bull. Math. Biol. 2013, 75, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar]
- Kussell, E.; Leibler, S. Phenotypic diversity, population growth, and information in fluctuating environments. Science 2005, 309, 2075–2078. [Google Scholar] [CrossRef]
- Belete, M.K.; Balázsi, G. Optimality and adaptation of phenotypically switching cells in fluctuating environments. Phys. Rev. E 2015, 92, 062716. [Google Scholar] [CrossRef] [PubMed]
- Peccoud, J.; Ycart, B. Markovian modelling of gene product synthesis. Theor. Popul. Biol. 1995, 48, 222–234. [Google Scholar] [CrossRef]
- Ramos, A.F.; Innocentini, G.C.P.; Forger, F.M.; Hornos, J.E.M. Symmetry in biology: From genetic code to stochastic gene regulation. IET Syst. Biol. 2010, 4, 311–329. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Peskin, C.S.; Tranchina, D.; Vargas, D.Y.; Tyagi, S. Stochastic mRNA Synthesis in Mammalian Cells. PLoS Biol. 2006, 4, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- Innocentini, G.C.P.; Hornos, J.E.M. Modeling stochastic gene expression under repression. J. Math. Biol. 2007, 55, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Shahrezaei, V.; Swain, P.S. Analytical distributions for stochastic gene expression. Proc. Natl. Acad. Sci. USA 2008, 105, 17256–17261. [Google Scholar] [CrossRef]
- Paulsson, J. Models of stochastic gene expression. Phys. Life Rev. 2005, 2, 157–175. [Google Scholar] [CrossRef]
- Munsky, B.; Neuert, G.; van Oudenaarden, A. Using Gene Expression Noise to Understand Gene Regulation. Science 2012, 336, 183–187. [Google Scholar] [CrossRef]
- Abramowitz, M.; Stegun, I.A. (Eds.) Handbook of Mathematical Functions: With Formulas, Graphs, and Mathematical Tables, 10th ed.; National Bureau of Standards Applied Mathematics Series; U.S. Department of Commerce, National Bureau of Standards: Washington, DC, USA, 1972; Volume 55.
- Thattai, M.; van Oudenaarden, A. Intrinsic noise in gene regulatory networks. Proc. Natl. Acad. Sci. USA 2001, 98, 8614–8619. [Google Scholar] [CrossRef]
- Zenklusen, D.; Larson, D.R.; Singer, R.H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 2008, 15, 1263. [Google Scholar] [CrossRef]
- Yvinec, R.; da Silva, L.G.S.; Prata, G.N.; Reinitz, J.; Ferreira Ramos, A. Bursting on a two state stochastic model for gene transcription in Drosophila embryos. bioRxiv 2017. [Google Scholar] [CrossRef]
- Manu, S.S.; Spirov, A.V.; Gursky, V.V.; Janssens, H.; Kim, A.R.; Radulescu, O.; Vanario-Alonso, C.E.; Sharp, D.H.; Samsonova, M.; Reinitz, J. Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. Plos Biol. 2009, 7, e1000049. [Google Scholar] [CrossRef] [PubMed]
- Bothma, J.P.; Garcia, H.G.; Ng, S.; Perry, M.W.; Gregor, T.; Levine, M. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. eLife 2015, 4, e07956. [Google Scholar] [CrossRef]
- Savageau, M.A. Comparison of classical and autogenous systems of regulations in inducible operons. Nature 1974, 252, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Becskei, A.; Serrano, L. Engineering stability in gene networks by autoregulation. Nature 2000, 405, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Nevozhay, D.; Adams, R.M.; Murphy, K.F.; Josic, K.; Balázsi, G. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 5123–5128. [Google Scholar] [CrossRef]
- Ramos, A.F.; Hornos, J.E.M.; Reinitz, J. Gene regulation and noise reduction by coupling of stochastic processes. Phys. Rev. E 2015, 91, 020701(R). [Google Scholar] [CrossRef]
- Cao, Z.; Grima, R. Analytical distributions for detailed models of stochastic gene expression in eukaryotic cells. Proc. Natl. Acad. Sci. USA 2020, 117, 4682–4692. [Google Scholar] [CrossRef]
- Ochiai, H.; Sugawara, T.; Sakuma, T.; Yamamoto, T. Stochastic promoter activation affects Nanog expression variability in mouse embryonic stem cells. Sci. Rep. 2014, 4, 1–9. [Google Scholar] [CrossRef]
- Senecal, A.; Munsky, B.; Proux, F.; Ly, N.; Braye, F.E.; Zimmer, C.; Mueller, F.; Darzacq, X. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 2014, 8, 75–83. [Google Scholar] [CrossRef]
- Halpern, K.B.; Tanami, S.; Landen, S.; Chapal, M.; Szlak, L.; Hutzler, A.; Nizhberg, A.; Itzkovitz, S. Bursty gene expression in the intact mammalian liver. Mol. Cell 2015, 58, 147–156. [Google Scholar] [CrossRef]
- Skinner, S.O.; Xu, H.; Nagarkar-Jaiswal, S.; Freire, P.R.; Zwaka, T.P.; Golding, I. Single-cell analysis of transcription kinetics across the cell cycle. Elife 2016, 5, e12175. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A.J.; Johnsson, P.; Hagemann-Jensen, M.; Hartmanis, L.; Faridani, O.R.; Reinius, B.; Segerstolpe, Å.; Rivera, C.M.; Ren, B.; Sandberg, R. Genomic encoding of transcriptional burst kinetics. Nature 2019, 565, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Skinner, S.O.; Sokac, A.M.; Golding, I. Stochastic kinetics of nascent RNA. Phys. Rev. Lett. 2016, 117, 128101. [Google Scholar] [CrossRef] [PubMed]
- Milo, R.; Jorgensen, P.; Moran, U.; Weber, G.; Springer, M. BioNumbers—the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2009, 38, D750–D753. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gama, L.R.; Giovanini, G.; Balázsi, G.; Ramos, A.F. Binary Expression Enhances Reliability of Messaging in Gene Networks. Entropy 2020, 22, 479. https://doi.org/10.3390/e22040479
Gama LR, Giovanini G, Balázsi G, Ramos AF. Binary Expression Enhances Reliability of Messaging in Gene Networks. Entropy. 2020; 22(4):479. https://doi.org/10.3390/e22040479
Chicago/Turabian StyleGama, Leonardo R., Guilherme Giovanini, Gábor Balázsi, and Alexandre F. Ramos. 2020. "Binary Expression Enhances Reliability of Messaging in Gene Networks" Entropy 22, no. 4: 479. https://doi.org/10.3390/e22040479
APA StyleGama, L. R., Giovanini, G., Balázsi, G., & Ramos, A. F. (2020). Binary Expression Enhances Reliability of Messaging in Gene Networks. Entropy, 22(4), 479. https://doi.org/10.3390/e22040479






