Investigation of Linear and Nonlinear Properties of a Heartbeat Time Series Using Multiscale Rényi Entropy
Abstract
1. Introduction
1.1. Heartbeat Interval Time Series
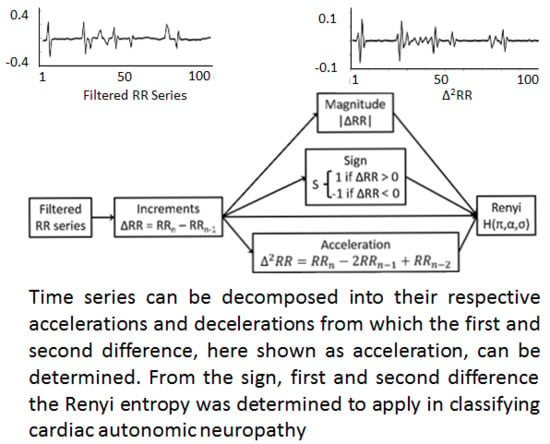
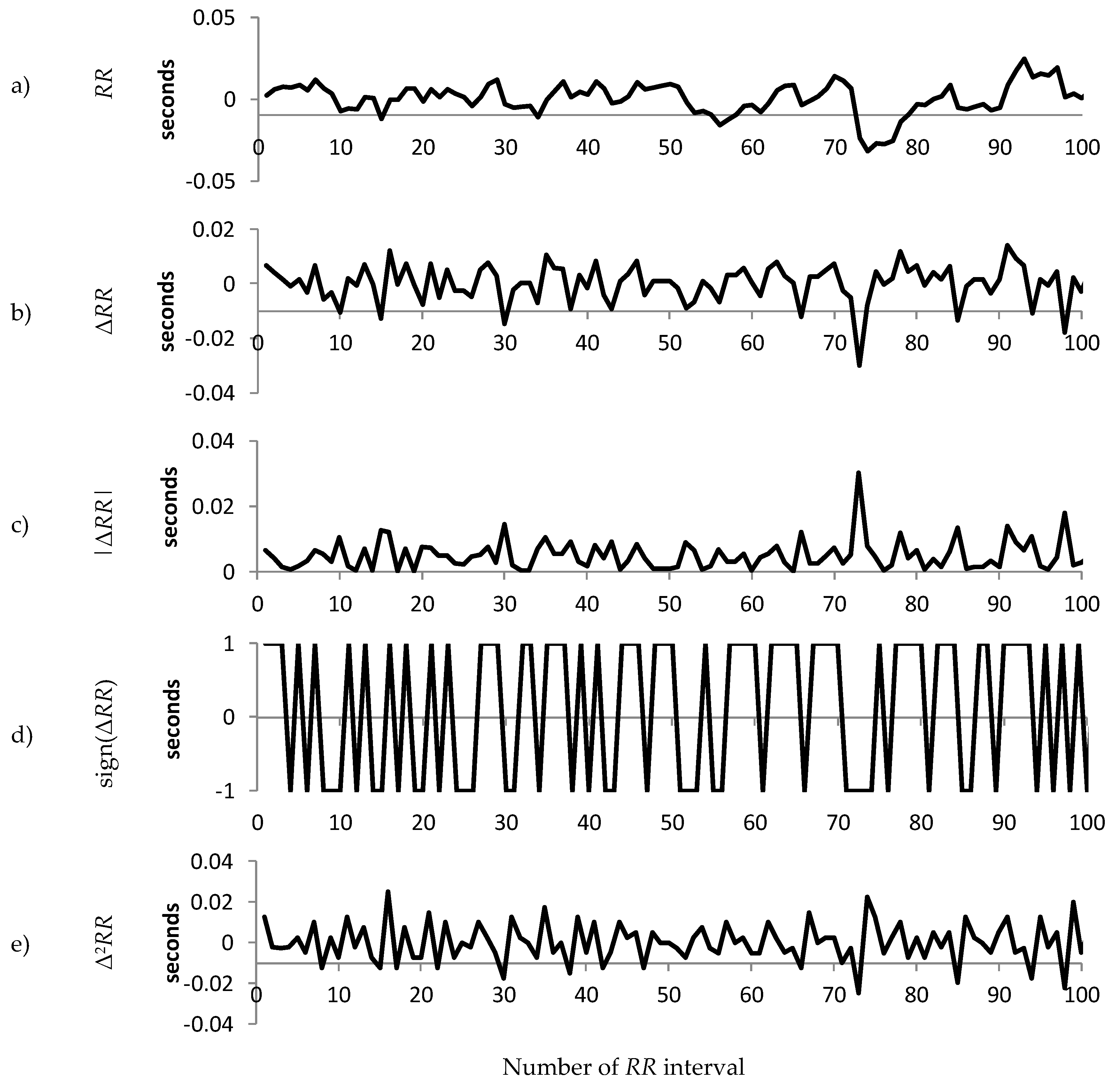
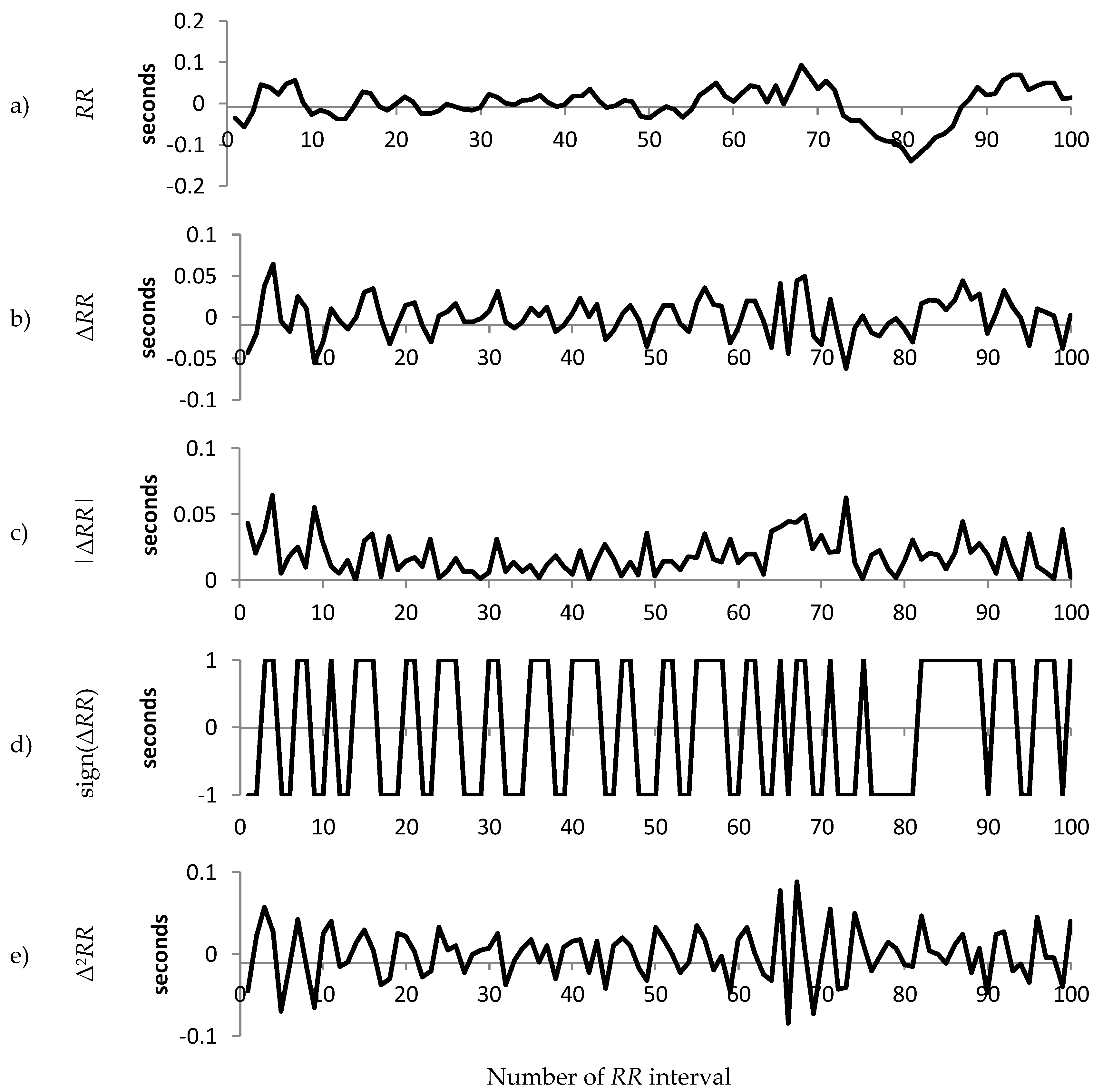
1.2. Decomposition of the RR Interval Time Series
1.3. The Rényi Entropy
2. Methods
2.1. Patient Selection
2.2. ECG Recording and Obtaining the RR Intervals
2.3. Decomposition
- Rényi entropy calculated from a sequence of the magnitude of the difference in RR intervals
- Rényi entropy calculated from a sequence of the sign of the difference in RR intervals
- Rényi entropy calculated from a sequence of the acceleration of RR intervals
2.4. Calculating the Multiscale Rényi (MSRen) Entropy
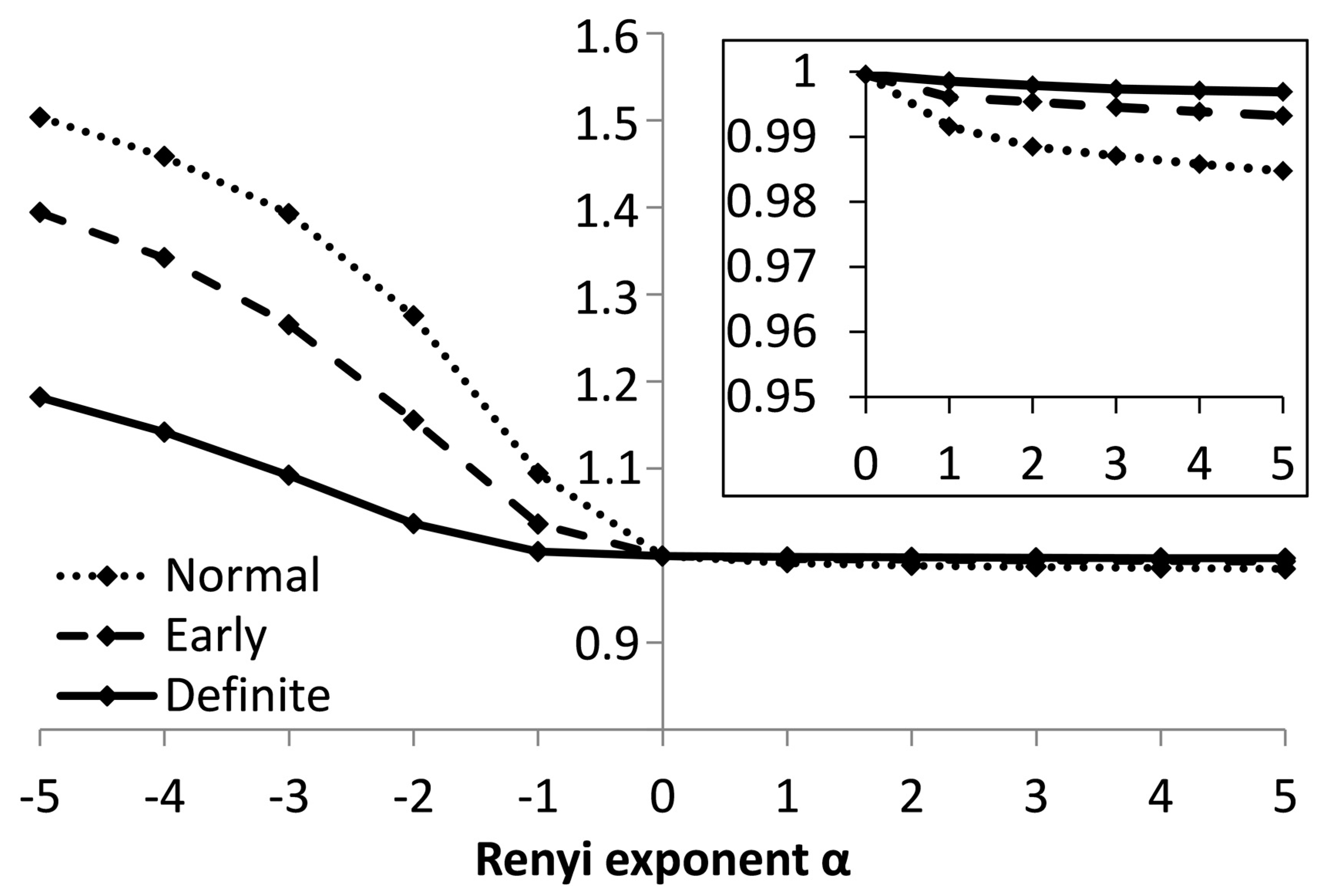
3. Results
4. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Goldberger, A.L.; Amaral, L.A.N.; Hausdorff, J.M.; Ivanov, P.C.; Peng, C.-K.; Stanley, H.E. Fractal dynamics in physiology: Alterations with disease and aging. Proc. Natl. Acad. Sci. USA 2002, 99, 2466–2472. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Goldberger, A.L.; Peng, C.-K. Multiscale entropy analysis of biological signals. Phys. Rev. E 2005, 71, 021906. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.K.; Havlin, S.; Stanley, H.E.; Goldberger, A.L. Quantification of scaling exponents and cross over phenomena in nonstationary heartbeat time series analysis. Chaos 1995, 5, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.C.; Rosenblum, M.G.; Peng, C.K.; Mietus, J.; Havlin, S.; Stanley, H.E.; Goldberger, A.L. Scaling behaviour of heartbeat intervals obtained by wavelet-based time-series analysis. Nature 1996, 383, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Agelink, M.W.; Malessa, R.; Baumann, B.; Majewski, T.; Akila, F.; Zeit, T.; Ziegler, D. Standardized tests of heart rate variability: Normal ranges obtained from 309 healthy humans, and effects of age, gender and heart rate. Clin. Auton. Res. 2001, 11, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Bellavere, F.; Balzani, I.; De Masi, G.; Carraro, M.; Carenza, P.; Cobelli, C.; Thomaseth, K. Power spectral analysis of heart-rate variations improves assessment of diabetic cardiac autonomic neuropathy. Diabetes 1992, 41, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Yeragani, V.K.; Srinivasan, K.; Vempati, S.; Pohl, R.; Balon, R. Fractal dimension of heart rate time series: An effective measure of autonomic function. J. Appl. Physiol. 1993, 75, 2429–2438. [Google Scholar] [CrossRef][Green Version]
- Malik, M.; Camm, J. HRV Variability; Futura Publishing Co.: Armonk, NY, USA, 1995. [Google Scholar]
- Electrophysiology, Task Force of the European Society of Cardiology the North American Society of Pacing. Special report: Heart rate variability standards of measurement, physiological interpretation, and clinical use. Circulation 1996, 93, 1043–1065. [Google Scholar] [CrossRef]
- Teich, M.C.; Lowen, S.B.; Jost, B.M.; Vibe-Rheymer, K. Heart Rate Variability: Measures and Models; IEEE Press: New York, NY, USA, 2001. [Google Scholar]
- Khandoker, A.H.; Jelinek, H.F.; Moritani, T.; Palaniswami, M. Association of cardiac autonomic neuropathy with alteration of sympatho-vagal balance through heart rate variability analysis. Med. Eng. Phys. 2010, 32, 161–167. [Google Scholar] [CrossRef]
- Pikkujämsä, S.M.; Mäkikallio, T.H.; Sourander, L.B.; Räihä, I.J.; Puukka, P.; Skyttä, J.; Peng, C.K.; Goldberger, A.L.; Huikuri, H.V. Cardiac interbeat interval dynamics from childhood to senescence: Comparison of conventional and new measures based on fractals and chaos theory. Circulation 1999, 100, 393–399. [Google Scholar] [CrossRef]
- Schmitt, D.T.; Ivanov, P.C. Fractal scale-invariant and nonlinear properties of cardiac dynamics remain stable with advanced age: A new mechanistic picture of cardiac control in healthy elderly. Am. J. Physiol. 2007, 293, 1923–1937. [Google Scholar] [CrossRef] [PubMed]
- Burr, R.L. Interpretation of normalised specrtal heart rate variability in sleep research: A critical review. Sleep 2007, 30, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Vanoli, E.; Adamson, P.B.; Ba, L.; Pinna, G.D.; Lazzara, R.; Orr, W.C. Heart rate variability during specific sleep stages. A comparison of healthy subjects with patients after myocardial infarction. Circulation 1995, 91, 1918–1922. [Google Scholar] [CrossRef] [PubMed]
- Hautala, A.J.; Makikallio, T.H.; Kiviniemi, A.; Laukkanen, R.T.; Nissila, S.; Huikuri, H.V.; Tulppo, M.P. Cardiovascular autonomic function correlates with the response to aerobic training in healthy sedentary subjects. Am. J. Physiol. Heart Circ. Physiol. 2003, 285, H1747–H1752. [Google Scholar] [CrossRef] [PubMed]
- Jelinek, H.F.; Huang, Z.Q.; Khandoker, A.H.; Chang, D.; Kiat, H. Cardiac rehabilitation outcomes following a 6-week program of PCI and CABG Patients. Front. Physiol. 2013, 4, 302. [Google Scholar] [CrossRef] [PubMed]
- Kiviniemi, A.M.; Tulppo, M.P.; Eskelinen, J.J.; Savolainen, A.M.; Kapanen, J.; Heinonen, I.H.; Huikuri, H.V.; Hannukainen, J.C.; Kalliokoski, K.K. Cardiac autonomic fucntion and high-intensity interval training in middle-aged men. Med. Sci. Sports Exerc. 2014, 46, 1960–1967. [Google Scholar] [CrossRef] [PubMed]
- La Rovere, M.; Mortara, A.; Sandrone, G.; Lombardi, F. Autonomic nervous system adaptations to short-term exercise training. Chest 1992, 101, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Soares-Miranda, L.; Sandercock, G.; Valente, H.; Vale, S.; Santos, R.; Mota, J. Vigorous physical activity and vagal modulation in young adults. Eur. J. Cardiovasc. Prevent. Rehab. 2009, 16, 705–711. [Google Scholar] [CrossRef]
- Tulppo, M.P.; Mäkikallio, T.H.; Seppänen, T.; Laukkanen, R.T.; Huikuri, H.V. Vagal modulation of heart rate during exercise: Effects of age and physical fitness. Am. J. Physiol. Heart Circ. Physiol. 1998, 274, H424–H429. [Google Scholar] [CrossRef]
- McLachlan, C.S.; Ocsan, R.; Spence, I.; Hambly, B.; Matthews, S.; Wang, L.; Jelinek, H.F. Increased total heart rate variability and enhanced cardiac vagal autonomic activity in healthy humans with sinus bradycardia. In Baylor University Medical Center Proceedings; Taylor & Francis: Oxford, UK, 2010; Volume 23, pp. 368–370. [Google Scholar]
- Mäkikallio, T.H.; Huikuri, H.V.; Hintze, U.; Videbæk, J.; Mitrani, R.D.; Castellanos, A.; Myerburg, R.J.; Møller, M.; DIAMOND Study Group. Fractal analysis and time- and frequency-domain measures of heart rate variability as predictors of mortality in patients with heart failure. Am. J. Cardiol. 2001, 87, 178–182. [Google Scholar]
- Huikuri, H.V.; Valkama, J.O.; Airaksinen, K.E.; Seppänen, T.; Kessler, K.M.; Takkunen, J.T.; Myerburg, R.J. Frequency domain measures of heart rate variability before the onset of nonsustained and sustained ventricular tachycardia in patients with coronary artery disease. Circulation 1993, 87, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Khandoker, A.H.; Jelinek, H.F.; Palaniswami, M. Heart rate variability and complexity in people with diabetes associated cardiac autonomic neuropathy. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; pp. 4696–4699. [Google Scholar]
- Kemp, A.H.; Quintana, D.S.; Felmingham, K.L.; Matthews, S.; Jelinek, H.F. Heart rate variability in unmedicated depressed patients without comorbid cardiovascular disease. PLoS ONE 2012, 7, e30777. [Google Scholar] [CrossRef] [PubMed]
- Carney, R.M.; Freedland, K.E. Depression and heart rate variability in patients with coronary artery disease. Clev. Clin. J. Med. 2009, 76, S13–S17. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, R.; Citi, L.; Valenza, G.; Guerrisi, M.; Orsolini, S.; Tessa, C.; Diciotti, S.; Toschi, N. Increased instability of heartbeat dynamics in Parkinson’s disease. In Computing in Cardiology; IEEE: Piscataway Township, NJ, USA, 2013; Volume 40, pp. 89–92. [Google Scholar]
- Kallio, M.; Suominen, K.; Bianchi, A.M.; Mäkikallio, T.; Haapaniemi, T.; Astafiev, S.; Sotaniemi, K.A.; Myllylä, V.V.; Tolonen, U. Comparison of heart rate variability analysis methods in patients with Parkinson’s disease. Med. Biol. Eng. Comput. 2002, 40, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Vinik, A.I.; Erbas, T.; Casellini, C.M. Diabetic cardiac autonomic neuropathy, inflammtion and cariovascular disease. J. Diabetes Investig. 2013, 4, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Charles, M.; Fleischer, J.; Witte, D.R.; Ejskjaer, N.; Borch-Johnsen, K.; Lauritzen, T.; Sandbaek, A. Impact of early detection and treatment of diabetes on the 6-year prevalence of cardiac autonomic neuropathy in people with screen-detected diabetes: ADDITION-Denmark, a cluster-randomised study. Diabetiologia 2013, 56, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Hurst, H.E. Long-term storage capacity of reservoirs. Trans. Am. Soc. Civ. Eng. 1951, 116, 770–808. [Google Scholar]
- Ashkenazy, Y.; Ivanov, P.C.; Havlin, S.; Peng, C.K.; Goldberger, A.L.; Stanley, H.E. Magnitude and sign correlations in heartbeat fluctuations. Phys. Rev. Lett. 2001, 86, 1900–1903. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, Y.; Ivanov, P.C.; Havlin, S.; Peng, C.K.; Yamamoto, Y.; Goldberger, A.L.; Stanley, H.E. Decomposition of heartbeat time series: Scaling analysis of the sign sequence. Comput. Cardiol. 2000, 27, 139–142. [Google Scholar]
- Ashkenazy, Y.; Lewkowicz, M.; Levitan, J.; Moelgaard, H.; Thomsen, P.E.B.; Saermark, K. Discrimination of the healthy and sick cardiac autonomic nervous system by a new wavelet analysis of heartbeat intervals. Fractals 1998, 6, 197–203. [Google Scholar] [CrossRef]
- Jelinek, H.F.; Tarvainen, M.P.; Cornforth, D.J. Renyi entropy in the identification of cardiac autonomic neuropathy in diabetes. Comput. Cardiol. 2012, 39, 909–911. [Google Scholar]
- Kurths, J.; Voss, A.; Saparin, P.; Witt, A.; Kleiner, H.J.; Wessel, N. Quantitative analysis of heart rate variability. Chaos 1995, 5, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Lake, D.E. Renyi entropy measures of heart rate Gaussianity. IEEE Trans. Biomed. Eng. 2006, 53, 21–27. [Google Scholar] [CrossRef]
- Voss, A.; Schulz, S.; Schroeder, R.; Baumert, M.; Caminal, P. Methods derived from nonlinear dynamics for analysing heart rate variability. Phil. Trans. Math. Phys. Eng. Sci. 2009, 367, 277–296. [Google Scholar] [CrossRef] [PubMed]
- Wessel, N.; Schumann, A.; Schirdewan, A.; Voss, A.; Kurths, J. Entropy measures in heart rate variability data. In International Symposium on Medical Data Analysis; Springer: Berlin/Heidelberg, Germany, 2000; pp. 78–87. [Google Scholar]
- Wessel, N.; Voss, A.; Malberg, H.; Ziehmann, C.; Voss, H.U.; Schirdewan, A.; Meyerfeldt, U.; Kurths, J. Nonlinear analysis of complex phenomena in cardiological data. Herzschr. Elektrophys. 2000, 11, 159–173. [Google Scholar] [CrossRef]
- Pincus, S. Approximate entropy as a measure of system complexity. Proc. Nat. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef] [PubMed]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef]
- Grassberger, P. Finite sample corrections to entropy and dimension estimates. Phys. Lett. A 1988, 128, 369–373. [Google Scholar] [CrossRef]
- Grassberger, P.; Procaccia, I. Measuring the strangeness of strange attractors. Physica 1983, 9, 189–208. [Google Scholar]
- Eckmann, J.P.; Ruelle, D. Ergodic theory of chaos and strange attractors. In The Theory of Chaotic Attractors; Springer: New York, NY, USA, 1985; pp. 273–312. [Google Scholar]
- Ivanov, P.C.; Amaral, L.A.N.; Goldberger, A.L.; Havlin, S.; Rosenblum, M.G.; Struzik, Z.R.; Stanley, H.E. Multifractality in human heartbeat dynamics. Nature 1999, 399, 461–465. [Google Scholar] [CrossRef]
- Costa, M.; Goldberger, A.L.; Peng, C.-K. Multiscale entropy analysis of complex physiological time series. Phys. Rev. Lett. 2002, 89, 068102. [Google Scholar] [CrossRef]
- Cornforth, D.; Tarvainen, M.; Jelinek, H.F. How to Calculate Renyi Entropy from Heart Rate Variability, and Why it Matters for Detecting Cardiac Autonomic Neuropathy. Front. Bioeng. Biotechnol. 2014, 2, 34. [Google Scholar] [CrossRef] [PubMed]
- Rényi, A. On measures of information and entropy. In Proceedings of the Fourth Berkeley Symposium on Mathematics, Statistics and Probability; The Regents of the University of California: Oakland, CA, USA, 1961; pp. 547–561. [Google Scholar]
- Xu, Y.; Ma, Q.D.Y.; Schmitt, D.T.; Bernaola-Galván, P.; Ivanov, P.C. Effects of coarse-graining on the scaling behavior of long-range correlated and anti-correlated signals. Phys. A Stat. Mech. Appl. 2011, 390, 4057–4072. [Google Scholar] [CrossRef] [PubMed]
- Jelinek, H.F.; Wilding, C.; Tinley, P. An innovative multi-disciplinary diabetes complications screening programme in a rural community: A description and preliminary results of the screening. Aust. J. Prim. Health 2006, 12, 14–20. [Google Scholar] [CrossRef]
- Spallone, V.; Bellavere, F.; Scionti, L.; Maule, S.; Quadri, R.; Bax, G.; Melga, P.; Viviani, G.L.; Esposito, K.; Morganti, R.; et al. Recommendations for the use of cardiovascular tests in diagnosing diabetic autonomic neuropathy. Nutr. Metab. Cardiovasc. Dis. 2011, 21, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Pop-Busui, R.; Evans, G.W.; Gerstein, H.C.; Fonseca, V.; Fleg, J.L.; Hoogwerf, B.J.; Genuth, S.; Grimm, R.H.; Corson, M.A.; Prineas, R. The ACCORD Study Group. Effects of cardiac autonomic dysfunction on mortality risk in the Action to Control Cardiovascular Risk in Diabetes (ACCORD) Trial. Diabetes Care 2010, 33, 1578–1584. [Google Scholar] [CrossRef]
- Flynn, A.C.; Jelinek, H.F.; Smith, M.C. Heart rate variability analysis: A useful assessment tool for diabetes associated cardiac dysfunction in rural and remote areas. Aust. J. Rural Health 2005, 13, 77–82. [Google Scholar] [CrossRef]
- Tarvainen, M.P.; Ranta-Aho, P.O.; Karjalainen, P.A. An advanced detrending method with application to HRV analysis. IEEE Trans. Biomed. Eng. 2002, 49, 172–175. [Google Scholar] [CrossRef]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale entropy analysis: A new measure of complexity loss in heart failure. J. Electrocardiol. 2003, 36, 39–40. [Google Scholar] [CrossRef]
- Gao, J.; Gurbaxani, B.M.; Hu, J.; Heilman, K.J.; Emauele, V.A.; Lewis, G.F.; Davila, M.; Unger, E.R.; Lin, J.M.S. Multiscale analysis of heart rate variability in nonstationary environments. Front. Physiol. 2013, 4, 119. [Google Scholar] [CrossRef]
- Saul, J.P.; Albrecht, P.; Berger, R.D.; Cohen, R.J. Analysis of long term heart rate variability: Methods, 1/f scaling and implications. Pharmacology 1988, 14, 419–422. [Google Scholar]
- Kobayashi, M.; Musha, T. 1/f fluctuation of heart beat period. IEEE Trans. Biomed. Eng. 1982, 29, 456–457. [Google Scholar] [CrossRef] [PubMed]
- Struzik, Z.R.; Hayano, J.; Sakata, S.; Kwak, S.; Yamamoto, Y. 1/f Scaling in heartrate requires antagonistic autonomic control. Phys. Rev. E 2004, 70, 050901. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Gao, J.; Tung, W.-W.; Cao, Y. Multiscale analysis of heart rate variability: A comparison of different complexity measures. Ann. Biomed. Eng. 2010, 38, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Kiyono, A.; Struzik, Z.R.; Aoyagi, N.; Yamamoto, Y. Multiscale probability density function analysis: Non-Gaussian and scale-invariant fluctuations of healthy human HRV. IEEE Trans. Biomed. Eng. 2006, 53, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Thurner, S.; Feurstein, M.C.; Teich, M.C. Multiresolution wavelet analysis of heartbeat intervals discriminates healthy patients from those with cardiac pathology. Phys. Rev. Lett. 1998, 80, 1544–1547. [Google Scholar] [CrossRef]
- Krstacic, G.; Krstacic, A.; Smalcelj, A.; Milicic, D.; Jembrek-Gostovic, M. The “Chaos Theory” and nonlinear dynamics in heart rate variability analysis: Does it work in short-time series in patients with coronary heart disease? Ann. Noninvasive Electrocardiol. 2007, 12, 130–136. [Google Scholar] [CrossRef]
- Ho, K.K.; Moody, G.B.; Peng, C.K.; Mietus, J.E.; Larson, M.G.; Levy, D.; Goldberger, A.L. Predicting survival in heart failure case and control subjects by use of fully automated methods for deriving nonlinear and conventional indices of heart rate dynamics. Circulation 1997, 96, 842–848. [Google Scholar] [CrossRef]
- Laitio, T.; Jalonen, J.; Kuusela, T.; Scheinin, H. The role of heart rate variability in risk stratification for adverse postoperative cardiac events. Anesth. Analg. 2007, 105, 1548–1560. [Google Scholar] [CrossRef]
- Goldberger, A.L. Non-linear dynamics for clinicians: Chaos theory, fractals, and complexity at the bedside. Lancet 1996, 347, 1312–1314. [Google Scholar] [CrossRef]
- Bellavere, F.; Bosello, G.; Fedele, D.; Cardone, C.; Ferri, M. Diagnosis and management of diabetic autonomic neuropathy. BMJ 1983, 287, 61. [Google Scholar] [CrossRef] [PubMed]



| Test | π = 1 | π = 2 | π = 4 | π = 8 | π = 16 | |
|---|---|---|---|---|---|---|
| α = 1 | NE | 0.0001 | <0.0001 | <0.0001 | 0.0001 | 0.002 |
| ED | 0.004 | 0.006 | 0.02 | n.s. | n.s. | |
| DN | <0.0001 | <0.0001 | 0.0002 | 0.002 | 0.03 | |
| α = 2 | NE | 0.0002 | <0.0001 | <0.0001 | <0.0001 | 0.0007 |
| ED | 0.002 | 0.004 | 0.01 | 0.05 | n.s. | |
| DN | <0.0001 | <0.0001 | 0.0001 | 0.001 | 0.02 | |
| α = 3 | NE | 0.0002 | <0.0001 | <0.0001 | <0.0001 | 0.0004 |
| ED | 0.003 | 0.005 | 0.009 | 0.004 | 0.2 | |
| DN | <0.0001 | <0.0001 | <0.0001 | 0.001 | 0.01 | |
| α = 4 | NE | 0.0003 | <0.0001 | <0.0001 | <0.0001 | 0.0003 |
| ED | 0.002 | 0.005 | 0.009 | n.s. | n.s. | |
| DN | <0.0001 | <0.0001 | <0.0001 | 0.0009 | 0.007 | |
| α = 5 | NE | 0.0003 | <0.0001 | <0.0001 | <0.0001 | 0.0002 |
| ED | 0.002 | 0.005 | 0.009 | 0.02 | n.s. | |
| DN | <0.0001 | <0.0001 | <0.0001 | 0.0007 | 0.006 |
| Test | π = 1 | π = 2 | π = 4 | π = 8 | π = 16 | |
|---|---|---|---|---|---|---|
| α = 1 | NE | 0.003 | 0.0005 | 0.0002 | 0.0003 | 0.0005 |
| ED | n.s. | 0.03 | 0.01 | 0.02 | n.s. | |
| DN | 0.00147 | 0.0002 | <0.0001 | 0.0005 | 0.004 | |
| α = 2 | NE | 0.007 | 0.0008 | 0.0003 | 0.0004 | 0.0004 |
| ED | n.s. | 0.02 | 0.009 | 0.02 | 0.05 | |
| DN | 0.002 | 0.0003 | <0.0001 | 0.0003 | 0.001 | |
| α = 3 | NE | 0.01 | 0.001 | 0.0003 | 0.0005 | 0.0003 |
| ED | n.s. | 0.03 | 0.007 | 0.02 | 0.04 | |
| DN | 0.002 | 0.0004 | <0.0001 | 0.0002 | 0.001 | |
| α = 4 | NE | 0.01 | 0.0009 | 0.0004 | 0.0005 | 0.0003 |
| ED | n.s. | 0.03 | 0.007 | 0.02 | 0.03 | |
| DN | 0.003 | 0.0004 | <0.0001 | 0.0002 | 0.0009 | |
| α = 5 | NE | 0.01 | 0.001 | 0.0004 | 0.0006 | 0.0002 |
| ED | n.s. | 0.03 | 0.007 | 0.02 | 0.03 | |
| DN | 0.0038 | 0.0004 | <0.0001 | 0.0001 | 0.0006 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jelinek, H.F.; Cornforth, D.J.; Tarvainen, M.P.; Khalaf, K. Investigation of Linear and Nonlinear Properties of a Heartbeat Time Series Using Multiscale Rényi Entropy. Entropy 2019, 21, 727. https://doi.org/10.3390/e21080727
Jelinek HF, Cornforth DJ, Tarvainen MP, Khalaf K. Investigation of Linear and Nonlinear Properties of a Heartbeat Time Series Using Multiscale Rényi Entropy. Entropy. 2019; 21(8):727. https://doi.org/10.3390/e21080727
Chicago/Turabian StyleJelinek, Herbert F., David J. Cornforth, Mika P. Tarvainen, and Kinda Khalaf. 2019. "Investigation of Linear and Nonlinear Properties of a Heartbeat Time Series Using Multiscale Rényi Entropy" Entropy 21, no. 8: 727. https://doi.org/10.3390/e21080727
APA StyleJelinek, H. F., Cornforth, D. J., Tarvainen, M. P., & Khalaf, K. (2019). Investigation of Linear and Nonlinear Properties of a Heartbeat Time Series Using Multiscale Rényi Entropy. Entropy, 21(8), 727. https://doi.org/10.3390/e21080727