Mechanisms of Transmission and Adaptation of tet(X4)-Positive IncHI1 Plasmids in XDR Escherichia coli from Pet Dogs: The Role of trhC, rsp, and the Tra1 Region
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Antimicrobial Susceptibility Testing
2.3. Conjugation and S1-PFGE
2.4. WGS and Analysis
2.5. Conjugation Frequencies
2.6. Plasmid Stability
2.7. Nucleotide Sequence Accession Numbers
3. Results
3.1. Characterization of tet(X4)-Positive E. coli Strains T28R and T16R
3.2. Analysis of Plasmids in T28R and T16R
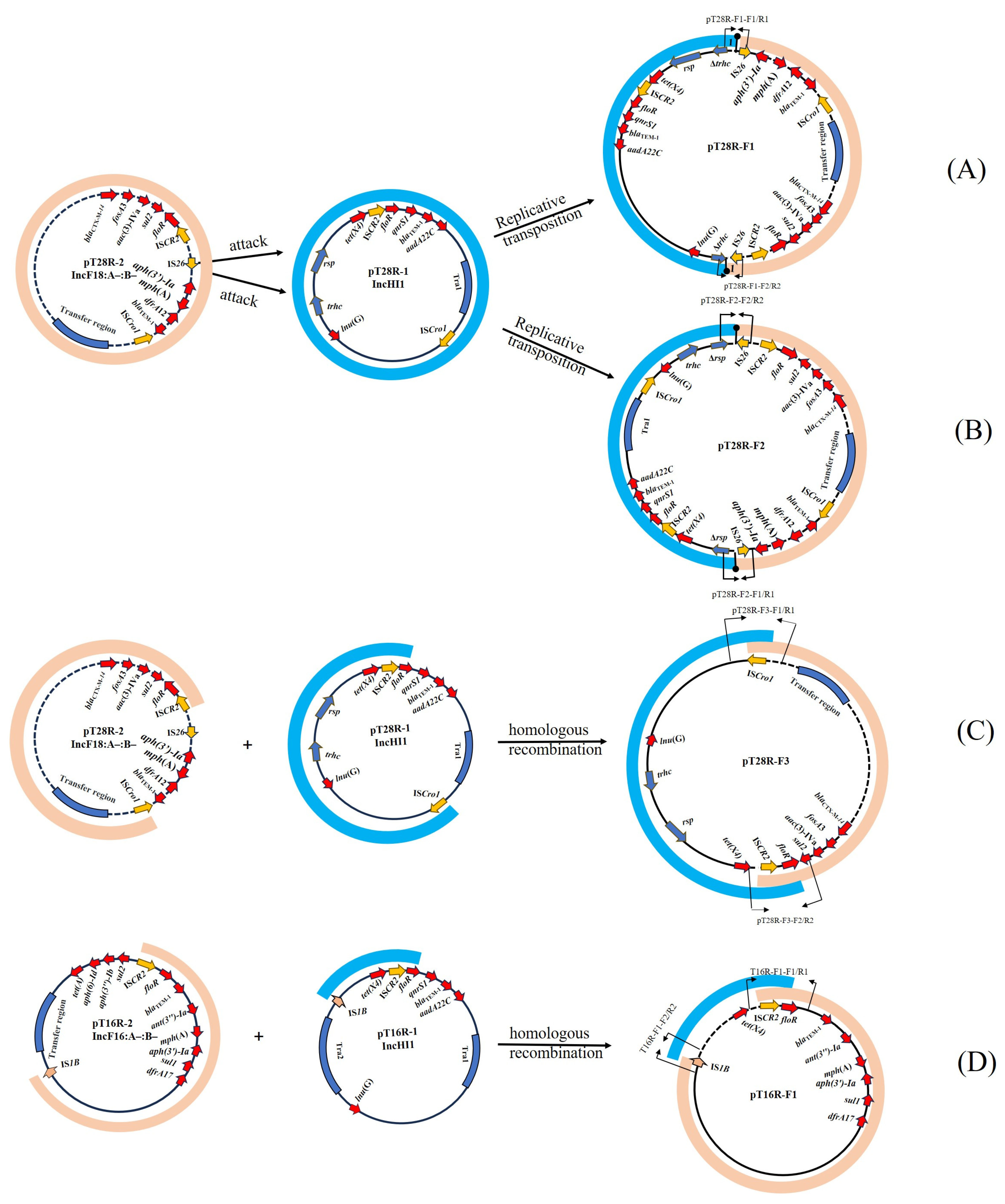
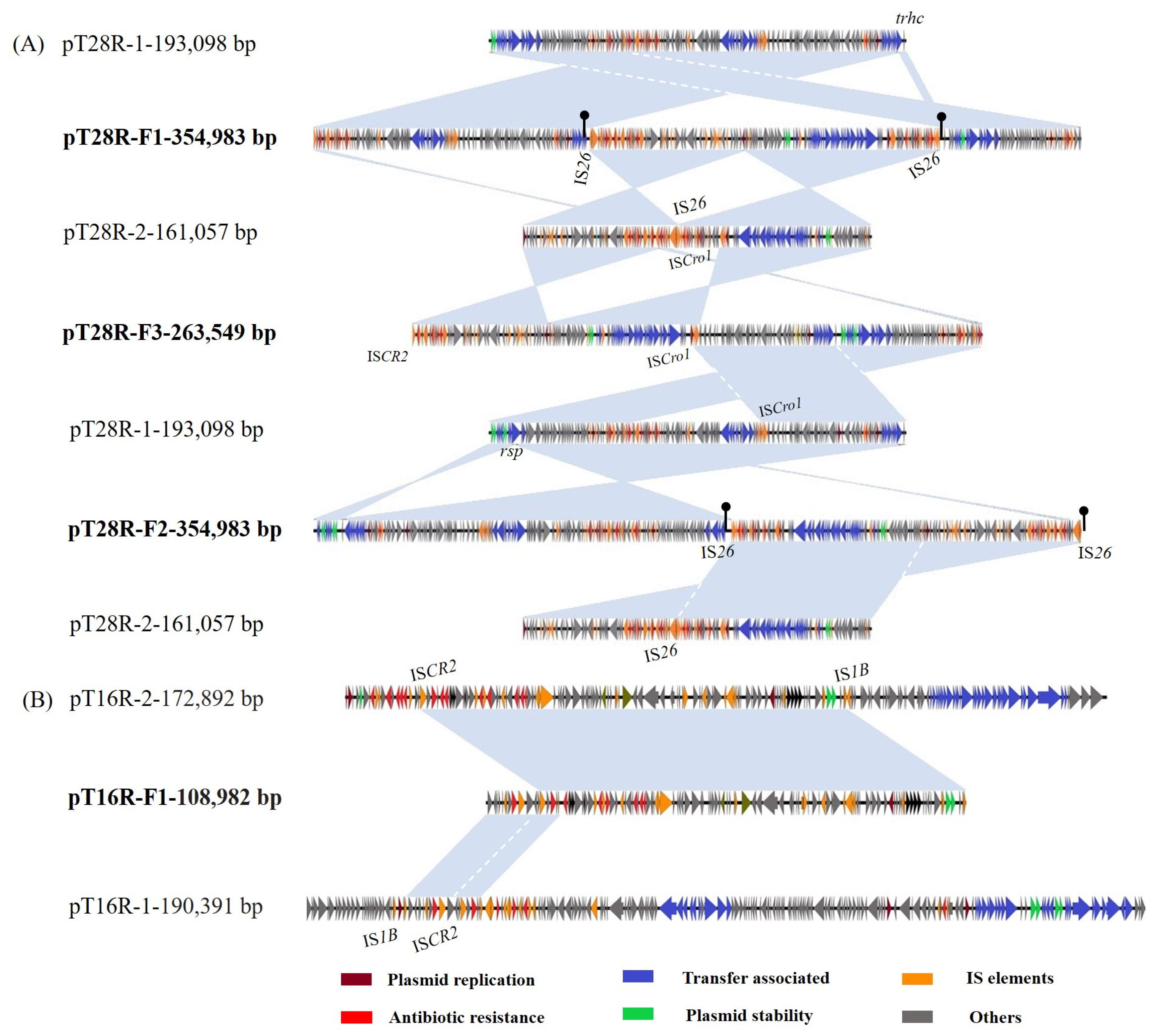
3.3. Multimodal Transmission of tet(X4) in a IncHI1 Plasmid Mediated by IS Elements
3.4. Biological Features of Fusion Plasmids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Peng, K.; Li, Y.; Liu, Y.; Wang, Z. Exploring tet(X)-bearing tigecycline-resistant bacteria of swine farming environments. Sci. Total Environ. 2020, 733, 139306. [Google Scholar] [CrossRef]
- Dai, S.; Liu, D.; Han, Z.; Wang, Y.; Lu, X.; Yang, M.; Zhang, Y. Mobile tigecycline resistance gene tet(X4) persists with different animal manure composting treatments and fertilizer receiving soils. Chemosphere 2022, 307, 135866. [Google Scholar] [CrossRef]
- Sun, C.; Cui, M.; Zhang, S.; Wang, H.; Song, L.; Zhang, C.; Zhao, Q.; Liu, D.; Wang, Y.; Shen, J.; et al. Plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from food-producing animals, China, 2008–2018. Emerg. Microbes Infect. 2019, 8, 1524–1527. [Google Scholar] [CrossRef]
- Feng, J.; Su, M.; Li, K.; Ma, J.; Li, R.; Bai, L.; Wang, X.; Wang, J.; Yang, Z. Extensive spread of tet(X4) in multidrug-resistant Escherichia coli of animal origin in western China. Veter- Microbiol. 2022, 269, 109420. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; He, R.; Qin, M.; Yang, Y.; Chen, J.; Feng, Y.; Liang, X.; Deng, W.; Ding, X.; Qin, L.-N.; et al. Identification of plasmid-mediated tigecycline-resistant gene tet(X4) in Enterobacter cloacae from pigs in China. Microbiol. Spectr. 2022, 10, e0206421. [Google Scholar] [CrossRef]
- Ma, J.; Wang, J.; Yang, H.; Su, M.; Li, R.; Bai, L.; Feng, J.; Huang, Y.; Yang, Z.; Tang, B. IncHI1 plasmids mediated the tet(X4) gene spread in Enterobacteriaceae in porcine. Front. Microbiol. 2023, 14, 1128905. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Cai, P.; Lu, Y.; Sun, R.-Y.; Cao, M.-T.; Xu, X.-L.; Webber, M.A.; Jiang, H.-X. IncHI1 plasmids are epidemic vectors that mediate transmission of tet(X4) in Escherichia coli isolated from China. Front. Microbiol. 2023, 14, 1153139. [Google Scholar] [CrossRef]
- Yu, Y.; Cui, C.-Y.; Kuang, X.; Chen, C.; Wang, M.-G.; Liao, X.-P.; Sun, J.; Liu, Y.-H. Prevalence of tet(X4) in Escherichia coli From Duck Farms in Southeast China. Front. Microbiol. 2021, 12, 716393. [Google Scholar] [CrossRef]
- Couturier, A.; Virolle, C.; Goldlust, K.; Berne-Dedieu, A.; Reuter, A.; Nolivos, S.; Yamaichi, Y.; Bigot, S.; Lesterlin, C. Real-time visualisation of the intracellular dynamics of conjugative plasmid transfer. Nat. Commun. 2023, 14, 294. [Google Scholar] [CrossRef]
- Wang, Q.; Lei, C.; Cheng, H.; Yang, X.; Huang, Z.; Chen, X.; Ju, Z.; Zhang, H.; Wang, H. Widespread dissemination of plasmid-mediated tigecycline resistance gene tet(X4) in Enterobacterales of porcine origin. Microbiol. Spectr. 2022, 10, e0161522. [Google Scholar] [CrossRef] [PubMed]
- Allain, M.; Mahérault, A.C.; Gachet, B.; Martinez, C.; Condamine, B.; Magnan, M.; Kempf, I.; Denamur, E.; Landraud, L. Dissemination of IncI plasmid encoding blaCTX-M-1 is not hampered by its fitness cost in the pig’s gut. Antimicrob. Agents Chemother. 2023, 67, e0011123. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Dong, N.; Chan, E.W.-C.; Chen, S. Transmission of ciprofloxacin resistance in Salmonella mediated by a novel type of conjugative helper plasmids. Emerg. Microbes Infect. 2019, 8, 857–865. [Google Scholar] [CrossRef]
- He, D.; Zhu, Y.; Li, R.; Pan, Y.; Liu, J.; Yuan, L.; Hu, G. Emergence of a hybrid plasmid derived from IncN1-F33:A−:B− and mcr-1-bearing plasmids mediated by IS26. J. Antimicrob. Chemother. 2019, 74, 3184–3189. [Google Scholar] [CrossRef]
- Wang, X.; Tang, B.; Liu, G.; Wang, M.; Sun, J.; Tan, R.; Pan, T.; Qu, J.; Liu, J.; Ou, H.-Y.; et al. Transmission of nonconjugative virulence or resistance plasmids mediated by a self-transferable IncN3 plasmid from carbapenem-resistant Klebsiella pneumoniae. Microbiol. Spectr. 2022, 10, e0136422. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Xie, M.; Chan, E.W.-C.; Chen, S. Delineation of ISEcp1 and IS26-mediated plasmid fusion processes by MinION single-molecule long-read sequencing. Front. Microbiol. 2022, 12, 796715. [Google Scholar] [CrossRef]
- Shan, X.; Yang, M.; Wang, N.; Schwarz, S.; Li, D.; Du, X.-D. Plasmid fusion and recombination events that occurred during conjugation of poxtA-Carrying plasmids in Enterococci. Microbiol. Spectr. 2022, 10, e0150521. [Google Scholar] [CrossRef]
- Li, R.; Lu, X.; Peng, K.; Liu, Y.; Xiao, X.; Wang, Z. Reorganization of mcr-1-bearing large MDR plasmids resolved by nanopore sequencing. J. Antimicrob. Chemother. 2020, 75, 1645–1647. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; He, D.-D.; Zhang, M.-K.; Pan, Y.-S.; Wu, H.; Yuan, L.; Liu, J.-H.; Hu, G.-Z. The formation of two Hybrid plasmids mediated by IS26 and Tn6952 in Salmonella enterica serotype enteritidis. Front. Microbiol. 2021, 12, 676574. [Google Scholar] [CrossRef]
- Gu, Y.; Lü, Z.; Cao, C.; Sheng, H.; Li, W.; Cui, S.; Li, R.; Lü, X.; Yang, B. Cunning plasmid fusion mediates antibiotic resistance genes represented by ESBLs encoding genes transfer in foodborne Salmonella. Int. J. Food Microbiol. 2021, 355, 109336. [Google Scholar] [CrossRef]
- Gibert, M.; Juárez, A.; Zechner, E.L.; Madrid, C.; Balsalobre, C. TrhR, TrhY and HtdA, a novel regulatory circuit that modulates conjugation of the IncHI plasmids. Mol. Microbiol. 2014, 94, 1146–1161. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.E.; Brose, E.C. Characterization of incompatibility group HI1 plasmids from Salmonella typhi by restriction endonuclease digestion and hybridization of DNA probes for Tn3, Tn9, and Tn10. Can. J. Microbiol. 1985, 31, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Sherburne, C.K.; Lawley, T.D.; Gilmour, M.W.; Blattner, F.R.; Burland, V.; Grotbeck, E.; Rose, D.J.; Taylor, D.E. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 2000, 28, 2177–2186. [Google Scholar] [CrossRef]
- Gunton, J.E.; Gilmour, M.W.; Alonso, G.; Taylor, D.E. Subcellular localization and functional domains of the coupling protein, TraG, from IncHI1 plasmid R27. Microbiology 2005, 151, 3549–3561. [Google Scholar] [CrossRef] [PubMed]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, e0021321. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Li, R.; Xie, M.; Dong, N.; Lin, D.; Yang, X.; Wong, M.H.Y.; Chan, E.W.-C.; Chen, S. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. GigaScience 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfigure: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Chen, C.; Wu, X.-T.; He, Q.; Chen, L.; Cui, C.-Y.; Zhang, Y.; Chen, S.-H.; Liao, X.-P.; Liu, Y.-H.; Sun, J. Complete sequence of a tet(X4)-harboring IncX1 plasmid, pYY76-1-2, in Escherichia coli from a cattle sample in China. Antimicrob. Agents Chemother. 2019, 63, e01528-19. [Google Scholar] [CrossRef]
- Wise, M.G.; Estabrook, M.A.; Sahm, D.F.; Stone, G.G.; Kazmierczak, K.M. Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014-2016 as part of the INFORM global surveillance program. PLoS ONE 2018, 13, e0195281. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Walsh, T.R.; Liu, D.; Shen, Z.; Zhang, R.; Yin, W.; Yao, H.; Li, J.; Shen, J. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob. Agents Chemother. 2017, 61, e02233-16. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Liu, Z.; Li, J.; Yin, W.; Lei, L.; Wu, C.; Shen, J. Characterization of NDM-1-producing carbapenemase in Acinetobacter spp. and E. coli isolates from diseased pigs. Front. Agric. Sci. Eng. 2015, 2, 223–229. [Google Scholar] [CrossRef]
- He, D.; Liu, L.; Guo, B.; Wu, S.; Chen, X.; Wang, J.; Zeng, Z.; Liu, J.-H. Chromosomal location of the fosA3 and blaCTX-M genes in Proteus mirabilis and clonal spread of Escherichia coli ST117 carrying fosA3-positive IncHI2/ST3 or F2:A-:B- plasmids in a chicken farm. Int. J. Antimicrob. Agents 2017, 49, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Chen, S. First detection of conjugative plasmid-borne fosfomycin resistance gene fosA3 in Salmonella isolates of food origin. Antimicrob. Agents Chemother. 2015, 59, 1381–1383. [Google Scholar] [CrossRef]
- He, K.; Li, W.; Zhao, B.; Xu, H.; Pan, Y.; He, D.; Hu, G.; Wu, H.; Yuan, L. Spreading Advantages of Coresident Plasmids blaCTX-M-Bearing IncFII and mcr-1-Bearing IncI2 in Escherichia coli. Microbiol. Spectr. 2022, 10, e0170621. [Google Scholar] [CrossRef]
- Liu, Y.; Qiao, Z.; Ma, Y.; Wang, M.; Hu, G.; Li, E. Molecular characterization of the tet(M)-carrying transposon Tn7124 and plasmids in Escherichia coli isolates recovered from swine. Front. Veter-Sci. 2024, 11, 1430398. [Google Scholar] [CrossRef]
- Yang, X.; Dong, N.; Chan, E.W.-C.; Zhang, R.; Chen, S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 2021, 29, 65–83. [Google Scholar] [CrossRef]
- Heaton, M.P.; Discotto, L.F.; Pucci, M.J.; Handwerger, S. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-response plasmid. Gene 1996, 171, 9–17. [Google Scholar] [CrossRef]
- Carraro, N.; Matteau, D.; Luo, P.; Rodrigue, S.; Burrus, V. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 2014, 10, e1004714. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, J.; Wang, M.; Liu, M.; Liu, G.; Qu, H.; Liu, J.; Deng, Z.; Sun, J.; Ou, H.-Y.; et al. Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 2021, 13, 119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Wu, C.; Zhou, P.; Zhang, J.; Xiong, Z.; Zhou, Y.; Yu, F. Characterization of hypervirulent and carbapenem-resistant K. pneumoniae isolated from neurological patients. Infect. Drug Resist. 2023, 16, 403–411. [Google Scholar] [CrossRef]
- Partridge, S.R.; Zong, Z.; Iredell, J.R. Recombination in IS26 and Tn2 in the evolution of multiresistance regions carrying blaCTX-M-15 on conjugative IncF plasmids from Escherichia coli. Antimicrob. Agents Chemother. 2011, 55, 4971–4978. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.-M.; Lopez-Cerero, L.; García-Duque, A.; Rodriguez-Baño, J.; Pascual, A. Interplay between IncF plasmids and topoisomerase mutations conferring quinolone resistance in the Escherichia coli ST131 clone: Stability and resistance evolution. Eur. J. Clin. Microbiol. Infect. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Chen, L. The significance of epidemic plasmids in the success of multidrug-resistant drug pandemic extraintestinal pathogenic Escherichia coli. Infect. Dis. Ther. 2023, 12, 1029–1041. [Google Scholar] [CrossRef]
- Han, C.-G.; Shiga, Y.; Tobe, T.; Sasakawa, C.; Ohtsubo, E. Structural and functional characterization of IS679 and IS66-family elements. J. Bacteriol. 2001, 183, 4296–4304. [Google Scholar] [CrossRef] [PubMed]
- Petty, N.K.; Bulgin, R.; Crepin, V.F.; Cerdeño-Tárraga, A.M.; Schroeder, G.N.; Quail, M.A.; Lennard, N.; Corton, C.; Barron, A.; Clark, L.; et al. The citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J. Bacteriol. 2009, 192, 525–538. [Google Scholar] [CrossRef]
- Alvarez-Fraga, L.; Phan, M.-D.; Goh, K.G.K.; Nhu, N.T.K.; Hancock, S.J.; Allsopp, L.P.; Peters, K.M.; Forde, B.M.; Roberts, L.W.; Sullivan, M.J.; et al. Differential Afa/Dr fimbriae expression in the multidrug-resistant Escherichia coli ST131 Clone. mBio 2022, 13, e0351921. [Google Scholar] [CrossRef]
- Lawley, T.D.; Gilmour, M.W.; Gunton, J.E.; Standeven, L.J.; Taylor, D.E. Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J. Bacteriol. 2002, 184, 2173–2180. [Google Scholar] [CrossRef]
- Rooker, M.M.; Sherburne, C.; Lawley, T.D.; Taylor, D.E. Characterization of the Tra2 region of the IncHI1 plasmid R27. Plasmid 1999, 41, 226–239. [Google Scholar] [CrossRef]
- Lawley, T.D.; Gilmour, M.W.; Gunton, J.E.; Tracz, D.M.; Taylor, D.E. Functional and mutational analysis of conjugative transfer region 2 (Tra2) from the IncHI1 plasmid R27. J. Bacteriol. 2003, 185, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, M.W.; Taylor, D.E. A Subassembly of R27-encoded transfer proteins is dependent on TrhC nucleoside triphosphate-binding motifs for function but not formation. J. Bacteriol. 2004, 186, 1606–1613. [Google Scholar] [CrossRef] [PubMed]
- Hüttener, M.; Prieto, A.; Aznar, S.; Bernabeu, M.; Glaría, E.; Valledor, A.F.; Paytubi, S.; Merino, S.; Tomás, J.; Juárez, A. Expression of a novel class of bacterial Ig-like proteins is required for IncHI plasmid conjugation. PLoS Genet. 2019, 15, e1008399. [Google Scholar] [CrossRef] [PubMed]
- Millan, A.S.; Heilbron, K.; MacLean, R.C. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J. 2014, 8, 601–612. [Google Scholar] [CrossRef]
- Pinto, U.M.; Pappas, K.M.; Winans, S.C. The ABCs of plasmid replication and segregation. Nat. Rev. Microbiol. 2012, 10, 755–765. [Google Scholar] [CrossRef]
| Plasmid | 28 °C | 37 °C | ||||
|---|---|---|---|---|---|---|
| Mean | No. | Range | Mean | No. | Range | |
| pT28R-1 | 2.28 × 10−3 | 3 | 4.37 × 10−4~4.2 × 10−3 | 2.7 × 10−7 | 3 | 4.4 × 10−7~5.2 × 10−8 |
| pT28R-2 | 9.5 × 10−3 | 3 | 2.23 × 10−2~5.15 × 10−3 | 1.6 × 10−2 | 3 | 1.1 × 10−2~4.47 × 10−3 |
| pT28R-F1 | 3.65 × 10−5 | 3 | 1.2 × 10−5~5.35 × 10−5 | 3.84 × 10−5 | 3 | 2.13 × 10−5~5.3 × 10−5 |
| pT28R-F2 | 2.45 × 10−5 | 3 | 9.2 × 10−6~4.32 × 10−5 | 3.62 × 10−5 | 3 | 4.1 × 10−6~5.2 × 10−5 |
| pT28R-F3 | 1.57 × 10−4 | 3 | 6.23 × 10−4~2.7 × 10−3 | 3.14 × 10−4 | 3 | 2.1 × 10−4~7.3 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, P.; Wang, Q.; Wang, L.; Zheng, M.; Feng, Y.; Xu, Y.; Yuan, L.; Hu, G.; Pan, Y.; He, D. Mechanisms of Transmission and Adaptation of tet(X4)-Positive IncHI1 Plasmids in XDR Escherichia coli from Pet Dogs: The Role of trhC, rsp, and the Tra1 Region. Vet. Sci. 2025, 12, 418. https://doi.org/10.3390/vetsci12050418
Ding P, Wang Q, Wang L, Zheng M, Feng Y, Xu Y, Yuan L, Hu G, Pan Y, He D. Mechanisms of Transmission and Adaptation of tet(X4)-Positive IncHI1 Plasmids in XDR Escherichia coli from Pet Dogs: The Role of trhC, rsp, and the Tra1 Region. Veterinary Sciences. 2025; 12(5):418. https://doi.org/10.3390/vetsci12050418
Chicago/Turabian StyleDing, Pengyun, Qianqian Wang, Liangliang Wang, Mengxiang Zheng, Yiming Feng, Yakun Xu, Li Yuan, Gongzheng Hu, Yushan Pan, and Dandan He. 2025. "Mechanisms of Transmission and Adaptation of tet(X4)-Positive IncHI1 Plasmids in XDR Escherichia coli from Pet Dogs: The Role of trhC, rsp, and the Tra1 Region" Veterinary Sciences 12, no. 5: 418. https://doi.org/10.3390/vetsci12050418
APA StyleDing, P., Wang, Q., Wang, L., Zheng, M., Feng, Y., Xu, Y., Yuan, L., Hu, G., Pan, Y., & He, D. (2025). Mechanisms of Transmission and Adaptation of tet(X4)-Positive IncHI1 Plasmids in XDR Escherichia coli from Pet Dogs: The Role of trhC, rsp, and the Tra1 Region. Veterinary Sciences, 12(5), 418. https://doi.org/10.3390/vetsci12050418





