1. Introduction
Land cover classification based on satellite imagery is one of the most common and well-researched machine learning (ML) tasks in the Earth Observation (EO) community. In Europe, the Copernicus Sentinel-2 missions provide large amounts of global coverage EO data. With 5-day revisit times, Sentinel-2 generated a total of 9.4 PB of satellite imagery by April 2020 [
1]. The total amount of EO data available through the Copernicus services is estimated to exceed 150 PB. The data represent an opportunity for building solutions in various domains (agriculture, water management, biology or law enforcement); however, computationally efficient methods that yield sufficient results are needed to deal with this big data source. The most important issue when trying to maximize the accuracy of statistical learning methods is effective feature engineering. With many features, the algorithms become slower and consume more resources. In this paper, we present FASTENER (FeAture SelecTion ENabled by EntRopy,
https://github.com/JozefStefanInstitute/FASTENER) genetic algorithm for efficient feature selection, especially in EO tasks.
EO data sets are characterized by a large number of instances (millions or even billions) and a fair number of features (hundreds). These characteristics differ from the usual feature selection data sets that often contain up to several hundred instances and usually thousands of features. FASTENER exploits entropy-based measures to converge to a (near) optimal set of features for statistical learning faster than other algorithms. The algorithm can be used in many scenarios and can, for example, complement multiple-source data fusion and extensive feature generation in the fields of natural language processing, DNA microarray analysis or the Internet of Things [
2]. FASTENER has been tested on a land cover classification EO data set from Slovenia. Additionally, we have tested the algorithm against 25 general feature selection data sets.
The main evaluation use case presented in this paper is based on our previous contributions [
3,
4] regarding crop (land cover) classification using Sentinel-2 data from the European Space Agency (ESA). The scientific contributions of our work are as follows:
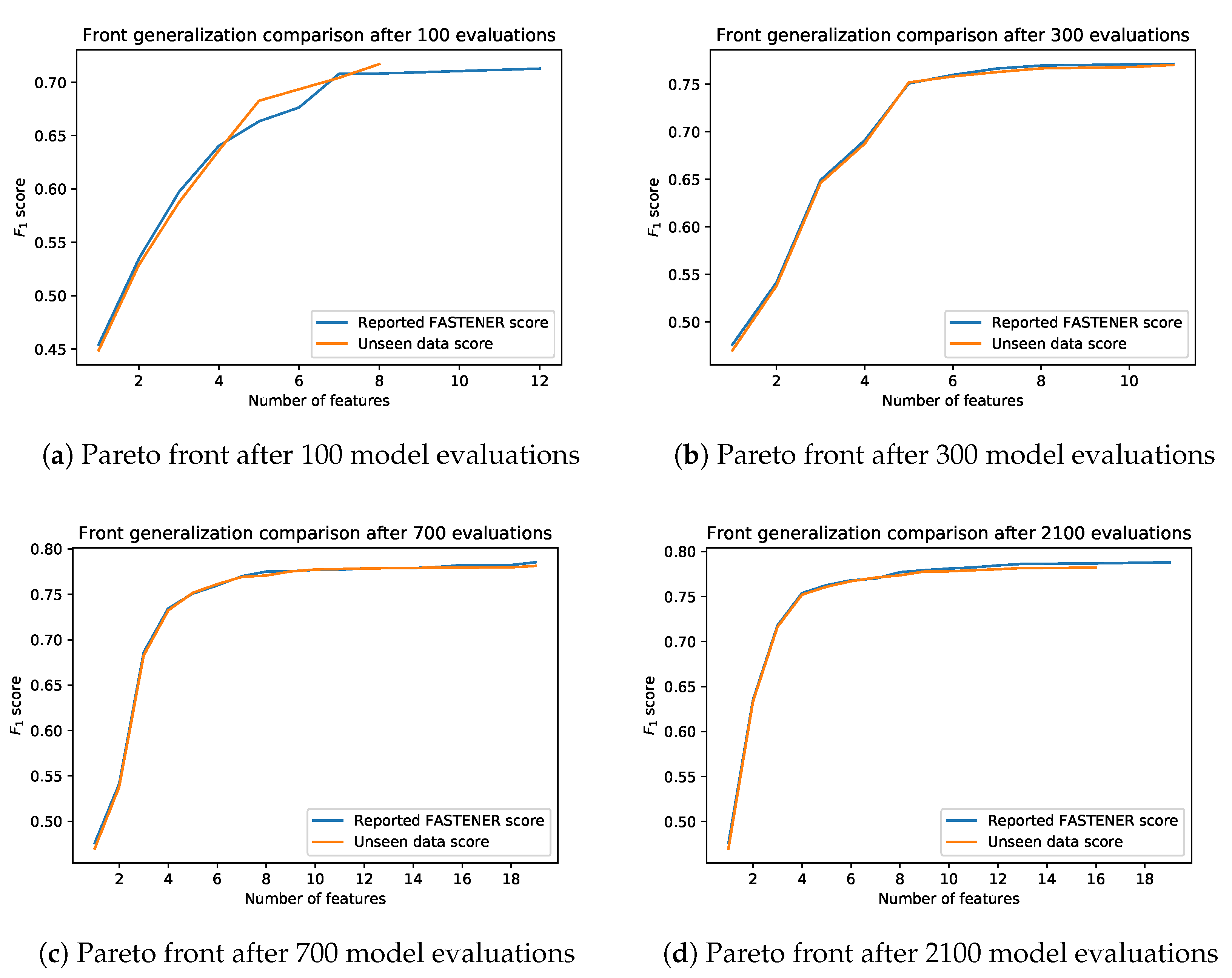
A novel genetic algorithm for feature selection based on entropy (FASTENER). Such an algorithm reduces the number of features while preserving (or even improving) the accuracy of the classification. The algorithm is particularly useful for data sets containing a large number of instances and hundreds of features. A reduced number of features reduces learning and inference times of classification algorithms as well as the time needed to derive the features. By using an entropy based approach, the algorithm converges to the (near) optimal subset faster than competing methods.
Improvement of the state-of-the-art in feature selection in the field of Earth Observation. FASTENER yields better result than current state-of-the-art algorithms in remote sensing. The algorithm has been tested and compared to other methods within the scope of the land-cover classification problem.
Usage of pre-trained models for information gain calculation for feature selection in Earth Observation scenarios. Usually, it is computationally expensive to train a machine learning model. The inference phase is much faster. FASTENER exploits the pre-trained models in order to estimate the information gain of certain features by inferring from data sets with randomly permuted values of the evaluated feature. This approach improves the convergence speed of the algorithm.
The paper is structured as follows. In the next section, related work regarding feature selection algorithms as well as state-of-the-art in feature selection in Earth Observation is described.
Section 4 contains a thorough description of the data used in the experiments and describes our algorithm.
Section 5 describes while
Section 6 discusses the experimental results and compares FASTENER to other algorithms based on NSGA-II. Finally, our conclusions are presented in
Section 7.
3. FASTENER—A Genetic Algorithm for Feature Selection
The presented algorithm is based on the POSS genetic algorithm and the Pareto ensemble pruning proposed in [
27,
28]. The main objective of the algorithm is to select an optimal subset of available features from a large feature set (
). The problem could be represented mathematically as finding the optimal (or near-optimal) binary vector
x of an indexed set
. Finding the optimal subset is NP-complete. Each element in
x corresponds to a particular feature that is included or excluded in our decision model. The optimality of such a set is measured by a scoring function, which we will denote by
(accuracy measure of model
M on a subset of features
x). Some standard examples of such measures in machine learning classification problems include precision, recall, accuracy and
score. In our experiments, the
score was used. We also adopt the notation
to denote the number of bits set in
x that directly corresponds to the number of selected features. Mathematically, for a fixed classification model
M and the number of features
k, we search for such an
x where the following maximum is reached:
As in POSS, the function creates a two-dimensional Pareto front. In one dimension, we evaluate k, the number of selected features, and in the other dimension, we present A score, which represents the optimality (e.g., accuracy) of such a subset. In this space, the ordering of instances is defined as follows. The pair dominates the pair if and only if and . Intuitively, the smaller set (by cardinality) of features provides classification results, which are at least as good as for the larger set. We denote such a relation by . The relation is obviously transitive but does not induce linear ordering. Pairs that are not comparable through such a relation are of particular interest, as they lie on the Pareto front. Feature subsets that lie on the Pareto front (not strongly dominated by other subsets) are the desired subsets. They provide the best classification power using the smallest number of features. The final result of a feature selection algorithm is the final Pareto front. The final Pareto front can be seen as an accumulation of the best results for a fixed k. The features with the best performance can then be selected from the front automatically (with a certain cut-off threshold), manually or by using additional information (e.g., time to calculate the features). The results clearly show that the Pareto front “plateaus” after the inclusion of some features and this fact can easily be used to automatically select the best number of features.
Our modification of the POSS algorithm combines Pareto front searching with a genetic algorithm to incorporate additional statistical information when recombining genes. The main ingredient in the algorithm is the concept of an item; a subset of features and scored item, which is an item with an assigned score corresponding to such a subset of features. The size of an item denoted by corresponds to the number of features selected. The algorithm works by successively evaluating items, combining them, and updating the Pareto front.
Each subset of features can be represented directly as a binary number, where set bits correspond to the included (selected) features and unset bits correspond to excluded ones. This allows for a natural representation of genes for such an item with a binary string or bit-set and also gives a natural human-readable representation as an arbitrary sized integer. Apart from this, many later needed operations can be quickly implemented as simple operations on a binary string. The size of an item corresponds to the number of set bits, the bit-wise and operation between two items corresponds to the genes that are common to both of them, while the bit-wise xor returns all different selected features.
In our implementation, the integer representation of item genes is treated in a little-endian way, as this allows easier extension of feature sets and keeps integer representations valid even when adding new features. For example, in a set of six features with 32 possible feature combinations, binary string 11010 represents a subset containing features with indices 0, 1 and 3 and is represented by a decimal number 11. Even if the number of features is increased, such a subset would still be represented by the number 11, albeit its binary string representation would be extended by zeros on the right side.
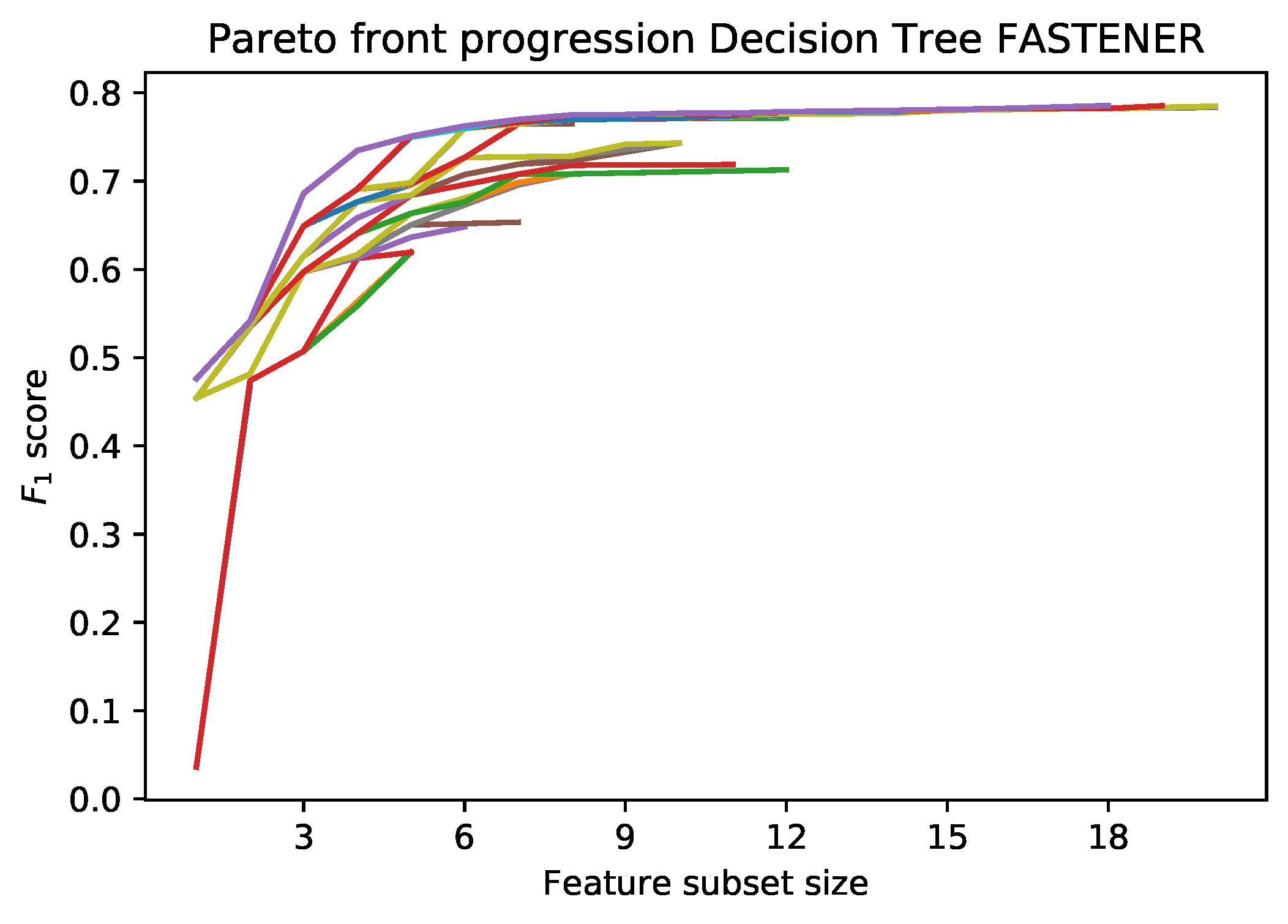
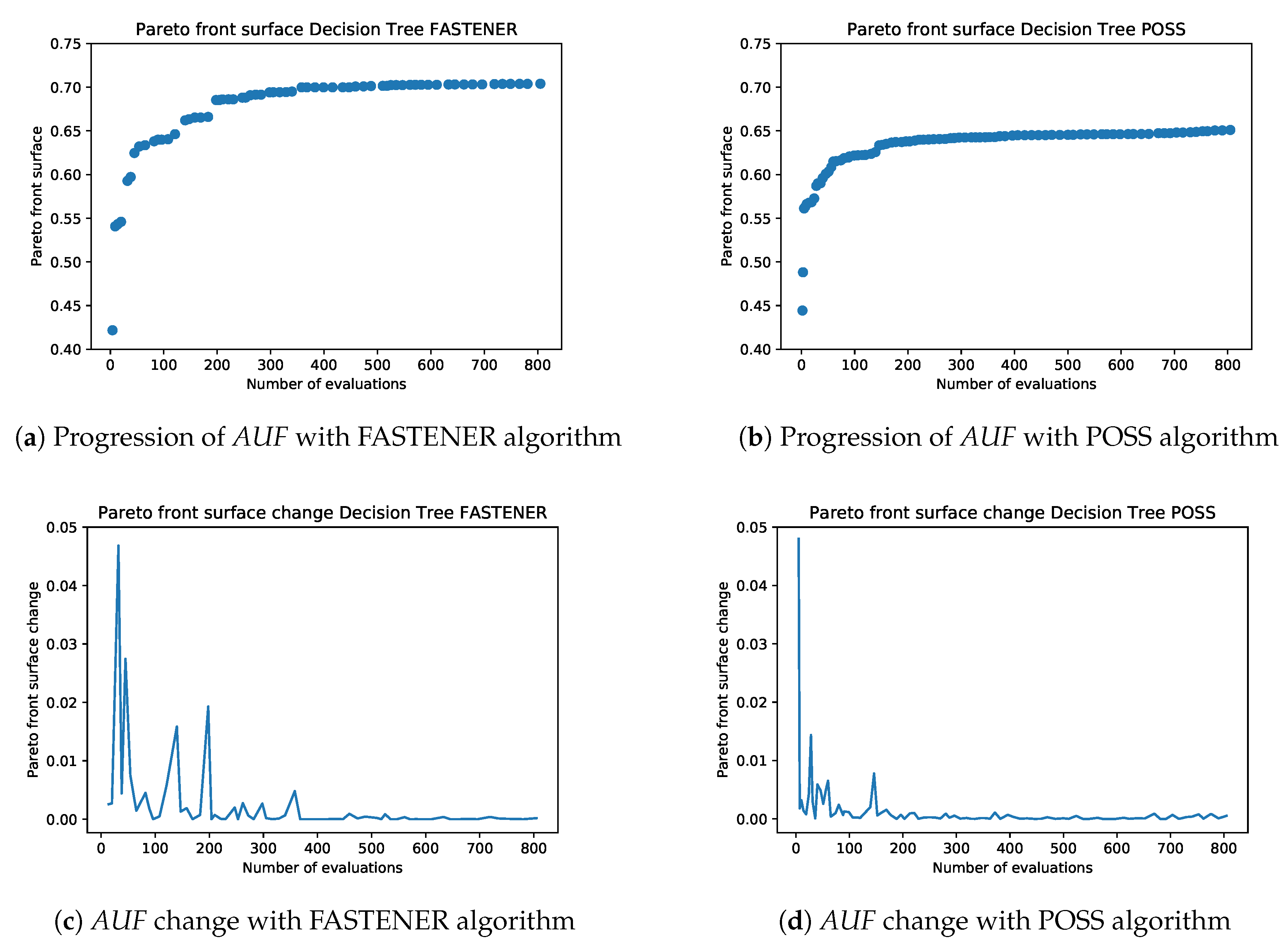
An example of the incremental Pareto front update is shown in
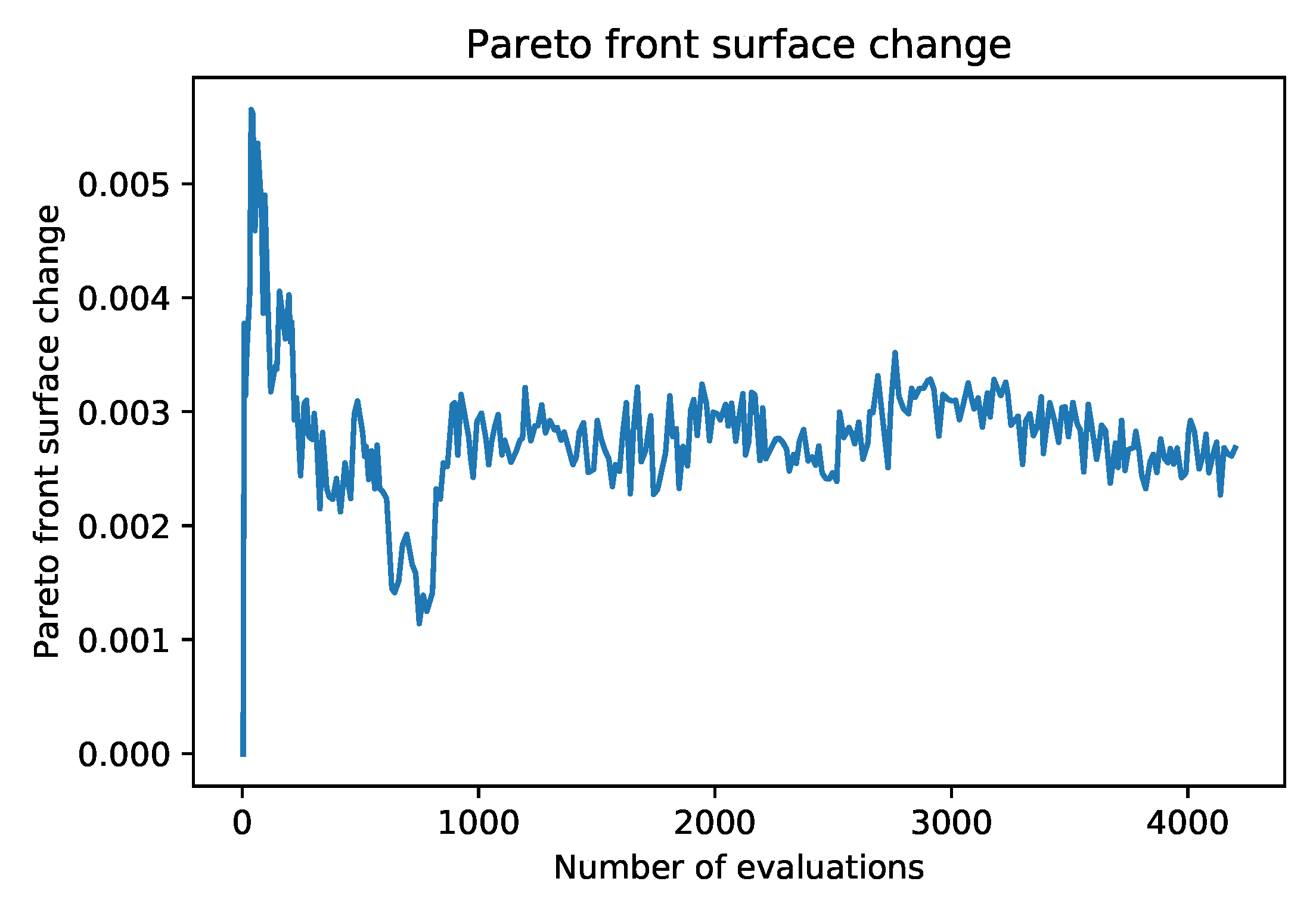
Figure 1. Each subsequent generation of the FASTENER algorithm produces a Pareto front that dominates the Pareto fronts from previous generations (in rare cases, where no improvement is made, the Pareto fronts of two subsequent generations may be the same). The main objective of the algorithm is to converge as quickly as possible to a theoretically limiting front (in terms of fitness function evaluations).
3.1. Algorithm
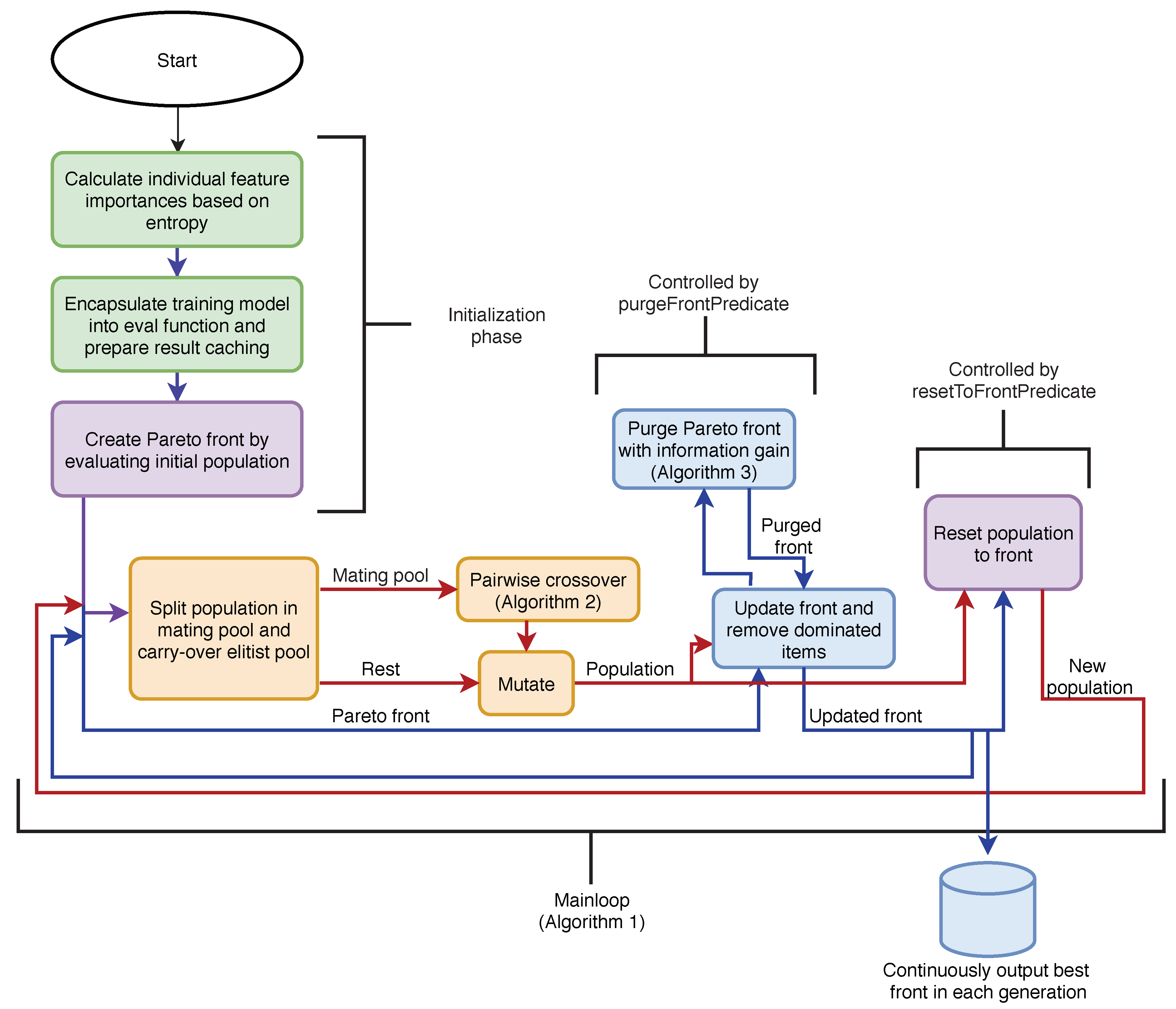
The algorithm stores the current Pareto-optimal items and progresses in generations. For each it keeps an item with the best score A. If there is no such item this means either that it is strictly dominated by another item or that the algorithm has not yet evaluated an item this size. In our implementation, the Pareto front is implemented as a simple dictionary. Apart from the Pareto front, the current population is kept in a separate set. The decoupling of the current population from the Pareto front allows the algorithm to “explore” different combinations more easily, while the Pareto front continues to be updated at each iteration.
As usual, in each generation, the current population of items is mated, a random genetic mutation is introduced and newly acquired items are evaluated. For each evaluation, the Pareto front is updated. If a new item is strictly better than an item on the front with the same size, the newly evaluated item is placed on the Pareto front. After all new items have been evaluated, the front is purged by removing items that are strictly dominated by other items. The pseudo-code for the basic algorithm is presented in Algorithm 1. The algorithm’s flow diagram is depicted in
Figure 2.
To prevent the search phase of the algorithm from diverging (the population as a whole is not controlled by the scoring function), the population is periodically reset to the current Pareto optimal items. In the presented algorithm, this decision is based only on the generation number; however, our implementation also considers the rate with which the running Pareto front is being updated between generations.
| Algorithm 1 Basic algorithm |
| Require: |
Initial population pop Evaluation function evalF Number of iterations K Mating pool selection strategy selectPool Crossover strategy crossover Mutation strategy mutate Predicate resetTOFront Predicate purgeFrontPredicate Front purging procedure purgeFront
- 1:
function FeatureSubsetSelection - 2:
front ← pop - 3:
for gen = 1 to K do - 4:
pool ← selectPool(pop) - 5:
pool ← crossover(pool) - 6:
pop ← mutate(pop ∪ pool) - 7:
pop ← evaluate(pop, evalF) - 8:
front ← front ∪ pop ▹ Add evaluated population to Pareto front - 9:
front ← removeDominated(front) ▹ Remove unoptimal items from Pareto front - 10:
if purgeFrontPredicate(gen) then - 11:
front ← purgeFront(front) - 12:
end if - 13:
if resetToFront(gen) then - 14:
pop ← front - 15:
end if - 16:
end for - 17:
return front - 18:
end function
|
3.2. Mutation
An important part when considering gene manipulation and modification is to keep the overall objective in mind. We want to find the optimal subset of features with good accuracy characteristics. The ultimate goal is to obtain a subset with a size much smaller than N. In preliminary experiments, the number of features with a sufficiently predictive power was around , which means a significant reduction in both training time and in data preparation time (which is true especially for EO data sets). Mutation and crossover methods should therefore be tailored to help maintain, reduce or only slightly increase the size of items.
The most interesting parts of the algorithm are the crossover and mutate procedures. The procedure for the mutation is adopted from the POSS algorithm and is a simple random bit mutation. Each random bit in the gene is flipped independently with a probability . Thus a new item is generated from each of the items selected for mating and, most importantly, the expected value of set bits is kept approximately the same. Formally, the expected value of the number of flips is 1, and therefore the mutations gravitate slowly toward x-s where . Since the desired range of features in which we can expect satisfactory score results is much lower than the asymptotic behaviour of mutations, this type of mutation serves as a size increaser. Mutation procedures therefore slowly increase and add new features to be considered. Furthermore, varying the probability of flipping is an easy way to control the speed of the introduction of new features. Setting the mutation rate to for and varying a with generation number and changes in the Pareto front updates can significantly increase the search space. In addition, all further analyses are still valid even if the mutation is set as an arbitrary function only dependent on N and heuristic data about mutations (independent of bit-index and bit status), called mutation function.
3.3. Crossover
The crossover procedure is designed to reduce the size of the newly produced item. The mating pool selection used for the algorithm is a simple random mating pool with a fixed size. Two different items from the mating pool are combined using the crossoverPair procedure. The crossoverPair procedure itself is presented in Algorithm 2 and requires 4 input parameters: two items to be mated, a scaling function for the number of non-intersected genes, and a set of entropy information for each individual feature. Without loss of generality, we may assume that .
Lemma 1. Let ITEM1 and ITEM2 be a random independent subset of where the probability of each gene being set is some positive function . The expected size of an intermediate gene conditional on the sizes of its parent genes is We can similarly derive conditional expectation for size change.
Lemma 2. The probability of an item having size k, assuming that each feature is selected independently with probability is Lemma 3. The probability of an intermediate item having size k conditional on sizes of its parents is similarlySince all features are equally likely to be selected, conditional probability is independent of scaling function f. Lemma 4. By linearity of expectation and assumption that and we can easily derive Remark 1. The assumption that each feature is selected independently with probability should not be confused with each subset of features selected with equal probability. In our case, this is beneficial because smaller sets are intrinsically better, which are easily controlled by the mutation function f.
| Algorithm 2 Randomized crossover with information gain weighting |
| Require: |
- 1:
function crossooverPair - 2:
if |item1| < |item2| then - 3:
swap(item1, item2) - 4:
end if - 5:
intermediate ← item & item2 ▹ Intersection of genes - 6:
rest ← item1 ⊕ item2 ▹ Bitwise XOR, features present in exactly one item - 7:
addNum ← onGeneScaling(abs(|item1| - |item2|)) - 8:
▹ Scale the number of new features according to provided function - 9:
additional ← rSelect(addNum, informationEntropy, rest) - 10:
▹ Randomly select addNum features, weighted by precalculated entropy - 11:
mated ← intermediate | additional ▹ Add selected features - 12:
return mated - 13:
end function
|
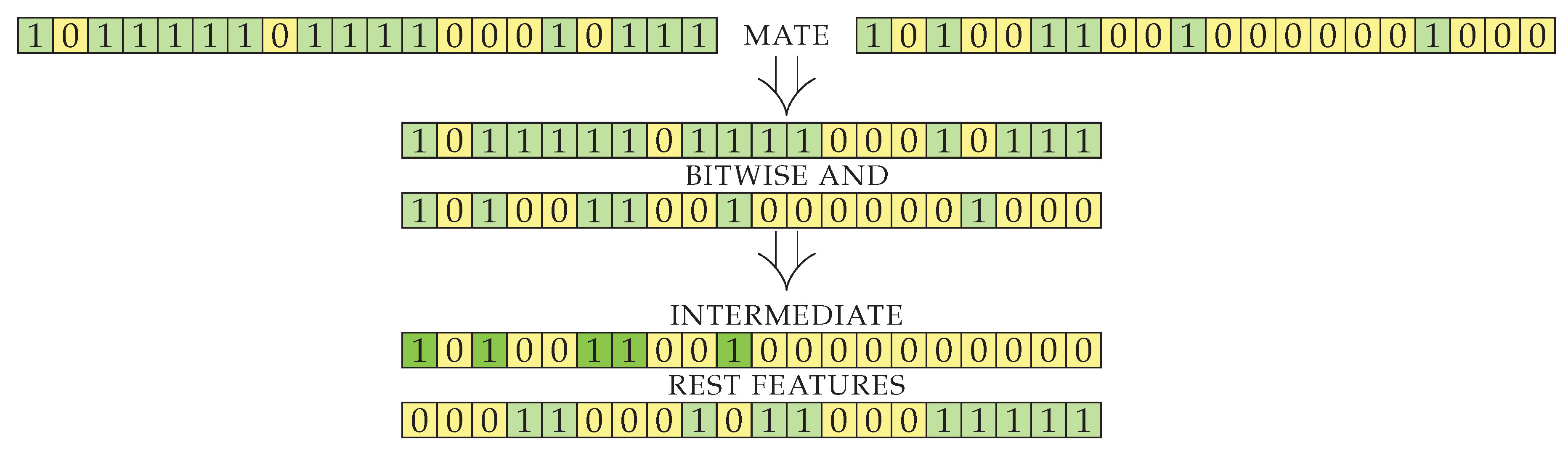
After crossover, the resulting genes initially contain features that were selected in both parents. The intersection of features is a good starting point, as it seems to produce good results during the running of the algorithm. Since the implementation of the genes is a simple bit string, the intermediate result is simply bitwise logical
and of its parents’ genes. Another way to combine the parents would be to produce offspring with the union of genes. The latter approach produces offspring that is too large (contains too many selected features) and is thus unsuitable for our objective, where we want to select a sufficiently small feature set. The size of the intermediate result obtained by the intersection (line 5) is smaller than or equal to the size of
item2 (smallest of the parents), but can be much smaller, depending on the input sets (see Lemma 3). With the intersection, the number of genes decreases too rapidly (see Lemma 4) and disposes of useful information. We therefore use additional genes presented in exactly one of the parents (generated by element-wise exclusive
or) to construct an additional set of features with good information gain. The visualization for the first part of
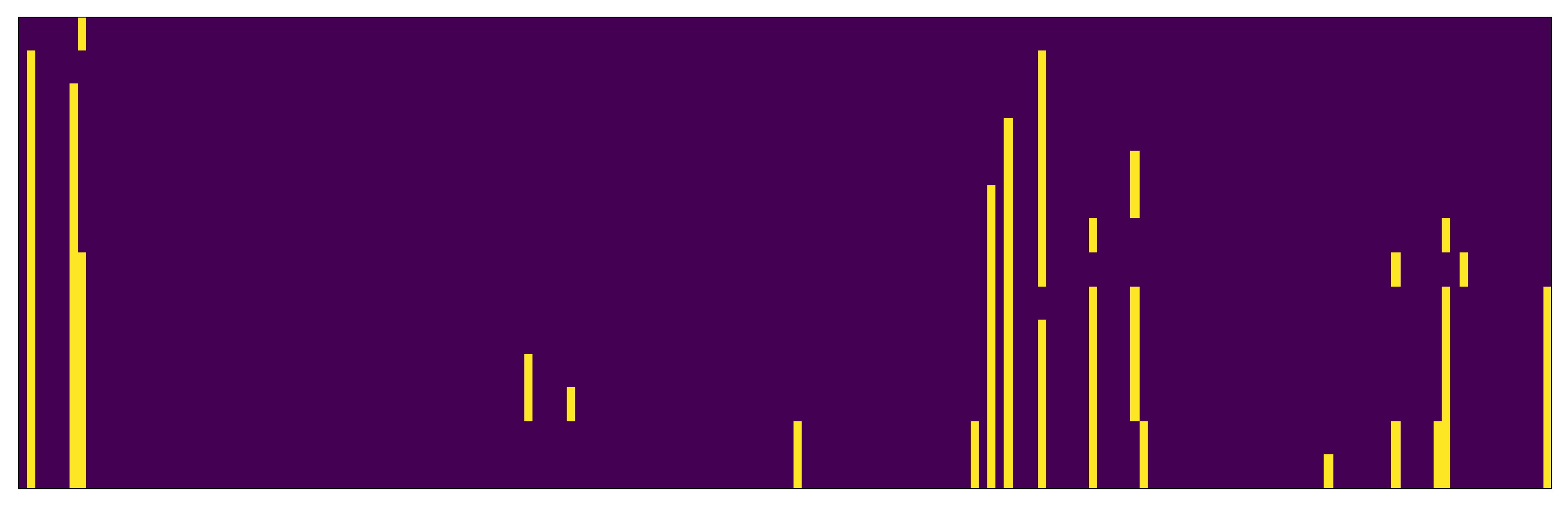
crossoverPair procedure on an example case is shown in
Figure 3. The genes of two items to be mated are split into two indexed sets: an intermediate set containing genes from both parents and a set called
rest containing genes to be used for the enrichment of intersected genes.
Enrichment of the intermediate set of genes is done as follows. First, the number of enrichment genes is calculated using the onGeneScaling function. This function is used to control the size of the final offspring. From Lemma 4, it follows that the expected size of the rest (features) set, with the size of the largest parent being constant, is linear to the size of the smaller parent (item2). Adding all genes from the rest set will result in an offspring too large (and diverge toward the whole feature space being selected), selecting none of the genes will diverge towards very small sets. In our experiments, we used the function (for onGeneScaling) that scaled the amount of features appropriately for our data set. For a larger number of features, the functions and with a sufficiently small base a are a good choice. As with most parameters, onGeneScaling can be swapped during the run-time and made dependent on the conditions of front updates and statistics.
The second part of the mating algorithm selects the best performing features using heuristics based on information gain. From all features in the rest set,
addNum are randomly selected (using a weighted random selection, where the weight of each feature is its individual precalculated information gain). The final result is an item with genes from the intersection set and selected genes from rest set. In the implementation, the results are simply combined with element-wise OR. The heuristics for the information gain in our algorithm is based on mutual information gain. The
scikit-learn implementation [
29] of mutual information gain is used for each feature in the train set.
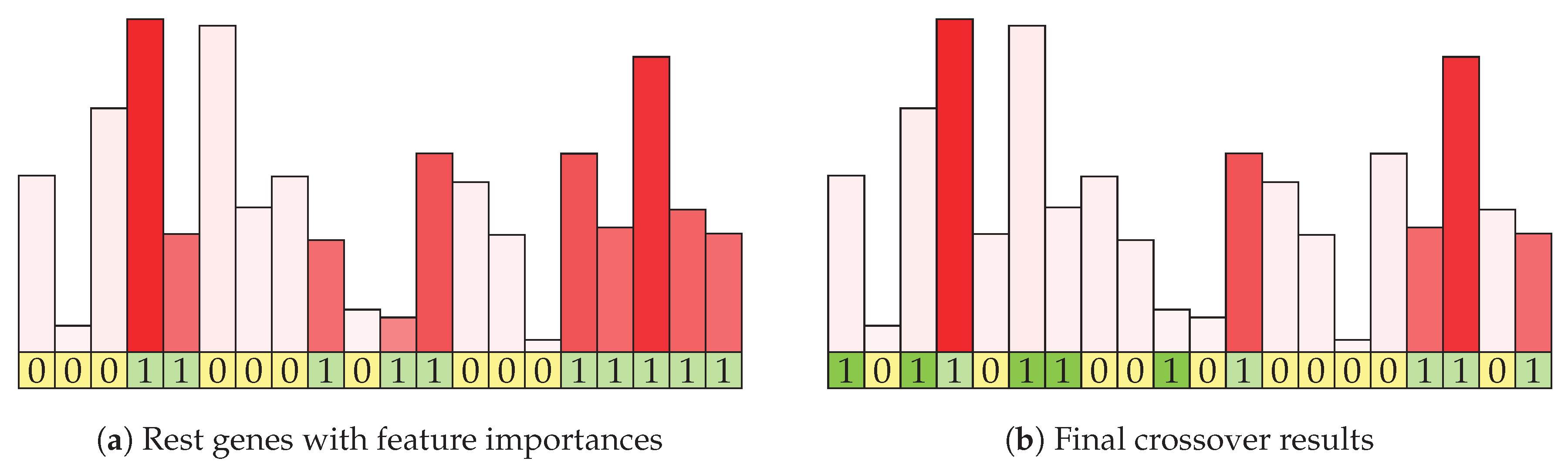
Figure 4a shows the continuation of the example mating algorithm from the previous figure. Each of the initial features is represented with its information gain, which is shown by the height and intensity of the shade of the column above it. Columns for features in the rest set are shown in red (and the corresponding genes are green in the bottom row). Of the eight features in the rest features row, five are selected using weighted random function.
Figure 4b shows the visualization of the result (offspring) of the mating procedure. The genes selected by intersection are marked with dark green shading, while the genes selected by information enrichment are shown in light green. The corresponding information gain statistics in shown with column color intensity. In this example, two genes of size 14 and 6 were combined and they produced a result with size 10, which may be further increased by the mutation strategy. The resulting item contains genes that are present in both parents and were therefore rated as good in the previous evaluation. It is enriched with a limited number of genes that are present in either parent, weighted according to their information gain.
3.4. Purging Features from Pareto Front Using Information Gain
An initial version of the algorithm presented in Algorithm 1 has been further improved to exploit pre-trained models for the calculation of the information gain and to purge non-relevant features from items on the Pareto front. For each item added to the front, a model trained on its genes is stored. Since the number of elements is small compared to the learning data, storing the models results in virtually no additional effort in run-time and is a useful tool for evaluating the optimization over time. In the same way as when the population is reset to the Pareto front, another predicate is introduced to indicate when the Pareto front should be purged using the information criterion. The gene purging procedure is presented in Algorithm 3.
| Algorithm 3 Gene purging algorithm |
| Require: |
- 1:
functionpurgeParetoFront - 2:
newFront = ← Dict.empty ▹ Empty dictionary representing new Pareto Front - 3:
for item in front do - 4:
if |item| == 1 then - 5:
continue ▹ Items with only one feature cannot be reduced - 6:
end if - 7:
baseResult ← item.result ▹ Score acquired by using all features from item - 8:
scoreDecrease ← [] ▹ Score decrease for each feature - 9:
for geneInd in item.genes WHERE ISSET(item.genes[geneInd]) do ▹ Check only set genes - 10:
newScore ← evalF(item.model, item.genes, geneInd) ▹ Evaluate existing trained model on test data set, where values of feature geneInd are randomly shuffled - 11:
scoreDecrease[geneInd] ← baseResult - newScore - 12:
end for - 13:
bestInd ← argmin(scoreDecrease) ▹ Take the gene which showed smallest score decrease when shuffled - 14:
newItem ← Item(genes=unsetBit(item.genes, bestInd)) ▹ Create new item where the gene with the smallest score decrease is unset - 15:
newFront[|newItem|] ← Evaluate(newItem, evalF) ▹ Evaluate newly created item (train and test model) - 16:
end for - 17:
front ← front ∪ newFront ▹ Update current front with newly calculated items - 18:
front ←removeDominated(front) ▹ Remove unoptimal items from Pareto front - 19:
return front - 20:
end function
|
The main idea of purging the items on the Pareto front with information criterion is to discard unneeded features from items that already perform well. For each item on the Pareto front, we try to discard a feature that makes the least contribution to the item’s score. The obvious way to do this would be to transverse through all the features used in the prediction model, remove superfluous features one by one, and train and re-evaluate such a model. For each item, a new model must be trained and re-evaluated. This is time-consuming and offers only minor improvements. Our proposal is to use a heuristic approach to select a feature to be removed. For each such feature, an existing model corresponding to the item is used (saving time by avoiding the training of a new model) and evaluated according to the previously used test data set. For each selected feature, its values are randomly shuffled across the test data set. The motivation for such heuristics is that the shuffling values of a feature with strong predictive power has a much stronger effect on the resulting score than shuffling values of a feature with only low predictive power. In this way, we can provide heuristics for estimating the predictive power of features in a model-agnostic way using only a linear number of model calls. Such heuristics provide a noticeable acceleration and can be used for any type of model. The approach targets black box models with a high ratio between training and inference time/resource consumption—e.g., gradient boosting or complex neural networks. For each newly shuffled feature in an item, a change in the base score is recorded (line 11). Interestingly, sometimes the change can even be negative if the included features are detrimental to the overall prediction procedure. This is proving to be an effective method for quickly sorting out bad features introduced by mutations at the end of simulations when the exploration is limited to the feature space previously explored.
The feature whose shuffling has the most detrimental effect is taken and a new item is constructed whose genes are the same as those of the original element, except that the resulting feature is omitted (lines 13 and 14). This procedure can be further generalized. For example, first could be scaled with some function (for example, ) and then the feature to be removed could be sampled by these new weights from the set. Special care should be taken that the scaling function is monotonous (and preferably a bijection) since negative values can occur and should not be bundled with positive ones. If a random selection is used, the final probabilities must also be re-scaled correctly due to possible negative weights. After the feature to be removed is selected, the new item is evaluated and all new items are brought to the front. The front should also be purged to take into account possible new features on the front.
The most important heuristic optimization used by the front purging functions is based on the fact that only a new model is built with the features with the most promising importance score (as inferred by a model when feature values are shuffled). The time required for this operation is linear in the number of features. For each feature, the model requires one evaluation (inference) on the training set. The implementation of the gene purging can be further optimized so that each feature is selected only once. Under the reasonable assumption of sufficiently deterministic training and evaluation, it is easy to see that evaluating an item and (potential) addition of a feature to the front is an idempotent operation. Removing the same feature from the same item on the front several times is not a reasonable change and therefore another feature could be considered for removal. An appropriate threshold for reducing the score may also be introduced so that features with very high predictive power are never removed.