Abstract
Inflammatory bowel diseases (IBDs) are a group of chronic gastrointestinal inflammatory diseases with unknown etiology. There is a combination of well documented factors in their pathogenesis, including intestinal microbiota dysbiosis. The symbiotic microbiota plays important functions in the host, and the loss of beneficial microbes could favor the expansion of microbial pathobionts. In particular, the bloom of potentially harmful Proteobacteria, especially Enterobacteriaceae, has been described as enhancing the inflammatory response, as observed in IBDs. Herein, we seek to investigate the contribution of Enterobacteriaceae to IBD pathogenesis whilst considering the continuous expansion of the literature and data. Despite the mechanism of their expansion still remaining unclear, their expansion could be correlated with the increase in nitrate and oxygen levels in the inflamed gut and with the bile acid dysmetabolism described in IBD patients. Furthermore, in several Enterobacteriaceae studies conducted at a species level, it has been suggested that some adherent-invasive Escherichia coli (AIEC) play an important role in IBD pathogenesis. Overall, this review highlights the pivotal role played by Enterobacteriaceae in gut dysbiosis associated with IBD pathogenesis and progression.
1. Introduction
Inflammatory bowel diseases (IBDs) are a heterogeneous group of chronic, relapsing–remitting, gastrointestinal (GI) inflammatory diseases with various degrees of damage, promoting the development of local and extra-intestinal complications. IBDs include ulcerative colitis (UC) disease and Crohn’s disease (CD); the first is restricted to the colon, while the second can affect different parts of the digestive tract [1,2].
Despite the etiology of IBDs remaining unknown, it is reported that there is an involvement of the host’s genetics, gut microbiota, and immune system [3,4,5]. Genetically susceptible individuals seem to be more prone to developing IBDs as a consequence of their atypical immune response to the autologous gut microbiota, following exposure to different environmental factors and stimuli [6,7]. Indeed, recent studies have revealed that pathologic alterations in the GI microbiota activate a mucosal immune response, thus developing chronic intestinal inflammation. These alterations in the GI microbiota trigger the so-called dysbiosis. Thus, the disruption of the intestinal eubiotic status can be considered a cause rather than simply a consequence of the chronic GI inflammation in IBDs [8,9,10,11,12].
Many studies have reported an increase in the proportion of potentially harmful Proteobacteria, especially of the Enterobacteriaceae family, in IBD patients [2,13,14,15,16,17,18,19]. Therefore, it would appear that the host’s inflammatory response could be the trigger of gut microbiota imbalance, most likely caused by Enterobacteriaceae blooming, which leads to the persistence of IBD’s inflammatory state (Figure 1).
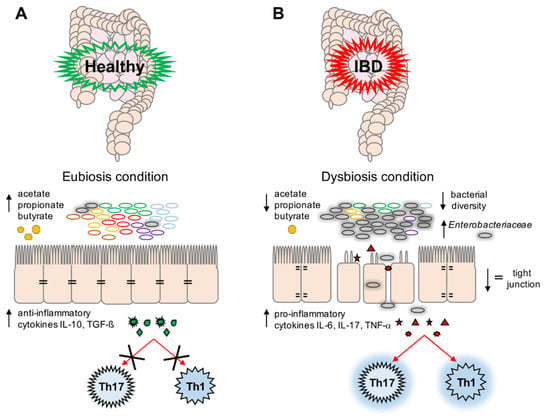
Figure 1.
A schematic representation of eubiosis (A) and dysbiosis (B) conditions. (A) Microbiota plays an important role in the maintenance of gut stability by: (i) producing short-chain fatty acids (SCFAs) (i.e., acetate, propionate, and butyrate), (ii) preventing the expansion in any microbial pathogens, and (iii) modulating the immune system (i.e., production of anti-inflammatory cytokines (Interleukin (IL)-10, Transforming Growth Factor (TGF)-β)) and decreasing the activation of T-helper cell (Th)17 and Th1 cells; (B) During dysbiotic conditions, bacteria belonging to the Enterobacteriaceae family could overgrow, leading to a decrease in bacterial diversity and in gut stability. This is reflected by a decrease in SCFA production, and a parallel increase in pro-inflammatory cytokines such as IL-6, IL-17, and tumor necrosis factor (TNF)-α, which activate Th17 and Th1 cells involved in the inflammation response. Moreover, a decrease in tight junctions and a subsequent loss of impermeability in the intestinal epithelium is observed.
The main aim of this review is to describe and clarify the overgrowth of Enterobacteriaceae in IBDs, especially focusing on their role during the inflammation process in relation to metabolism changes observed in the gut microbiota of IBD patients. The secondary aim is to provide an update on certain adherent-invasive Escherichia coli (AIEC) strains frequently isolated from IBD patients, and their potential role in both CD and UC disease, discussing the difficulties in the characterization at species level of Enterobacteriaceae by targeted metagenomic approaches.
2. The Role of Gut Bacterial Microbiota in Inflammatory Bowel Diseases
Within the GI microbiota, the most studied component is the bacterial microbiota. Humans have more than 200 different bacterial species, with highly variable abundance in the gut. Thirty percent of this population is shared by different subjects, generating the core microbiota [20,21,22].
The bacterial microbiota plays important functions in the host, such as (i) the modulation of the immune system [23], (ii) the secretion of enzymes involved in the digestion of substrates not fully accessible to the host [24], and (iii) competition with pathogenic microorganisms for the same ecological niche [25]. In this context, the functionality of the GI tract is ensured and maintained also by the gut microbiota, which plays a positive role in the preservation of the intestinal barrier [26] (Figure 1A).
In particular, eubiotic microbiota is necessary to maintain the host’s immune homeostasis by the induction and function of T cells. Alterations in the gut microbiota’s composition can lead to the imbalance of T-cell subpopulations, such as Th1, Th2, Th17, and Treg cells, causing gut inflammation [27].
Numerous studies support the involvement of the GI microbiota in the pathogenesis of IBDs. Indeed, characteristic dysbiosis is often observed in subjects with CD and UC [28,29,30,31]; often, the induction or the maintenance of remission in IBDs is frequently related to the use of antibiotics and probiotics [32,33]. Several studies have demonstrated changes in microbial richness and evenness, and the relative abundance of specific bacterial taxa in IBD patients with respect to healthy subjects [34,35,36,37,38,39,40,41,42,43]. In particular, at the phylum level, the proportion of both Firmicutes and Bacteroidetes appears decreased, while the proportion of Proteobacteria and Actinobacteria is increased [13,44,45,46].
As shown by Geveres and co-workers, an increase in Veillonellaceae, Pasteurellaceae, Enterobacteriaceae, and Fusobacteriaceae, and a decrease in Bacteroidales, Erysipelotrichales, and Clostridiales, were correlated with IBDs’ clinical severity [38]. Similarly, a study on pediatric patients with CD concluded that the presence and severity of inflammation depends on microbiota dysbiosis; nonetheless, this study found an independent correlation between dysbiosis and other factors such as diet and/or the usage of antibiotics [39].
In particular, patients suffering from IBDs are associated with a decline in the abundance of protective anaerobic commensal bacteria, such as Faecalibacterium prausnitzii, Clostridium spp., and Bacteroidetes fragilis. F. prausnitzii has been shown to have anti-inflammatory properties, including the ability to modulate the host’s mucosal immune response by the production of short-chain fatty acids (SCFAs) [40]. SCFAs, such as acetate, propionate, and butyrate, are a primary energy source for colonic epithelial cells [40]. Thus, the underrepresentation of this bacterium in IBDs results in a decrease in the beneficial effects of SCFAs, including the inhibition of pro-inflammatory cytokine expression, the production of mucin and antimicrobial peptides, and tight junction protein downregulation [2,22,32,40,45,46] (Figure 1B).
Moreover, the gut microbiota in IBDs is often characterized by an expansion of translocating facultative aerobic bacteria—possible pathobionts—particularly belonging to the family of Enterobacteriaceae, such as Escherichia and Shigella [13,38,47,48,49]. Pathobionts are defined as commensal microorganisms that might become pathogens in the setting of a specific environmental stimulus in genetically susceptible subjects [50].
Another bacterium abundantly found in patients suffering from UC and CD is Fusobacterium nucleatum [51]. This bacterium showed the ability to adhere to and invade the intestinal barrier, inducing aberrant inflammation and exacerbating colitis, both in human and animal models [52,53,54,55,56]. Moreover, it has been recently proposed that the detection of F. nucleatum in the fecal metagenome could be used as an early biomarker of gut dysbiosis in IBDs and colorectal cancer [57].
3. The Role of Other Gut Microbiota Inhabitants in Inflammatory Bowel Diseases
Although fewer studies have examined the role of fungi and viruses specifically in the propagation of inflammation in IBDs, both of them are ubiquitous components of the intestinal microbiota [36]. As an example, patients with IBDs show differences in the presence of certain fungi compared with non-IBD controls, and different studies have revealed that this fungal dysbiosis is associated with an increased ratio between Basidiomycota and Ascomycota at the phylum level, indicating the emerging need to characterize the gut microbiota in IBDs [11,22,36,58,59,60,61].
Among viruses, bacteriophages are the predominant elements of the so-called virome in healthy humans [22,36]. In this context, changes in bacteriophage composition in IBD patients have been described [62,63,64]. However, whether bacteriophages could display a direct role in IBDs still remains unclear and has yet to be determined [36,65].
4. The Enterobacteriaceae Overgrowth in Inflammatory Bowel Diseases
These bacteria belonging to the Enterobacteriaceae family are localized near the intestinal mucosa due to their relatively higher tolerance of oxygen dispersed by the epithelium [66].
These bacteria are the first colonizers in the aerobic GI tracts of newborns. In fact, they are facultative anaerobes, which deplete oxygen to create a reduced environment suitable for following the colonization of strict anaerobes [67]. Following this, bacterial oligosaccharide fermenters, such as Bifidobacterium, bloom in the GI tract because of breast milk intake. Subsequently, the introduction of a solid food diet rich in polysaccharides produces the expansion of anaerobes and polysaccharide fermenters such as Bacteroides, Clostridium, and Ruminococcus, and a simultaneous decrease in Bifidobacterium and Enterobacteriaceae [68,69].
In IBD mouse models, microbiota dysbiosis in response to inflammation has been described, with a large relative increase in Enterobacteriaceae [48,70,71,72]. These studies support the notion that Enterobacteriaceae seem to have a growth advantage over other members of the gut microbiota commensals in the inflamed GI mucosa.
Moreover, an increment in Enterobacteriaceae was also associated with chronic UC inflammation compared to an acute status, showing a positive correlation of this bacterial family with a severe disease stage [73].
Furthermore, in a CD pediatric cohort, Enterobacteriaceae enrichment has been associated with an aggressive disease course with a higher risk of treatment failure [74].
In IBD patients, the functions of the gut microbiota have also been found to be perturbed. As an example, bacterial amino acid biosynthesis and carbohydrate metabolism are diminished, whereas nutrient uptake is enhanced [2]. In these patients, bacterial genes such as redox tolerance, secretion systems, and adherence and/or invasion, are overrepresented and the pathways linked to the production of bacterial SCFAs are repressed [37].
In a mouse model of UC, the blooming of Enterobacteriaceae has been linked to tetrathionate respiration, a metabolic pathway promoting intestinal colonization by Salmonella enterica subsp. Typhimurium, and with benzoate degradation, a pathway linked to Enterobacteriaceae growth and virulence pathways [75]. To date, the link between functional and compositional microbiota changes and IBD pathogenesis remains unclear. Moreover, it remains to be discovered whether the differences in the microbiota originating prior to the initiation of the disease process are a mere consequence of a hyperactive immune response.
The mechanisms by which Enterobacteriaceae bloom in IBDs, particularly during inflammation, are not completely known. In order to explain this observation, the “oxygen hypothesis” has been formulated [76].
In physiological conditions, enterocytes reduce oxygen levels in the gut lumen by beta-oxidation processes, creating an anaerobic environment [77]. In IBD patients in which intestinal inflammation occurs, beta-oxidation is reduced, and the oxygen level is increased. This event leads to an increase in facultative aerobes such as Proteobacteria/Enterobacteriaceae and, as a consequence, intestinal dysbiosis [78,79].
Moreover, in inflammatory conditions, an increment in nitrate production by the host has been demonstrated [80,81,82]. The expression of the gene encoding nitric oxide synthase is inhibited by the SCFA butyrate, via the activation of the peroxisome proliferator-activated receptor (PPAR) [83]. Butyrate represents the major energy source for enterocytes and it is involved in the maintenance of colonic mucosal health [84]. Intestinal inflammation leads to a reduction in healthy butyrate-producing microbiota, which leads to an increase in nitrate production and then the blooming of Enterobacteriaceae [83]. Consequently, the increased levels of both oxygen and nitrate observed in IBD patients could lead to Enterobacteriaceae overgrowth.
5. Inflammation and Enterobacteriaceae
Enterobacteriaceae contain molecular components that directly enhance the inflammatory response. Microbe-associated molecular patterns (MAMPs) are molecules located on the bacterial surface. These molecules interact with the receptors on immune cells to trigger inflammation [85]. One of the most potent MAMPs is the endotoxin lipopolysaccharide (LPS), which interacts with Toll-Like Receptor (TLR) 4 on immune cells [86]. The lipid-A of LPS can be acylated with five or six acyl chains. Hexa-acylated LPS shows immunostimulant activity that is 100-fold more active than penta-acylated LPS [87]. Enterobacteriaceae, and, in general, Proteobacteria, possess a more immunostimulatory version of LPS. Moreover, Enterobacteriaceae present unmethylated immunostimulatory motifs that can induce an immune response through an interaction with TLR-9 [88]. The blooming of Enterobacteriaceae, which contain these motifs, leads to a more selective pressure and shift toward an Enterobacteriaceae-dominated community [89].
Some scientific evidence leads to the hypothesis that dysregulation in innate immune systems induces Proteobacteria overgrowth, which promotes intestinal inflammation [90]. Regarding this, it has been reported that mice lacking TLR-5 developed transmissible spontaneous colitis associated with an abnormal expansion of Enterobacteriaceae [91]. Moreover, spontaneous colitis has also been reported in interleukin (IL)-10-deficient mice, due to their intolerance of intestinal microbiota. Indeed, IL-10 is the main anti-inflammatory cytokine required for immune tolerance to naïve microbiota [92], and in this mouse model, the onset and progression of gut inflammation has been correlated with an increase in E. coli [92].
Several studies also reported the role of Enterobacteriaceae in inducing the secretion of IL-8, tumor necrosis factor (TNF)-α, and IL-1β, and in the disruption of intestinal mucosa tight junctions, followed by an increment in intestinal permeability and inflammation [93,94].
Furthermore, a positive correlation between the nucleotide-binding oligomerization domain (NOD)-2 risk allele and Enterobacteriaceae was described in intestinal biopsies from an adult IBD patient cohort [95]. Since NOD-2 is an intracellular bacterial-sensing receptor that drives inflammatory signaling, this evidence strengthens the hypothesis of the direct involvement of Enterobacteriaceae in inflammation.
6. Bile Acid Dysmetabolism and Enterobacteriaceae in Inflammatory Bowel Diseases
In IBDs, an impairment in the microbial bile acid (BA) metabolism has been demonstrated, with high levels of fecal-conjugated BAs and low levels of secondary BA production [96]. These impairments in deconjugation and transformation abilities were associated with microbiomes [96].
In physiological conditions, cholesterol and BAs regulate multiple metabolic processes in the host [97,98]. Hepatocytes synthesize primary BAs from cholesterol conjugated to glycine and taurine. These conjugated BAs are stored in the gall bladder and are involved in lipid absorption and fat emulsification [99]. Gut microbiota carry out numerous BA biotransformation reactions [99], including the hydrolysis of conjugated BAs and glycine or taurine by bile salt hydrolase (BSH), and BAs’ 7α/7β-dehydroxylation by a multistep biochemical pathway found only in anaerobic bacteria [100].
An impairment in the gut microbiota’s metabolism significantly harms BAs’ metabolism and, consequently, the host’s glucose and cholesterol homeostasis [99]. Moreover, a decrease in BAs seems to favor the blooming of pathogenic and pro-inflammatory members of Enterobacteriaceae [101]. These changes in the gut microbiota’s ability of BA modification may be a significant factor in the onset or progression of IBDs [102]. Furthermore, modified BAs display altered binding profiles for BA receptors (i.e., Farnesoid X receptor (FXR)), inferring that the dysbiosis of the gut microbiota’s characteristic of IBDs may alter the capacity for BA modification in this community [103]. Cirrhotic patients have demonstrated a positive correlation between BA dysmetabolism and Enterobacteriaceae overgrowth [104]. Moreover, in obese patients the administration of vancomycin increased the abundance in Proteobacteria and, concomitantly, decreased fecal secondary BAs, with a simultaneous postprandial increase in primary BAs in plasma [105]. Finally, Heinken and co-workers demonstrated through a computational model that the dysbiotic IBDs’ microbiota were enriched in pathogenic Escherichia spp., which highly contributed to the BAs’ deconjugation transformation [106].
Future studies should be undertaken to validate and strengthen the evidence that Enterobacteriaceae play a significant role in BA dysmetabolism in IBDs, and to explore if BA modification driven by dysbiosis might represent a marker of IBDs’ onset or progression.
7. The Role of Escherichia coli in Inflammatory Bowel Diseases
One of the well-studied members of the Enterobacteriaceae family is E. coli, which is frequently able to colonize the human intestine [107]. This bacterium has been proposed as a possible cause of the beginning of disease in IBD patients [108]; indeed, several studies have found an increased number of virulent E. coli strains isolated from IBD patients with respect to healthy controls [108,109].
In the genome of E. coli strains, there are three nitrate reductases and three nitric oxide reductases. Hence, non-fermentable nitrates can be used by E. coli as a substrate for nitrate respiration and converted into fermentable nitrates. As a consequence, the expansion of E. coli/Enterobacteriaceae in the lumen of the inflamed gut observed in IBD subjects could reflect the high levels of non-fermentable nitrate generated in the gut by the host’s inflammatory response [80]. Consequently, in human and animal models, the presence of E. coli strains in IBDs has been reported (Table 1). However, whether E. coli strains may specifically benefit from nitrate respiration during IBDs has yet to be studied [94,110].

Table 1.
Escherichia coli strains identified in human and in animal models of inflammatory bowel diseases (IBDs).
Most of the E. coli strains identified in IBD samples are AIEC [18,37,47,114,119]. AIEC members are (i) able to adhere to and invade epithelial cells utilizing a process dependent on actin microfilaments and microtubule recruitment; (ii) able to survive within the macrophage phagolysosome; (iii) able to induce the production of tumor necrosis factor (TNF) from infected macrophages; and, finally, lacking any defined invasive element [122,123]. The adaptive evolution of the genome of AIEC strains in a susceptible host and their ability to promote inflammation makes these microorganisms potential pathobionts [120,124,125,126].
Several studies have reported that different E. coli strains persist within epithelial cells and macrophages and selectively colonize the ileum of CD patients [111,112,113,114,120]. In particular, AIEC strains adhere to epithelial cells by the binding between the type 1 pili, expressed by bacteria, and the carcinoembryonic antigen-related cell adhesion molecule (CEACAM) 6, expressed on the apical surface of ileal epithelial cells [127]. Epithelial expression of CEACAM 6 is stimulated by interferon (IFN)-γ and TNF, which are overexpressed by macrophages harboring AIEC strains, generating a positive feedback loop [29,123].
E. coli strains from ileal biopsy from CD patients express several virulence genes mediating epithelial adherence and iron acquisition. Moreover, AIEC strains possess a wide arsenal of genes, such as fliC, OmpC, and others that regulate type 1 pilus expression and mediate epithelial cell adherence/invasion [29,128,129].
Further observations suggest that UC-associated AIEC could be distinct from those observed in CD, with greater invasive properties [115] or displaying different types of gene expression [121]. Although several studies confirmed the presence of AIEC strains in a higher proportion with respect to CD patients [116,118], others reported a lower prevalence compared with that of CD [115,117,119]. In the systematic review of Nadalian and co-workers, an increase in AIEC strains in UC patients emerged with respect to non-IBD controls, as well as the possible involvement of these bacterial strains in the pathogenesis of this disease [130].
Some studies of animal models suggest that the accumulation of AIEC and other E. coli strains in the gut could be a consequence of inflammation [17,72,92,131]. Moreover, several studies revealed that AIEC infection could induce changes in the gut microbiota [132,133]. Nowadays, it still remains unclear whether AIEC strains cause intestinal inflammation leading to IBDs or whether their blooming could be a consequence of inflammation, which leads to the aggravation of these diseases.
8. Identification of Enterobacteriaceae from Direct Samples
The number of genera and species in the Enterobacteriaceae family has markedly increased. Currently, this family comprises >60 genera and 250 species [134,135]. To better define the role of Enterobacteriaceae in gut inflammation or in intestinal microbiota dysbiosis, it is important to distinguish each family component at genus or species level. As reported in Table 1, the main studies in which Enterobacteriaceae species have been correlated to IBDs are based on bacterial identification using culture-based techniques. In fact, the low sequence variance in 16S rRNA partial gene sequencing, usually selected in 16S rRNA-based metagenomics, does not allow resolution at genus/species level [136,137].
Thus, it is mandatory to select other molecular targets that offer a superior discriminatory power of Enterobacteriaceae at the level of genus/species, especially for gut microbiota studies with targeted metagenomics approaches [138]. In the past, different genes, such as tuf, atpD, dnaJ, rpoB, infB, and gyrB, have been explored using molecular biology techniques [136,139,140,141,142].
Phylogenetic analyses of the tuf and atpD gene sequences have demonstrated controversial results in the identification of some Enterobacteriaceae species from isolated microbiological culture [136].
The dnaJ gene encodes the heat shock protein 40 and contains regions highly conserved in the Enterobacteriaceae family, suitable for broad-specificity primer design [143]. Despite this target demonstrating high performance in the unambiguous identification of Enterobacteriaceae species, it remains suboptimal for Enterobacteriaceae discrimination from direct specimens. In fact, the amplicon generated by polymerase chain reaction (PCR) is relatively long (758 base pair (bp)) for metagenomic-based approaches, which require short-length fragments [143].
The rpoB gene encodes the RNA polymerase subunit [139], and a partial region of this locus has been employed in describing nematode microbiota [144]. This study revealed that this target could be considered a highly appropriate marker for assessing the taxonomic structure of mock communities; however, further studies confirming this are required.
The infB gene is a universally distributed gene in all prokaryotes and encodes for translation initiation factor 2 (IF2) [140]. In Hedegaard and colleagues’ study, a partial sequence of infB was used to develop a method for identifying enterobacteria at species level. The authors concluded that infB can be used to both discriminate and classify strains of the same species [140].
The gyrB gene, which encodes the subunit B protein of DNA gyrase, has been previously used for describing phylogenetic relationships between Enterobacteriaceae [141]. A fragment of the gyrB gene (gyrBint) of 506 bp has been successfully employed to identify Enterobacteriaceae, proving its applicability in a clinical laboratory [145].
Nowadays, shotgun metagenomics approaches could allow gut microbiota taxonomic profiling, including Enterobacteriaceae, at species level, from direct samples [146,147,148]. However, sample contamination by high levels of the host’s DNA and the high cost of the analysis represent drawbacks of shotgun metagenomics with respect to targeted metagenomics [149]. Moreover, comparative studies on gut microbiota composition using both approaches have highlighted the differential depiction of bacterial profiles, although there was a consistent description of core microbiota [150,151]. Thus, both metagenomics approaches could be considered complementary in the genus-/species-level description of Enterobacteriaceae.
9. Concluding Remarks
There is a plethora of evidence on the contribution of microbiota dysbiosis to gut inflammation in IBDs. The loss of beneficial microbes that produce anti-inflammatory molecules favors Enterobacteriaceae as the environment for the expansion of pathogens and/or pathobionts. The proinflammatory nature of Enterobacteriaceae has been well established. These bacterial family members contain molecules such as MAMPs and LPS that directly enhance the inflammatory response of the host. Moreover, Enterobacteriaceae could also participate in intestinal dysmetabolism, altering the BA metabolism, formerly impaired in IBDs.
Amongst Enterobacteriaceae, AIEC strains colonize the intestinal mucosa of IBD patients. Several studies in animal models correlated the intestinal overgrowth of these bacteria to the gut inflammation trigger. However, the contribution of specific Enterobacteriaceae species or shifts in the IBD gut microbiota remains under debate.
In fact, is emerged the difficulty to discriminate by the 16S rRNA-based metagenomics the effective composition of Enterobacteriaceae in IBDs and to define the active role of each component in these diseases.
The identification of Enterobacteriaceae components and the deduction of their function in intestinal dysbiosis in IBDs represents a stimulating challenge, requiring expertise and effort in order to improve the knowledge and the comprehension of the fine mechanism of IBDs’ pathogenesis and progression.
Author Contributions
Conceptualization, V.B. and F.D.C.; writing—original draft preparation, V.B. and F.D.C.; writing—review and editing, V.B., L.P., F.S. and F.D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cosnes, J.; Gower-Rousseau, C.; Seksik, P.; Cortot, A. Epidemiology and Natural History of Inflammatory Bowel Diseases. Gastroenterology 2011, 140, 1785–1794. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the Intestinal Microbiome in Inflammatory Bowel Disease and Treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Cheon, J.H. Pathogenesis of Inflammatory Bowel Disease and Recent Advances in Biologic Therapies. Immune Netw. 2017, 17, 25. [Google Scholar] [CrossRef] [PubMed]
- Putignani, L.; Del Chierico, F.; Vernocchi, P.; Cicala, M.; Cucchiara, S.; Dallapiccola, B.; Dysbiotrack Study Group. Gut Microbiota Dysbiosis as Risk and Premorbid Factors of IBD and IBS Along the Childhood-Adulthood Transition. Inflamm. Bowel Dis. 2016, 22, 487–504. [Google Scholar] [CrossRef]
- Hold, G.L.; Smith, M.; Grange, C.; Watt, E.R.; El-Omar, E.M.; Mukhopadhya, I. Role of the Gut Microbiota in Inflammatory Bowel Disease Pathogenesis: What Have We Learnt in the Past 10 Years? World J. Gastroenterol. 2014, 20, 1192–1210. [Google Scholar] [CrossRef]
- Lo Presti, A.; Zorzi, F.; Del Chierico, F.; Altomare, A.; Cocca, S.; Avola, A.; De Biasio, F.; Russo, A.; Cella, E.; Reddel, S.; et al. Fecal and Mucosal Microbiota Profiling in Irritable Bowel Syndrome and Inflammatory Bowel Disease. Front. Microbiol. 2019, 10, 1655. [Google Scholar] [CrossRef]
- Strober, W.; Fuss, I.; Mannon, P. The Fundamental Basis of Inflammatory Bowel Disease. J. Clin. Investig. 2007, 117, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.J.; Sartor, R.B. Therapeutic Manipulation of the Microbiome in IBD: Current Results and Future Approaches. Curr. Treat. Options Gastroenterol. 2015, 13, 105–120. [Google Scholar] [CrossRef]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The Long-Term Stability of the Human Gut Microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- Lane, E.R.; Zisman, T.L.; Suskind, D.L. The Microbiota in Inflammatory Bowel Disease: Current and Therapeutic Insights. J. Inflamm. Res. 2017, 10, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.K.; Kamm, M.A.; Teo, S.M.; Inouye, M.; Wagner, J.; Kirkwood, C.D. Recent Advances in Characterizing the Gastrointestinal Microbiome in Crohn’s Disease: A Systematic Review. Inflamm. Bowel Dis. 2015, 21, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-Mediated Inflammation Disrupts the Intestinal Microbiota and Promotes the Overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 204. [Google Scholar] [CrossRef]
- Gophna, U.; Sommerfeld, K.; Gophna, S.; Doolittle, W.F.; Veldhuyzen van Zanten, S.J.O. Differences between Tissue-Associated Intestinal Microfloras of Patients with Crohn’s Disease and Ulcerative Colitis. J. Clin. Microbiol. 2006, 44, 4136–4141. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Medina, M.; Aldeguer, X.; Gonzalez-Huix, F.; Acero, D.; Garcia-Gil, L.J. Abnormal Microbiota Composition in the Ileocolonic Mucosa of Crohn’s Disease Patients as Revealed by Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis. Inflamm. Bowel Dis. 2006, 12, 1136–1145. [Google Scholar] [CrossRef]
- Wang, M.; Molin, G.; Ahrné, S.; Adawi, D.; Jeppsson, B. High Proportions of Proinflammatory Bacteria on the Colonic Mucosa in a Young Patient with Ulcerative Colitis as Revealed by Cloning and Sequencing of 16S RRNA Genes. Dig. Dis. Sci. 2007, 52, 620–627. [Google Scholar] [CrossRef]
- Carvalho, F.A.; Barnich, N.; Sauvanet, P.; Darcha, C.; Gelot, A.; Darfeuille-Michaud, A. Crohn’s Disease-Associated Escherichia coli LF82 Aggravates Colitis in Injured Mouse Colon via Signaling by Flagellin. Inflamm. Bowel Dis. 2008, 14, 1051–1060. [Google Scholar] [CrossRef]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD-What Role Do Proteobacteria Play? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef]
- Lavelle, A.; Lennon, G.; O’Sullivan, O.; Docherty, N.; Balfe, A.; Maguire, A.; Mulcahy, H.E.; Doherty, G.; O’Donoghue, D.; Hyland, J.; et al. Spatial Variation of the Colonic Microbiota in Patients with Ulcerative Colitis and Control Volunteers. Gut 2015, 64, 1553–1561. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The Abundance and Variety of Carbohydrate-Active Enzymes in the Human Gut Microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The Gut Microbiota Shapes Intestinal Immune Responses during Health and Disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Zuo, T.; Ng, S.C. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front. Microbiol. 2018, 9, 2247. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The Gut Flora as a Forgotten Organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced Diversity of Faecal Microbiota in Crohn’s Disease Revealed by a Metagenomic Approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-Phylogenetic Characterization of Microbial Community Imbalances in Human Inflammatory Bowel Diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef]
- Li, J.; Butcher, J.; Mack, D.; Stintzi, A. Functional Impacts of the Intestinal Microbiome in the Pathogenesis of Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015, 21, 139–153. [Google Scholar] [CrossRef]
- Lee, N.; Kim, W.-U. Microbiota in T-Cell Homeostasis and Inflammatory Diseases. Exp. Mol. Med. 2017, 49, e340. [Google Scholar] [CrossRef]
- Sartor, R.B. Therapeutic Manipulation of the Enteric Microflora in Inflammatory Bowel Diseases: Antibiotics, Probiotics, and Prebiotics. Gastroenterology 2004, 126, 1620–1633. [Google Scholar] [CrossRef]
- Sartor, R.B. Microbial Influences in Inflammatory Bowel Diseases. Gastroenterology 2008, 134, 577–594. [Google Scholar] [CrossRef]
- Rietdijk, S.T.; D’Haens, G.R. Recent Developments in the Treatment of Inflammatory Bowel Disease. J. Dig. Dis. 2013, 14, 282–287. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The Gut Microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium Prausnitzii Is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-Throughput Clone Library Analysis of the Mucosa-Associated Microbiota Reveals Dysbiosis and Differences between Inflamed and Non-Inflamed Regions of the Intestine in Inflammatory Bowel Disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Rausch, P.; Wang, J.; Skieceviciene, J.; Kiudelis, G.; Bhagalia, K.; Amarapurkar, D.; Kupcinskas, L.; Schreiber, S.; Rosenstiel, P.; et al. Geographical Patterns of the Standing and Active Human Gut Microbiome in Health and IBD. Gut 2016, 65, 238–248. [Google Scholar] [CrossRef]
- Halfvarson, J.; Brislawn, C.J.; Lamendella, R.; Vázquez-Baeza, Y.; Walters, W.A.; Bramer, L.M.; D’Amato, M.; Bonfiglio, F.; McDonald, D.; Gonzalez, A.; et al. Dynamics of the Human Gut Microbiome in Inflammatory Bowel Disease. Nat. Microbiol. 2017, 2, 17004. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut Microbiota and IBD: Causation or Correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jobin, C. Microbial Imbalance and Intestinal Pathologies: Connections and Contributions. Dis. Model. Mech. 2014, 7, 1131–1142. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef]
- Lewis, J.D.; Chen, E.Z.; Baldassano, R.N.; Otley, A.R.; Griffiths, A.M.; Lee, D.; Bittinger, K.; Bailey, A.; Friedman, E.S.; Hoffmann, C.; et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 2017, 22, 247. [Google Scholar] [CrossRef]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Doré, J. Low Counts of Faecalibacterium Prausnitzii in Colitis Microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef]
- Ahmad, M.S.; Krishnan, S.; Ramakrishna, B.S.; Mathan, M.; Pulimood, A.B.; Murthy, S.N. Butyrate and Glucose Metabolism by Colonocytes in Experimental Colitis in Mice. Gut 2000, 46, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg Induction by a Rationally Selected Mixture of Clostridia Strains from the Human Microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Altomare, A.; Putignani, L.; Del Chierico, F.; Cocca, S.; Angeletti, S.; Ciccozzi, M.; Tripiciano, C.; Dalla Piccola, B.; Cicala, M.; Guarino, M.P.L. Gut Mucosal-Associated Microbiota Better Discloses Inflammatory Bowel Disease Differential Patterns than Faecal Microbiota. Dig. Liver Dis. 2019, 51, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Xenoulis, P.G.; Palculict, B.; Allenspach, K.; Steiner, J.M.; Van House, A.M.; Suchodolski, J.S. Molecular-Phylogenetic Characterization of Microbial Communities Imbalances in the Small Intestine of Dogs with Inflammatory Bowel Disease. FEMS Microbiol. Ecol. 2008, 66, 579–589. [Google Scholar] [CrossRef]
- Kang, S.; Denman, S.E.; Morrison, M.; Yu, Z.; Dore, J.; Leclerc, M.; McSweeney, C.S. Dysbiosis of Fecal Microbiota in Crohn’s Disease Patients as Revealed by a Custom Phylogenetic Microarray. Inflamm. Bowel Dis. 2010, 16, 2034–2042. [Google Scholar] [CrossRef] [PubMed]
- Darfeuille-Michaud, A.; Neut, C.; Barnich, N.; Lederman, E.; Di Martino, P.; Desreumaux, P.; Gambiez, L.; Joly, B.; Cortot, A.; Colombel, J.F. Presence of Adherent Escherichia coli Strains in Ileal Mucosa of Patients with Crohn’s Disease. Gastroenterology 1998, 115, 1405–1413. [Google Scholar] [CrossRef]
- Håkansson, Å.; Tormo-Badia, N.; Baridi, A.; Xu, J.; Molin, G.; Hagslätt, M.-L.; Karlsson, C.; Jeppsson, B.; Cilio, C.M.; Ahrné, S. Immunological Alteration and Changes of Gut Microbiota after Dextran Sulfate Sodium (DSS) Administration in Mice. Clin. Exp. Med. 2015, 15, 107–120. [Google Scholar] [CrossRef]
- Boudeau, J.; Glasser, A.L.; Masseret, E.; Joly, B.; Darfeuille-Michaud, A. Invasive Ability of an Escherichia coli Strain Isolated from the Ileal Mucosa of a Patient with Crohn’s Disease. Infect. Immun. 1999, 67, 4499–4509. [Google Scholar] [CrossRef]
- Chow, J.; Tang, H.; Mazmanian, S.K. Pathobionts of the Gastrointestinal Microbiota and Inflammatory Disease. Curr. Opin. Immunol. 2011, 23, 473–480. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, Opportunist and Oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.; Kaplan, G.G.; Beck, P.L.; Rioux, K.; Panaccione, R.; Devinney, R.; Lynch, T.; Allen-Vercoe, E. Invasive Potential of Gut Mucosa-Derived Fusobacterium nucleatum Positively Correlates with IBD Status of the Host. Inflamm. Bowel Dis. 2011, 17, 1971–1978. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hong, X.L.; Sun, T.T.; Huang, X.W.; Wang, J.L.; Xiong, H. Fusobacterium nucleatum Exacerbates Colitis by Damaging Epithelial Barriers and Inducing Aberrant Inflammation. J. Dig. Dis. 2020, 21, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Ohkusa, T.; Sato, N.; Ogihara, T.; Morita, K.; Ogawa, M.; Okayasu, I. Fusobacterium varium Localized in the Colonic Mucosa of Patients with Ulcerative Colitis Stimulates Species-Specific Antibody. J. Gastroenterol. Hepatol. 2002, 17, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Ohkusa, T.; Yoshida, T.; Sato, N.; Watanabe, S.; Tajiri, H.; Okayasu, I. Commensal Bacteria Can Enter Colonic Epithelial Cells and Induce Proinflammatory Cytokine Secretion: A Possible Pathogenic Mechanism of Ulcerative Colitis. J. Med. Microbiol. 2009, 58, 535–545. [Google Scholar] [CrossRef]
- Bashir, A.; Miskeen, A.Y.; Hazari, Y.M.; Asrafuzzaman, S.; Fazili, K.M. Fusobacterium nucleatum, Inflammation, and Immunity: The Fire within Human Gut. Tumor Biol. 2016, 37, 2805–2810. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.-W.; Roh, T.-Y. Opportunistic Detection of Fusobacterium nucleatum as a Marker for the Early Gut Microbial Dysbiosis. BMC Microbiol. 2020, 20, 208. [Google Scholar] [CrossRef]
- Ott, S.J.; Kühbacher, T.; Musfeldt, M.; Rosenstiel, P.; Hellmig, S.; Rehman, A.; Drews, O.; Weichert, W.; Timmis, K.N.; Schreiber, S. Fungi and Inflammatory Bowel Diseases: Alterations of Composition and Diversity. Scand. J. Gastroenterol. 2008, 43, 831–841. [Google Scholar] [CrossRef]
- Mukhopadhya, I.; Hansen, R.; Meharg, C.; Thomson, J.M.; Russell, R.K.; Berry, S.H.; El-Omar, E.M.; Hold, G.L. The Fungal Microbiota of De-Novo Paediatric Inflammatory Bowel Disease. Microbes Infect. 2015, 17, 304–310. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal Microbiota Dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef]
- Sovran, B.; Planchais, J.; Jegou, S.; Straube, M.; Lamas, B.; Natividad, J.M.; Agus, A.; Dupraz, L.; Glodt, J.; Da Costa, G.; et al. Enterobacteriaceae Are Essential for the Modulation of Colitis Severity by Fungi. Microbiome 2018, 6, 152. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Brocal, V.; García-López, R.; Vázquez-Castellanos, J.F.; Nos, P.; Beltrán, B.; Latorre, A.; Moya, A. Study of the Viral and Microbial Communities Associated with Crohn’s Disease: A Metagenomic Approach. Clin. Transl. Gastroenterol. 2013, 4, e36. [Google Scholar] [CrossRef]
- Wagner, J.; Maksimovic, J.; Farries, G.; Sim, W.H.; Bishop, R.F.; Cameron, D.J.; Catto-Smith, A.G.; Kirkwood, C.D. Bacteriophages in Gut Samples from Pediatric Crohn’s Disease Patients: Metagenomic Analysis Using 454 Pyrosequencing. Inflamm. Bowel Dis. 2013, 19, 1598–1608. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-Specific Alterations in the Enteric Virome in Inflammatory Bowel Disease. Cell 2015, 160, 447–460. [Google Scholar] [CrossRef]
- Matsuoka, K.; Kanai, T. The Gut Microbiota and Inflammatory Bowel Disease. Semin. Immunopathol. 2015, 37, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.Y.; Inohara, N.; Nuñez, G. Mechanisms of Inflammation-Driven Bacterial Dysbiosis in the Gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef]
- Arrieta, M.-C.; Stiemsma, L.T.; Amenyogbe, N.; Brown, E.M.; Finlay, B. The Intestinal Microbiome in Early Life: Health and Disease. Front. Immunol. 2014, 5, 427. [Google Scholar] [CrossRef]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of Microbial Consortia in the Developing Infant Gut Microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 4578–4585. [Google Scholar] [CrossRef]
- Fallani, M.; Amarri, S.; Uusijarvi, A.; Adam, R.; Khanna, S.; Aguilera, M.; Gil, A.; Vieites, J.M.; Norin, E.; Young, D. Determinants of the Human Infant Intestinal Microbiota after the Introduction of First Complementary Foods in Infant Samples from Five European Centres. Microbiology 2011, 157, 1385–1392. [Google Scholar] [CrossRef]
- Ettreiki, C. Juvenile Ferric Iron Prevents Microbiota Dysbiosis and Colitis in Adult Rodents. World J. Gastroenterol. 2012, 18, 2619. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.; Eibach, D.; Kops, F.; Brenneke, B.; Woltemate, S.; Schulze, J.; Bleich, A.; Gruber, A.D.; Muthupalani, S.; Fox, J.G.; et al. Intestinal Microbiota Composition of Interleukin-10 Deficient C57BL/6J Mice and Susceptibility to Helicobacter Hepaticus-Induced Colitis. PLoS ONE 2013, 8, e70783. [Google Scholar] [CrossRef] [PubMed]
- Munyaka, P.M.; Rabbi, M.F.; Khafipour, E.; Ghia, J.-E. Acute Dextran Sulfate Sodium (DSS)-Induced Colitis Promotes Gut Microbial Dysbiosis in Mice. J. Basic Microbiol. 2016, 56, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Walujkar, S.A.; Dhotre, D.P.; Marathe, N.P.; Lawate, P.S.; Bharadwaj, R.S.; Shouche, Y.S. Characterization of Bacterial Community Shift in Human Ulcerative Colitis Patients Revealed by Illumina Based 16S RRNA Gene Amplicon Sequencing. Gut Pathog. 2014, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Olbjørn, C.; Cvancarova Småstuen, M.; Thiis-Evensen, E.; Nakstad, B.; Vatn, M.H.; Jahnsen, J.; Ricanek, P.; Vatn, S.; Moen, A.E.; Tannæs, T.M.; et al. Fecal Microbiota Profiles in Treatment-Naïve Pediatric Inflammatory Bowel Disease—Associations with Disease Phenotype, Treatment, and Outcome. Clin. Exp. Gastroenterol. 2019, 12, 37–49. [Google Scholar] [CrossRef]
- Rooks, M.G.; Veiga, P.; Wardwell-Scott, L.H.; Tickle, T.; Segata, N.; Michaud, M.; Gallini, C.A.; Beal, C.; van Hylckama-Vlieg, J.E.; Ballal, S.A.; et al. Gut Microbiome Composition and Function in Experimental Colitis during Active Disease and Treatment-Induced Remission. ISME J. 2014, 8, 1403–1417. [Google Scholar] [CrossRef] [PubMed]
- Rigottier-Gois, L. Dysbiosis in Inflammatory Bowel Diseases: The Oxygen Hypothesis. ISME J. 2013, 7, 1256–1261. [Google Scholar] [CrossRef]
- Rivera-Chávez, F.; Lopez, C.A.; Bäumler, A.J. Oxygen as a Driver of Gut Dysbiosis. Free Radic. Biol. Med. 2017, 105, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Hughes, E.R.; Winter, M.G.; Duerkop, B.A.; Spiga, L.; de Carvalho, T.F.; Zhu, W.; Gillis, C.C.; Büttner, L.; Smoot, M.P.; Behrendt, C.L. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe 2017, 21, 208–219. [Google Scholar] [CrossRef]
- Litvak, Y.; Byndloss, M.X.; Tsolis, R.M.; Bäumler, A.J. Dysbiotic Proteobacteria Expansion: A Microbial Signature of Epithelial Dysfunction. Curr. Opin. Microbiol. 2017, 39, 1–6. [Google Scholar] [CrossRef]
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-Derived Nitrate Boosts Growth of E. coli in the Inflamed Gut. Science 2013, 339, 708–711. [Google Scholar] [CrossRef]
- Lundberg, J.O.; Hellström, P.M.; Lundberg, J.M.; Alving, K. Greatly Increased Luminal Nitric Oxide in Ulcerative Colitis. Lancet 1994, 344, 1673–1674. [Google Scholar] [CrossRef]
- Enocksson, A.; Lundberg, J.; Weitzberg, E.; Norrby-Teglund, A.; Svenungsson, B. Rectal Nitric Oxide Gas and Stool Cytokine Levels during the Course of Infectious Gastroenteritis. Clin. Diagn. Lab. Immunol. 2004, 11, 250–254. [Google Scholar] [CrossRef]
- Byndloss, M.X.; Olsan, E.E.; Rivera-Chávez, F.; Tiffany, C.R.; Cevallos, S.A.; Lokken, K.L.; Torres, T.P.; Byndloss, A.J.; Faber, F.; Gao, Y. Microbiota-Activated PPAR-γ Signaling Inhibits Dysbiotic Enterobacteriaceae Expansion. Science 2017, 357, 570–575. [Google Scholar] [CrossRef]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.; Brummer, R. The Role of Butyrate on Colonic Function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef]
- Scales, B.S.; Dickson, R.P.; Huffnagle, G.B. A Tale of Two Sites: How Inflammation Can Reshape the Microbiomes of the Gut and Lungs. J. Leukoc. Biol. 2016, 100, 943–950. [Google Scholar] [CrossRef]
- Larsen, J.M.; Musavian, H.S.; Butt, T.M.; Ingvorsen, C.; Thysen, A.H.; Brix, S. Chronic Obstructive Pulmonary Disease and Asthma-associated Proteobacteria, but Not Commensal Prevotella Spp., Promote T Oll-like Receptor 2-independent Lung Inflammation and Pathology. Immunology 2015, 144, 333–342. [Google Scholar] [CrossRef]
- Brix, S.; Eriksen, C.; Larsen, J.M.; Bisgaard, H. Metagenomic Heterogeneity Explains Dual Immune Effects of Endotoxins. J. Allergy Clin. Immunol. 2015, 135, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Hemmi, H.; Takeuchi, O.; Kawai, T.; Kaisho, T.; Sato, S.; Sanjo, H.; Matsumoto, M.; Hoshino, K.; Wagner, H.; Takeda, K. A Toll-like Receptor Recognizes Bacterial DNA. Nature 2000, 408, 740–745. [Google Scholar] [CrossRef]
- Kant, R.; de Vos, W.M.; Palva, A.; Satokari, R. Immunostimulatory CpG Motifs in the Genomes of Gut Bacteria and Their Role in Human Health and Disease. J. Med. Microbiol. 2014, 63, 293–308. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.A.; Koren, O.; Goodrich, J.K.; Johansson, M.E.V.; Nalbantoglu, I.; Aitken, J.D.; Su, Y.; Chassaing, B.; Walters, W.A.; González, A.; et al. Transient Inability to Manage Proteobacteria Promotes Chronic Gut Inflammation in TLR5-Deficient Mice. Cell Host Microbe 2012, 12, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Maharshak, N.; Packey, C.D.; Ellermann, M.; Manick, S.; Siddle, J.P.; Huh, E.Y.; Plevy, S.; Sartor, R.B.; Carroll, I.M. Altered Enteric Microbiota Ecology in Interleukin 10-Deficient Mice during Development and Progression of Intestinal Inflammation. Gut Microbes 2013, 4, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Le Bouguénec, C.; Servin, A.L. Diffusely Adherent Escherichia coli Strains Expressing Afa/Dr Adhesins (Afa/Dr DAEC): Hitherto Unrecognized Pathogens. FEMS Microbiol. Lett. 2006, 256, 185–194. [Google Scholar] [CrossRef]
- Mirsepasi-Lauridsen, H.C.; Vallance, B.A.; Krogfelt, K.A.; Petersen, A.M. Escherichia coli Pathobionts Associated with Inflammatory Bowel Disease. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef]
- Knights, D.; Silverberg, M.S.; Weersma, R.K.; Gevers, D.; Dijkstra, G.; Huang, H.; Tyler, A.D.; van Sommeren, S.; Imhann, F.; Stempak, J.M.; et al. Complex Host Genetics Influence the Microbiome in Inflammatory Bowel Disease. Genome Med. 2014, 6. [Google Scholar] [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.-A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting Dysbiosis, Bile-Acid Dysmetabolism and Gut Inflammation in Inflammatory Bowel Diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Molinero, N.; Ruiz, L.; Sánchez, B.; Margolles, A.; Delgado, S. Intestinal Bacteria Interplay With Bile and Cholesterol Metabolism: Implications on Host Physiology. Front. Physiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Hegyi, P.; Maléth, J.; Walters, J.R.; Hofmann, A.F.; Keely, S.J. Guts and Gall: Bile Acids in Regulation of Intestinal Epithelial Function in Health and Disease. Physiol. Rev. 2018, 98, 1983–2023. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.-J.; Hylemon, P.B. Consequences of Bile Salt Biotransformations by Intestinal Bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile Salt Biotransformations by Human Intestinal Bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Alves, J.M.; Hylemon, P.B.; Bajaj, J.S. Cirrhosis, Bile Acids and Gut Microbiota: Unraveling a Complex Relationship. Gut Microbes 2013, 4, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, L.A.; Jones, B.V. Dysbiosis Modulates Capacity for Bile Acid Modification in the Gut Microbiomes of Patients with Inflammatory Bowel Disease: A Mechanism and Marker of Disease? Gut 2012, 61, 1642–1643. [Google Scholar] [CrossRef]
- Gadaleta, R.M.; Van Erpecum, K.J.; Oldenburg, B.; Willemsen, E.C.; Renooij, W.; Murzilli, S.; Klomp, L.W.; Siersema, P.D.; Schipper, M.E.; Danese, S. Farnesoid X Receptor Activation Inhibits Inflammation and Preserves the Intestinal Barrier in Inflammatory Bowel Disease. Gut 2011, 60, 463–472. [Google Scholar] [CrossRef]
- Kakiyama, G.; Pandak, W.M.; Gillevet, P.M.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; Takei, H.; Muto, A.; Nittono, H.; Ridlon, J.M.; et al. Modulation of the Fecal Bile Acid Profile by Gut Microbiota in Cirrhosis. J. Hepatol. 2013, 58, 949–955. [Google Scholar] [CrossRef]
- Vrieze, A.; Out, C.; Fuentes, S.; Jonker, L.; Reuling, I.; Kootte, R.S.; van Nood, E.; Holleman, F.; Knaapen, M.; Romijn, J.A.; et al. Impact of Oral Vancomycin on Gut Microbiota, Bile Acid Metabolism, and Insulin Sensitivity. J. Hepatol. 2014, 60, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Heinken, A.; Ravcheev, D.A.; Baldini, F.; Heirendt, L.; Fleming, R.M.T.; Thiele, I. Systematic Assessment of Secondary Bile Acid Metabolism in Gut Microbes Reveals Distinct Metabolic Capabilities in Inflammatory Bowel Disease. Microbiome 2019, 7. [Google Scholar] [CrossRef]
- Apperloo-Renkema, H.Z.; Van der Waaij, B.D.; Van der Waaij, D. Determination of Colonization Resistance of the Digestive Tract by Biotyping of Enterobacteriaceae. Epidemiol. Infect. 1990, 105, 355–361. [Google Scholar] [CrossRef]
- Burke, D.A.; Axon, A.T. Adhesive Escherichia coli in Inflammatory Bowel Disease and Infective Diarrhoea. BMJ 1988, 297, 102–104. [Google Scholar] [CrossRef]
- Giaffer, M.H.; Holdsworth, C.D.; Duerden, B.I. Virulence Properties of Escherichia coli Strains Isolated from Patients with Inflammatory Bowel Disease. Gut 1992, 33, 646–650. [Google Scholar] [CrossRef]
- Zhu, W.; Winter, M.G.; Byndloss, M.X.; Spiga, L.; Duerkop, B.A.; Hughes, E.R.; Büttner, L.; de Lima Romão, E.; Behrendt, C.L.; Lopez, C.A.; et al. Precision Editing of the Gut Microbiota Ameliorates Colitis. Nature 2018, 553, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Neut, C.; Bulois, P.; Desreumaux, P.; Membré, J.-M.; Lederman, E.; Gambiez, L.; Cortot, A.; Quandalle, P.; van Kruiningen, H.; Colombel, J.-F. Changes in the Bacterial Flora of the Neoterminal Ileum after Ileocolonic Resection for Crohn’s Disease. Am. J. Gastroenterol. 2002, 97, 939–946. [Google Scholar] [CrossRef]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.-L.; Barnich, N.; Bringer, M.-A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.-F. High Prevalence of Adherent-Invasive Escherichia coli Associated with Ileal Mucosa in Crohn’s Disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.M.; Campbell, B.J.; Hart, C.A.; Mpofu, C.; Nayar, M.; Singh, R.; Englyst, H.; Williams, H.F.; Rhodes, J.M. Enhanced Escherichia coli Adherence and Invasion in Crohn’s Disease and Colon Cancer. Gastroenterology 2004, 127, 80–93. [Google Scholar] [CrossRef]
- Baumgart, M.; Dogan, B.; Rishniw, M.; Weitzman, G.; Bosworth, B.; Yantiss, R.; Orsi, R.H.; Wiedmann, M.; McDonough, P.; Kim, S.G.; et al. Culture Independent Analysis of Ileal Mucosa Reveals a Selective Increase in Invasive Escherichia coli of Novel Phylogeny Relative to Depletion of Clostridiales in Crohn’s Disease Involving the Ileum. ISME J. 2007, 1, 403–418. [Google Scholar] [CrossRef]
- Sasaki, M.; Sitaraman, S.V.; Babbin, B.A.; Gerner-Smidt, P.; Ribot, E.M.; Garrett, N.; Alpern, J.A.; Akyildiz, A.; Theiss, A.L.; Nusrat, A.; et al. Invasive Escherichia coli Are a Feature of Crohn’s Disease. Lab. Investig. 2007, 87, 1042–1054. [Google Scholar] [CrossRef]
- Schippa, S.; Conte, M.P.; Borrelli, O.; Iebba, V.; Aleandri, M.; Seganti, L.; Longhi, C.; Chiarini, F.; Osborn, J.; Cucchiara, S. Dominant Genotypes in Mucosa-Associated Escherichia coli Strains from Pediatric Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2009, 15, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Raso, T.; Crivellaro, S.; Chirillo, M.G.; Pais, P.; Gaia, E.; Savoia, D. Analysis of Escherichia coli Isolated from Patients Affected by Crohn’s Disease. Curr. Microbiol. 2011, 63, 131–137. [Google Scholar] [CrossRef]
- Thomazini, C.M.; Samegima, D.A.G.; Rodrigues, M.A.M.; Victoria, C.R.; Rodrigues, J. High Prevalence of Aggregative Adherent Escherichia coli Strains in the Mucosa-Associated Microbiota of Patients with Inflammatory Bowel Diseases. Int. J. Med. Microbiol. 2011, 301, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Elliott, T.R.; Hudspith, B.N.; Wu, G.; Cooley, M.; Parkes, G.; Quiñones, B.; Randall, L.; Mandrell, R.E.; Fagerquist, C.K.; Brostoff, J.; et al. Quantification and Characterization of Mucosa-Associated and Intracellular Escherichia coli in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2013, 19, 2326–2338. [Google Scholar] [CrossRef]
- Conte, M.P.; Longhi, C.; Marazzato, M.; Conte, A.L.; Aleandri, M.; Lepanto, M.S.; Zagaglia, C.; Nicoletti, M.; Aloi, M.; Totino, V.; et al. Adherent-Invasive Escherichia coli (AIEC) in Pediatric Crohn’s Disease Patients: Phenotypic and Genetic Pathogenic Features. BMC Res. Notes 2014, 7, 748. [Google Scholar] [CrossRef]
- Prorok-Hamon, M.; Friswell, M.K.; Alswied, A.; Roberts, C.L.; Song, F.; Flanagan, P.K.; Knight, P.; Codling, C.; Marchesi, J.R.; Winstanley, C.; et al. Colonic Mucosa-Associated Diffusely Adherent AfaC+ Escherichia coli Expressing LpfA and Pks Are Increased in Inflammatory Bowel Disease and Colon Cancer. Gut 2014, 63, 761–770. [Google Scholar] [CrossRef]
- Rolhion, N.; Darfeuille-Michaud, A. Adherent-Invasive Escherichia coli in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2007, 13, 1277–1283. [Google Scholar] [CrossRef]
- Palmela, C.; Chevarin, C.; Xu, Z.; Torres, J.; Sevrin, G.; Hirten, R.; Barnich, N.; Ng, S.C.; Colombel, J.-F. Adherent-Invasive Escherichia coli in Inflammatory Bowel Disease. Gut 2018, 67, 574–587. [Google Scholar] [CrossRef] [PubMed]
- Miquel, S.; Peyretaillade, E.; Claret, L.; de Vallée, A.; Dossat, C.; Vacherie, B.; Zineb, E.H.; Segurens, B.; Barbe, V.; Sauvanet, P.; et al. Complete Genome Sequence of Crohn’s Disease-Associated Adherent-Invasive E. coli Strain LF82. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Nash, J.H.; Villegas, A.; Kropinski, A.M.; Aguilar-Valenzuela, R.; Konczy, P.; Mascarenhas, M.; Ziebell, K.; Torres, A.G.; Karmali, M.A.; Coombes, B.K. Genome Sequence of Adherent-Invasive Escherichia coli and Comparative Genomic Analysis with Other E. coli Pathotypes. BMC Genom. 2010, 11, 667. [Google Scholar] [CrossRef] [PubMed]
- Schippa, S.; Iebba, V.; Totino, V.; Santangelo, F.; Lepanto, M.; Alessandri, C.; Nuti, F.; Viola, F.; Di Nardo, G.; Cucchiara, S.; et al. A Potential Role of Escherichia coli Pathobionts in the Pathogenesis of Pediatric Inflammatory Bowel Disease. Can. J. Microbiol. 2012, 58, 426–432. [Google Scholar] [CrossRef]
- Barnich, N.; Carvalho, F.A.; Glasser, A.-L.; Darcha, C.; Jantscheff, P.; Allez, M.; Peeters, H.; Bommelaer, G.; Desreumaux, P.; Colombel, J.-F.; et al. CEACAM6 Acts as a Receptor for Adherent-Invasive E. coli, Supporting Ileal Mucosa Colonization in Crohn Disease. J. Clin. Investig. 2007, 117, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Boudeau, J.; Barnich, N.; Darfeuille-Michaud, A. Type 1 Pili-Mediated Adherence of Escherichia coli Strain LF82 Isolated from Crohn’s Disease Is Involved in Bacterial Invasion of Intestinal Epithelial Cells. Mol. Microbiol. 2001, 39, 1272–1284. [Google Scholar] [CrossRef]
- Rolhion, N.; Carvalho, F.A.; Darfeuille-Michaud, A. OmpC and the Sigma(E) Regulatory Pathway Are Involved in Adhesion and Invasion of the Crohn’s Disease-Associated Escherichia coli Strain LF82. Mol. Microbiol. 2007, 63, 1684–1700. [Google Scholar] [CrossRef] [PubMed]
- Nadalian, B.; Yadegar, A.; Houri, H.; Olfatifar, M.; Shahrokh, S.; Asadzadeh Aghdaei, H.; Suzuki, H.; Zali, M.R. Prevalence of the Pathobiont Adherent-Invasive Escherichia coli and Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. J. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Drouet, M.; Vignal, C.; Singer, E.; Djouina, M.; Dubreuil, L.; Cortot, A.; Desreumaux, P.; Neut, C. AIEC Colonization and Pathogenicity: Influence of Previous Antibiotic Treatment and Preexisting Inflammation. Inflamm. Bowel Dis. 2012, 18, 1923–1931. [Google Scholar] [CrossRef]
- Small, C.-L.N.; Reid-Yu, S.A.; McPhee, J.B.; Coombes, B.K. Persistent Infection with Crohn’s Disease-Associated Adherent-Invasive Escherichia coli Leads to Chronic Inflammation and Intestinal Fibrosis. Nat. Commun. 2013, 4, 1957. [Google Scholar] [CrossRef]
- Chassaing, B.; Koren, O.; Carvalho, F.A.; Ley, R.E.; Gewirtz, A.T. AIEC Pathobiont Instigates Chronic Colitis in Susceptible Hosts by Altering Microbiota Composition. Gut 2014, 63, 1069–1080. [Google Scholar] [CrossRef]
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-Based Phylogeny and Taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales Ord. Nov. Divided into the Families Enterobacteriaceae, Erwiniaceae Fam. Nov., Pectobacteriaceae Fam. Nov., Yersiniaceae Fam. Nov., Hafniaceae Fam. Nov., Morganellaceae Fam. Nov., and Budviciaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef]
- Alnajar, S.; Gupta, R.S. Phylogenomics and Comparative Genomic Studies Delineate Six Main Clades within the Family Enterobacteriaceae and Support the Reclassification of Several Polyphyletic Members of the Family. Infect. Genet. Evol. 2017, 54, 108–127. [Google Scholar] [CrossRef] [PubMed]
- Hong Nhung, P.; Ohkusu, K.; Mishima, N.; Noda, M.; Monir Shah, M.; Sun, X.; Hayashi, M.; Ezaki, T. Phylogeny and Species Identification of the Family Enterobacteriaceae Based on DnaJ Sequences. Diagn. Microbiol. Infect. Dis. 2007, 58, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S RRNA Gene Sequencing for Bacterial Identification in the Diagnostic Laboratory: Pluses, Perils, and Pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Saputra, D.; Rasmussen, S.; Larsen, M.V.; Haddad, N.; Sperotto, M.M.; Aarestrup, F.M.; Lund, O.; Sicheritz-Pontén, T. Reads2Type: A Web Application for Rapid Microbial Taxonomy Identification. BMC Bioinform. 2015, 16. [Google Scholar] [CrossRef]
- Mollet, C.; Drancourt, M.; Raoult, D. RpoB Sequence Analysis as a Novel Basis for Bacterial Identification. Mol. Microbiol. 1997, 26, 1005–1011. [Google Scholar] [CrossRef]
- Hedegaard, J.; de Steffensen, S.A.A.; Nørskov-Lauritsen, N.; Mortensen, K.K.; Sperling-Petersen, H.U. Identification of Enterobacteriaceae by Partial Sequencing of the Gene Encoding Translation Initiation Factor 2. Int. J. Syst. Evol. Microbiol. 1999, 49, 1531–1538. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dauga, C. Evolution of the GyrB Gene and the Molecular Phylogeny of Enterobacteriaceae: A Model Molecule for Molecular Systematic Studies. Int. J. Syst Evol. Microbiol. 2002, 52, 531–547. [Google Scholar] [CrossRef] [PubMed]
- Paradis, S.; Boissinot, M.; Paquette, N.; Bélanger, S.D.; Martel, E.A.; Boudreau, D.K.; Picard, F.J.; Ouellette, M.; Roy, P.H.; Bergeron, M.G. Phylogeny of the Enterobacteriaceae Based on Genes Encoding Elongation Factor Tu and F-ATPase β-Subunit. Int. J. Syst. Evol. Microbiol. 2005, 55, 2013–2025. [Google Scholar] [CrossRef] [PubMed]
- McLean, K.; Rosenthal, C.A.; Sengupta, D.; Owens, J.; Cookson, B.T.; Hoffman, N.G.; Salipante, S.J. Improved Species-Level Clinical Identification of Enterobacteriaceae through Broad-Range dnaJ PCR and Sequencing. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Ogier, J.-C.; Pagès, S.; Galan, M.; Barret, M.; Gaudriault, S. RpoB, a Promising Marker for Analyzing the Diversity of Bacterial Communities by Amplicon Sequencing. BMC Microbiol. 2019, 19. [Google Scholar] [CrossRef]
- Delmas, J.; Breysse, F.; Devulder, G.; Flandrois, J.-P.; Chomarat, M. Rapid Identification of Enterobacteriaceae by Sequencing DNA Gyrase Subunit B Encoding Gene. Diagn. Microbiol. Infect. Dis. 2006, 55, 263–268. [Google Scholar] [CrossRef]
- Lindgreen, S.; Adair, K.L.; Gardner, P.P. An Evaluation of the Accuracy and Speed of Metagenome Analysis Tools. Sci. Rep. 2016, 6, 19233. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, A.B.; Ounit, R.; Afshinnekoo, E.; Prill, R.J.; Hénaff, E.; Alexander, N.; Minot, S.S.; Danko, D.; Foox, J.; Ahsanuddin, S. Comprehensive Benchmarking and Ensemble Approaches for Metagenomic Classifiers. Genome Biol. 2017, 18, 182. [Google Scholar] [CrossRef]
- Leo, S.; Lazarevic, V.; Girard, M.; Gaïa, N.; Schrenzel, J.; de Lastours, V.; Fantin, B.; Bonten, M.; Carmeli, Y.; Rondinaud, E.; et al. Metagenomic Characterization of Gut Microbiota of Carriers of Extended-Spectrum Beta-Lactamase or Carbapenemase-Producing Enterobacteriaceae Following Treatment with Oral Antibiotics and Fecal Microbiota Transplantation: Results from a Multicenter Randomized Trial. Microorganisms 2020, 8, 941. [Google Scholar] [CrossRef]
- Vincent, A.T.; Derome, N.; Boyle, B.; Culley, A.I.; Charette, S.J. Next-Generation Sequencing (NGS) in the Microbiological World: How to Make the Most of Your Money. J. Microbiol. Methods 2017, 138, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Clooney, A.G.; Fouhy, F.; Sleator, R.D.; O’Driscoll, A.; Stanton, C.; Cotter, P.D.; Claesson, M.J. Comparing Apples and Oranges?: Next Generation Sequencing and Its Impact on Microbiome Analysis. PLoS ONE 2016, 11, e0148028. [Google Scholar] [CrossRef] [PubMed]
- Tessler, M.; Neumann, J.S.; Afshinnekoo, E.; Pineda, M.; Hersch, R.; Velho, L.F.M.; Segovia, B.T.; Lansac-Toha, F.A.; Lemke, M.; DeSalle, R. Large-Scale Differences in Microbial Biodiversity Discovery between 16S Amplicon and Shotgun Sequencing. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).