Metagenomic Analysis Reveals the Effects of Microplastics on Antibiotic Resistance Genes in Sludge Anaerobic Digestion
Abstract
1. Introduction
2. Material and Methods
2.1. Anaerobic Sludge Digestion Experiments
2.2. DNA Extraction
2.3. Metagenomic Sequencing
2.4. Bioinformatic Analysis
2.4.1. Annotation of ARGs
2.4.2. Identification of MGEs
2.4.3. Taxonomic Classification and Functional Annotation
2.4.4. Statistical Analysis and Visualization
3. Results and Discussion
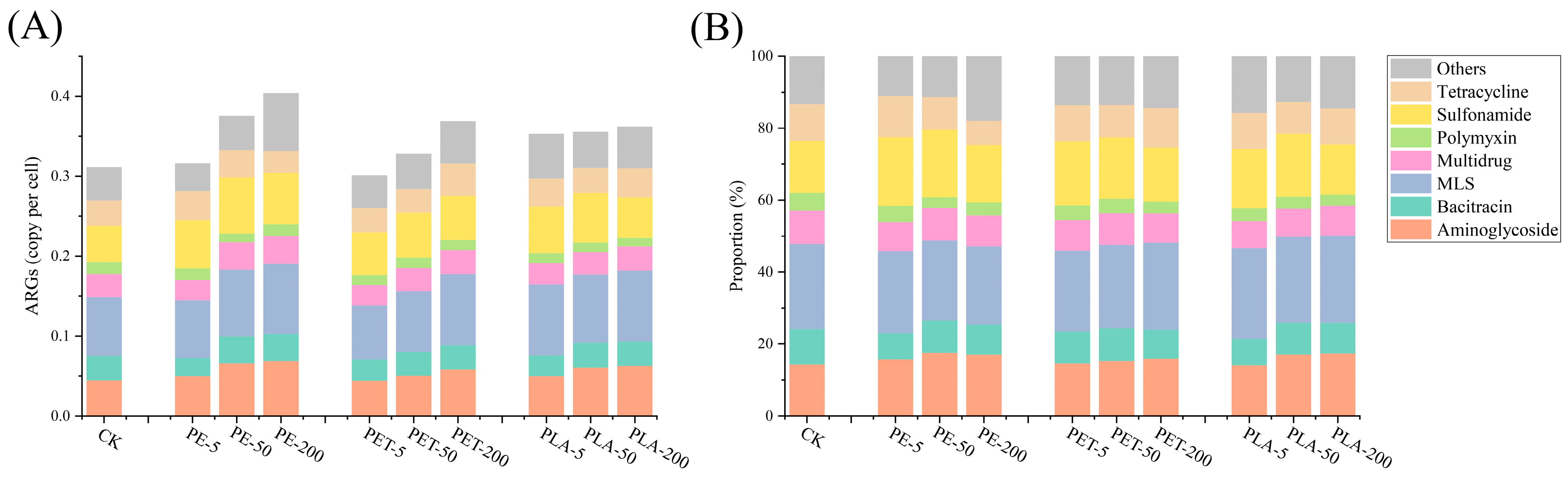
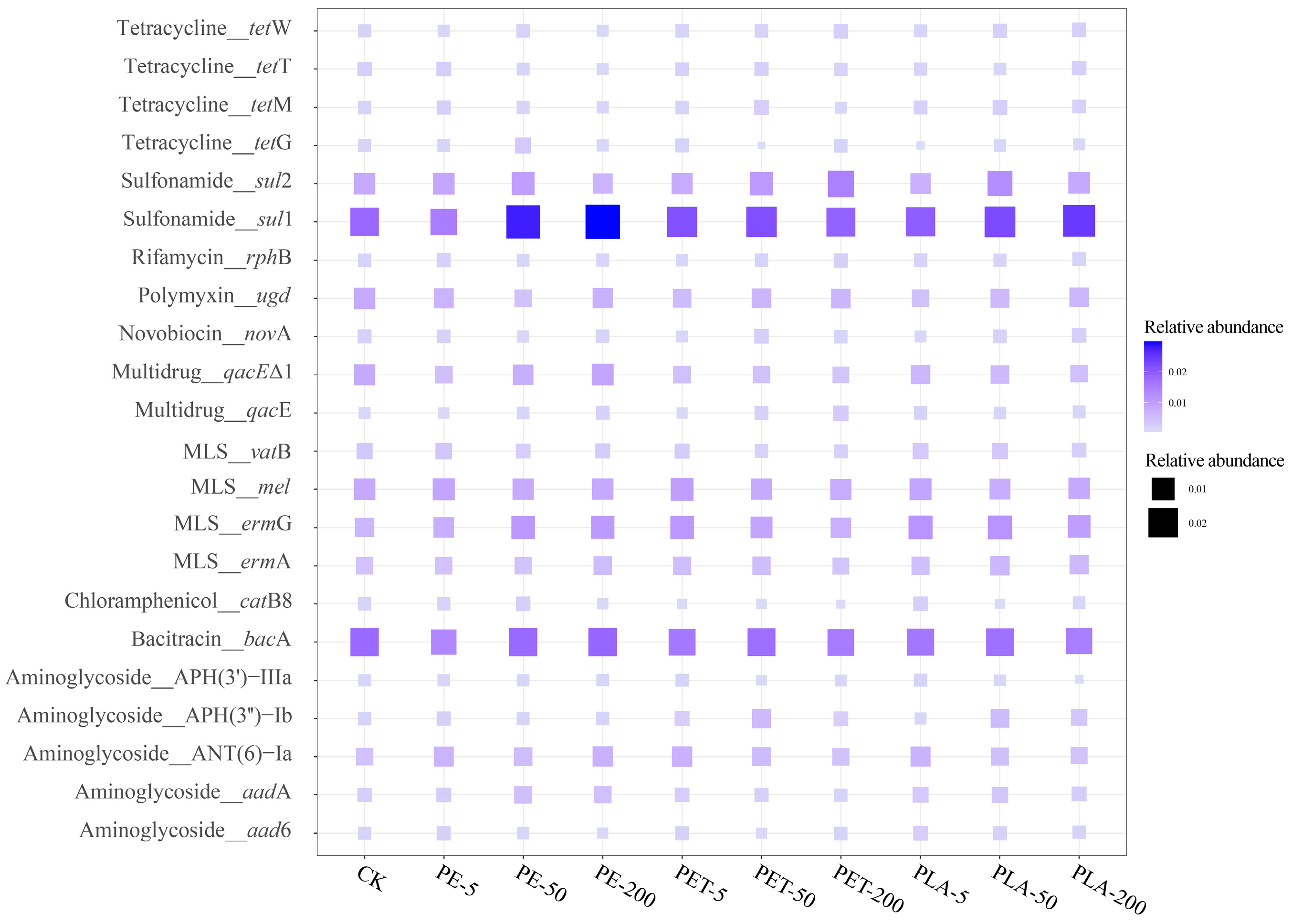
3.1. The Effects of MPs on ARG Abundances
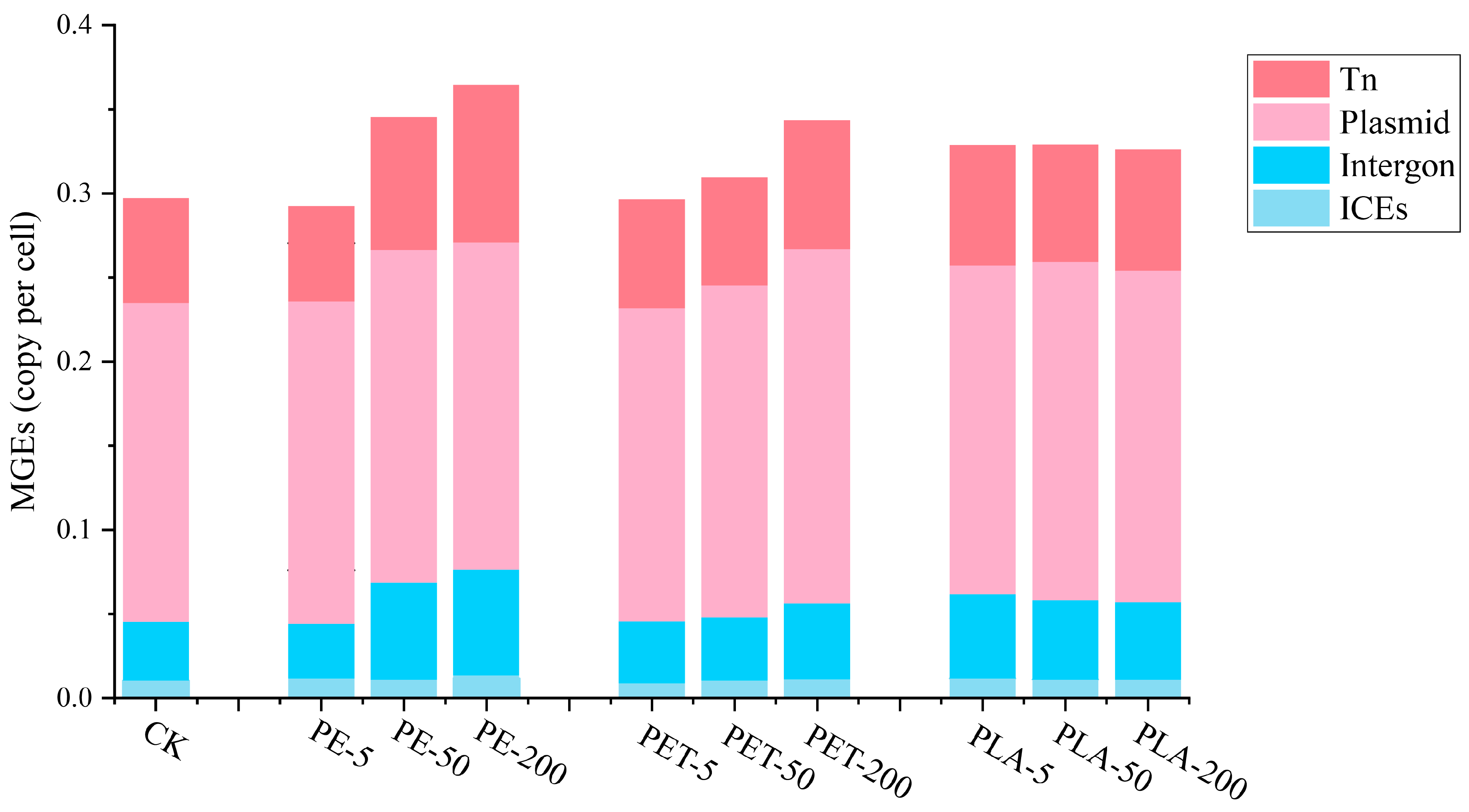
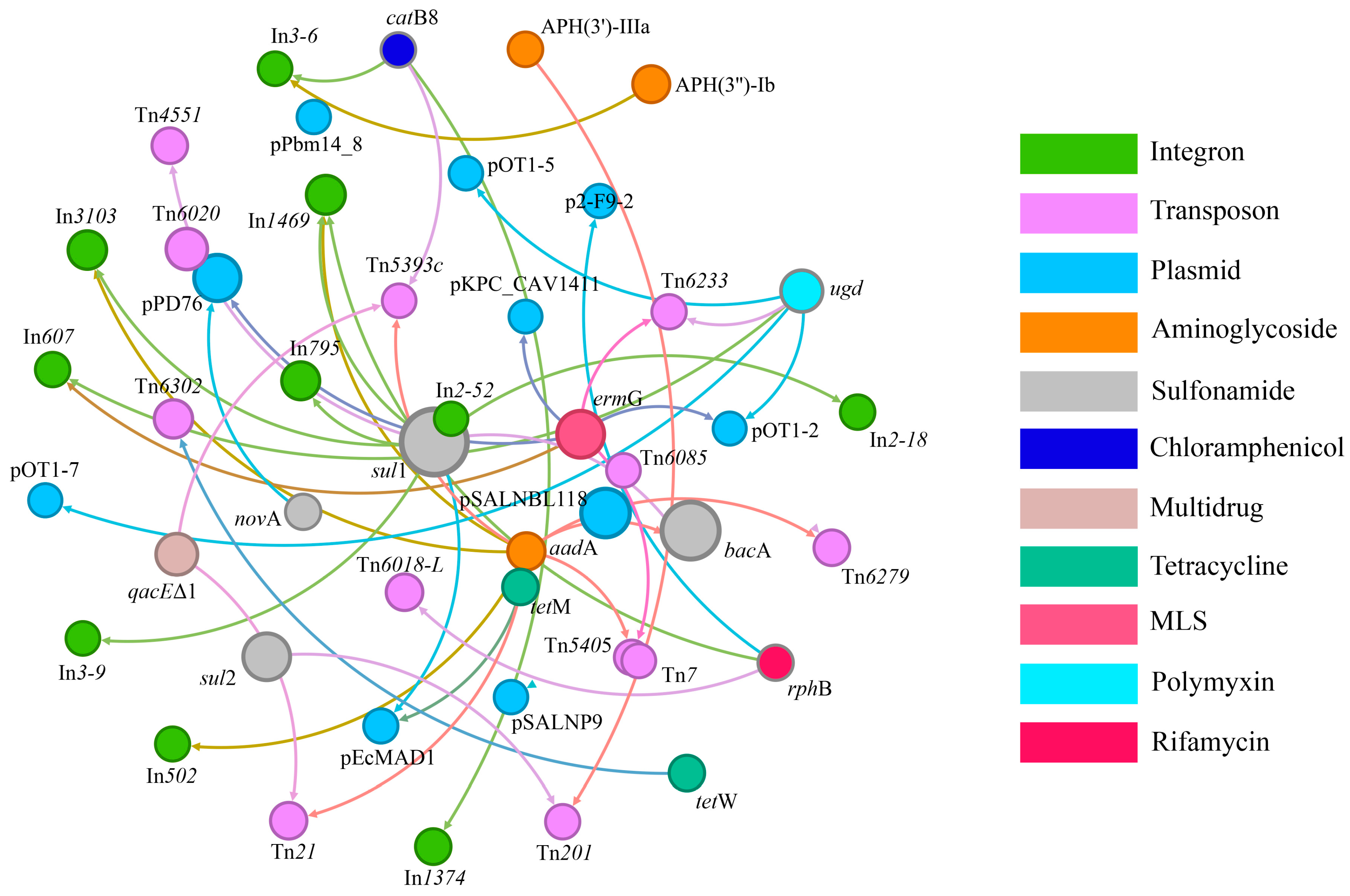
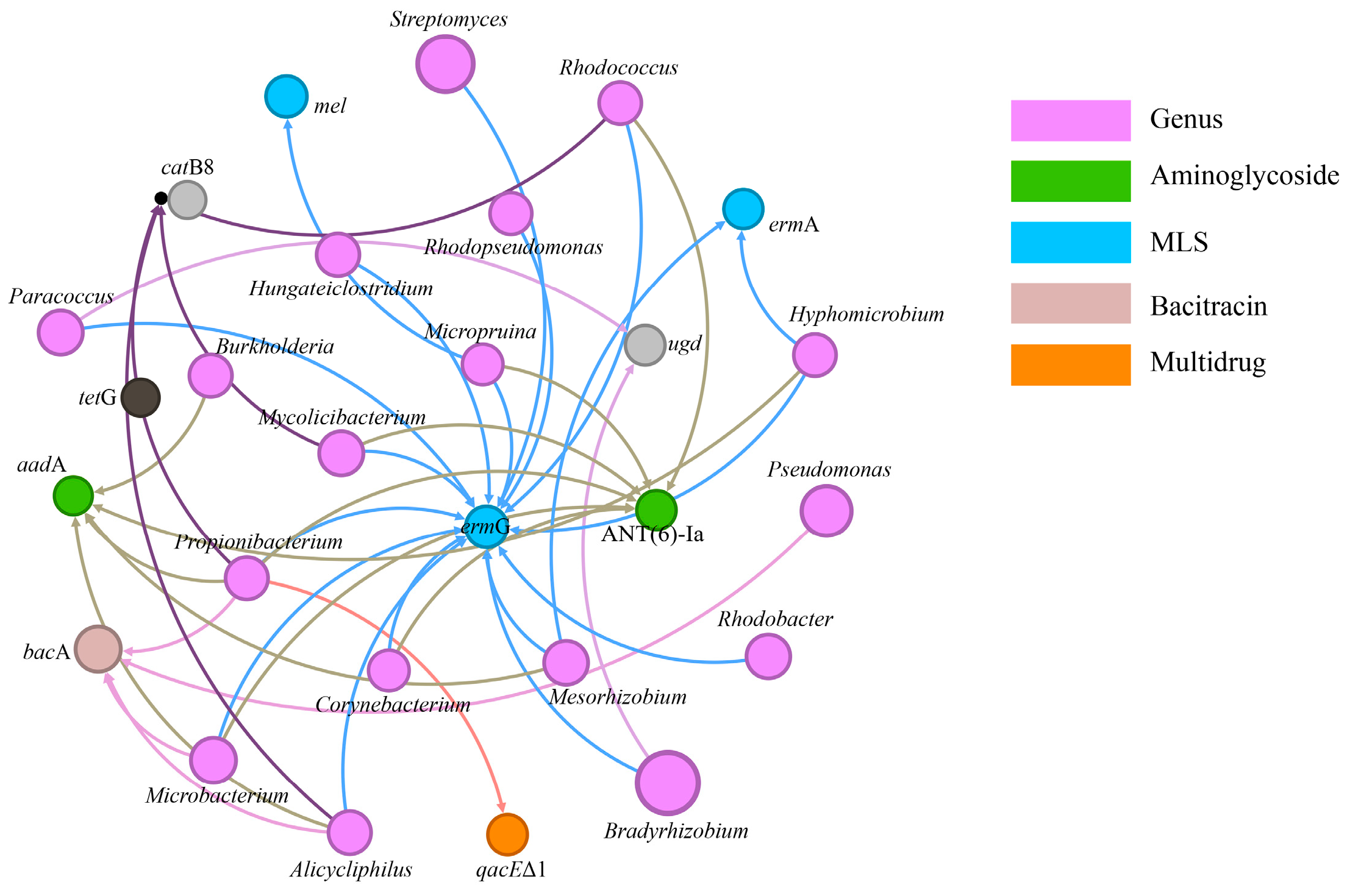
3.2. The Effects of MPs on MGE Abundances
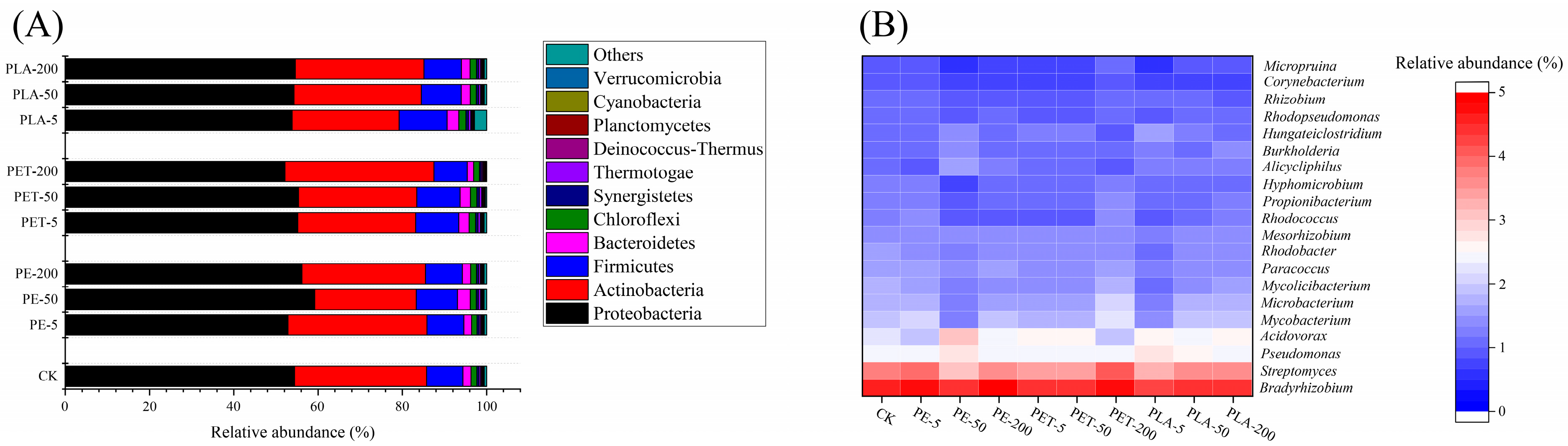
3.3. The Effects of MPs on Microbial Communities
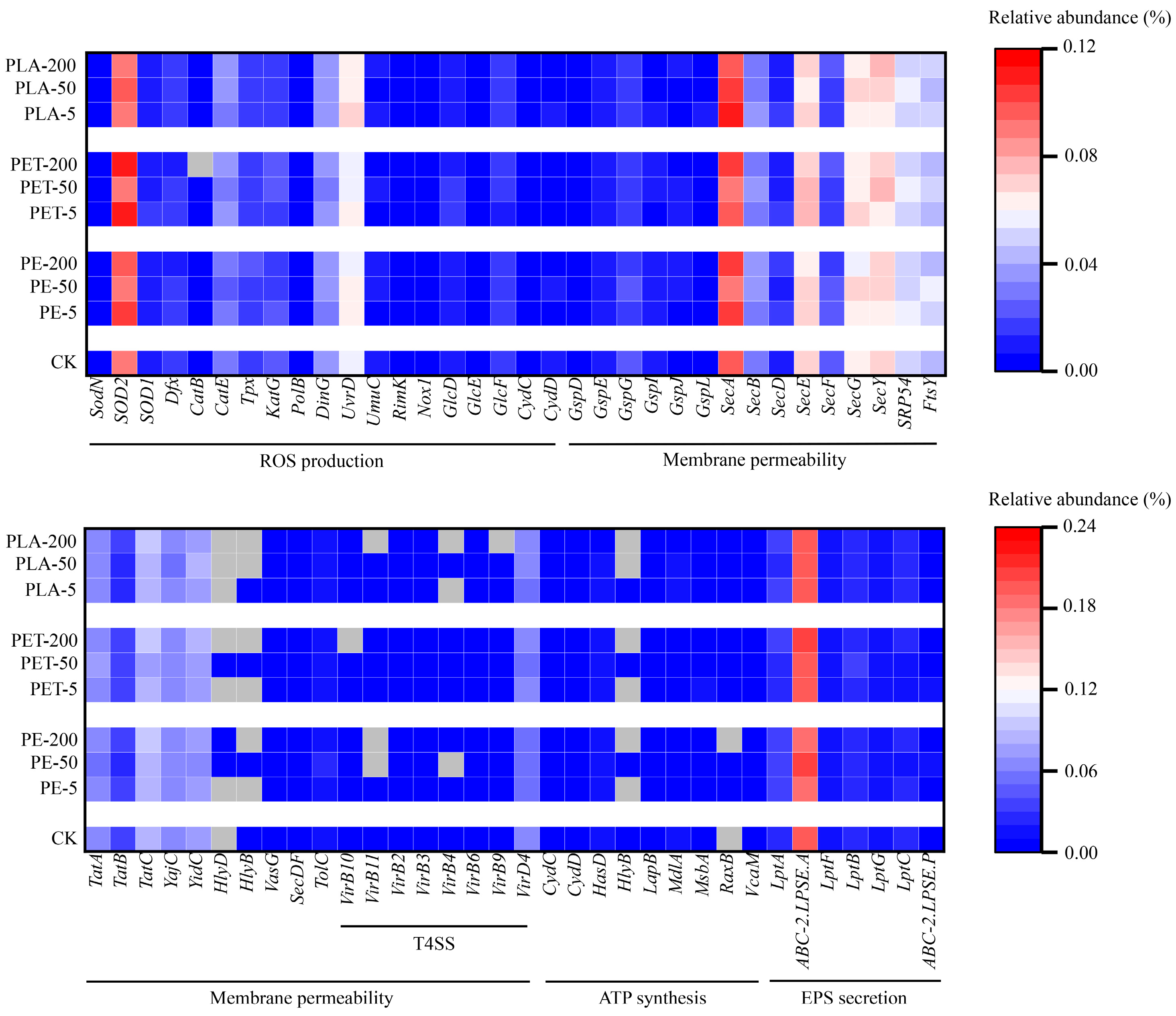
3.4. Changes in Functional Genes Revealed by Metagenomic Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Naghavi, M.; Vollset, S.E.; Ikuta, K.S.; Swetschinski, L.R.; Gray, A.P.; Wool, E.E.; Aguilar, G.R.; Mestrovic, T.; Smith, G.; Han, C. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef]
- Wang, Y.; Han, Y.P.; Li, L.; Liu, J.X.; Yan, X. Distribution, sources, and potential risks of antibiotic resistance genes in wastewater treatment plant: A review. Environ. Pollut. 2022, 310, 119870. [Google Scholar] [CrossRef] [PubMed]
- Nnorom, M.; Saroj, D.; Avery, L.; Hough, R.; Guo, B. A review of the impact of conductive materials on antibiotic resistance genes during the anaerobic digestion of sewage sludge and animal manure. J. Hazard Mater. 2023, 446, 130628. [Google Scholar] [CrossRef]
- Wang, J.L.; Mao, D.Q.; Mu, Q.H.; Luo, Y. Fate and proliferation of typical antibiotic resistance genes in five full-scale pharmaceutical wastewater treatment plants. Sci. Total Environ. 2015, 526, 366–373. [Google Scholar] [CrossRef]
- Xu, S.; Lu, W.J.; Qasim, M.Z. High-throughput characterization of the expressed antibiotic resistance genes in sewage sludge with transcriptional analysis. Ecotoxicol. Environ. Saf. 2020, 205, 111377. [Google Scholar] [CrossRef]
- Guo, J.H.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z.G. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.X.; Zhao, Y.Y.; Han, S.; Li, X.X. Regulatory strategies for inhibiting horizontal gene transfer of ARGs in paddy and dryland soil through computer-based methods. Sci. Total Environ. 2023, 856, 159096. [Google Scholar] [CrossRef] [PubMed]
- Tokuda, M.; Shintani, M. Microbial evolution through horizontal gene transfer by mobile genetic elements. Microb. Biotechnol. 2024, 17, e14408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Wang, L.; Halden, R.U.; Kannan, K. Polyethylene terephthalate and polycarbonate microplastics in sewage sludge collected from the United States. Environ. Sci. Technol. Lett. 2019, 6, 650–655. [Google Scholar] [CrossRef]
- Park, H.J.; Oh, M.J.; Kim, P.G.; Kim, G.; Jeong, D.H.; Ju, B.K.; Lee, W.S.; Chung, H.M.; Kang, H.J.; Kwon, J.H. National reconnaissance survey of microplastics in municipal wastewater treatment plants in Korea. Environ. Sci. Technol. 2020, 54, 1503–1512. [Google Scholar] [CrossRef]
- Patil, S.; Kamdi, P.; Chakraborty, S.; Das, S.; Bafana, A.; Krishnamurthi, K.; Sivanesan, S. Characterization and removal of microplastics in a sewage treatment plant from urban Nagpur, India. Environ. Monit. Assess. 2023, 195, 47. [Google Scholar] [CrossRef] [PubMed]
- Bucci, K.; Bayoumi, M.; Stevack, K.; Watson-Leung, T.; Rochman, C.M. Microplastics may induce food dilution and endocrine disrupting effects in fathead minnows (Pimephales promelas), and decrease offspring quality. Environ. Pollut. 2024, 345, 123551. [Google Scholar] [CrossRef] [PubMed]
- Mintenig, S.M.; Int-Veen, I.; Löder, M.G.J.; Primpke, S.; Gerdts, G. Identification of microplastic in effluents of waste water treatment plants using focal plane array-based micro-Fourier-transform infrared imaging. Water Res. 2017, 108, 365–372. [Google Scholar] [CrossRef]
- Jiang, J.H.; Wang, X.W.; Ren, H.Y.; Cao, G.L.; Xie, G.J.; Xing, D.F.; Liu, B.F. Investigation and fate of microplastics in wastewater and sludge filter cake from a wastewater treatment plant in China. Sci. Total Environ. 2020, 746, 141378. [Google Scholar] [CrossRef]
- Ahrendt, C.; Perez-Venegas, D.J.; Urbina, M.; Gonzalez, C.; Echeveste, P.; Aldana, M.; Pulgar, J.; Galbán-Malagón, C. Microplastic ingestion cause intestinal lesions in the intertidal fish Girella laevifrons. Mar. Pollut. Bull. 2020, 151, 110795. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.J.; Pan, J.; Li, M.; Li, Y.; Bartlam, M.; Wang, Y.Y. Selective enrichment of bacterial pathogens by microplastic biofilm. Water Res. 2019, 165, 114979. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.L.; Zhang, Z.J.; Zhu, J.D.; Shi, J.H.; Wei, H.W.; Xie, B.; Shi, H.H. Microplastics act as vectors for antibiotic resistance genes in landfill leachate: The enhanced roles of the long-term aging process. Environ. Pollut. 2021, 270, 116278. [Google Scholar] [CrossRef]
- Balasundaram, G.; Gahlot, P.; Ahmed, B.; Biswas, P.; Tyagi, V.K.; Svensson, K.; Kumar, V.; Kazmi, A.A. Advanced steam-explosion pretreatment mediated anaerobic digestion of municipal sludge: Effects on methane yield, emerging contaminants removal, and microbial community. Environ. Res. 2023, 238, 117195. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Li, H.; Zhu, L.; Yang, X.L.; Zhang, Q.S.; Wang, Y.L.; Wang, D.B. Effect and mechanism of benzalkonium bromide on short chain fatty acid production from anaerobic sludge fermentation process. J. Environ. Manag. 2023, 343, 118203. [Google Scholar] [CrossRef]
- Wei, W.; Huang, Q.S.; Sun, J.; Dai, X.H.; Ni, B.J. Revealing the mechanisms of polyethylene microplastics affecting anaerobic digestion of waste activated sludge. Environ. Sci. Technol. 2019, 53, 9604–9613. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.H.; Li, X.; Liu, H.; Zamyadi, A.; Guo, W.S.; Wen, H.T.; Gao, L.; Nghiem, L.D.; Wang, Q.L. Advancements in detection and removal of antibiotic resistance genes in sludge digestion: A state-of-art review. Bioresour. Technol. 2022, 344, 126197. [Google Scholar] [CrossRef]
- Calderon-Franco, D.; Sarelse, R.; Christou, S.; Pronk, M.; van Loosdrecht, M.C.M.; Abeel, T.; Weissbrodt, D.G. Metagenomic profiling and transfer dynamics of antibiotic resistance determinants in a full-scale granular sludge wastewater treatment plant. Water Res. 2022, 219, 118571. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.L.; Zheng, X.W.; Li, L.G.; Zhang, A.N.; Jiang, X.T.; Zhang, T. ARGs-OAP v3.0: Antibiotic-resistance gene database curation and analysis pipeline optimization. Engineering 2023, 27, 234–241. [Google Scholar] [CrossRef]
- Yin, X.L.; Chen, X.; Jiang, X.T.; Yang, Y.; Li, B.; Shum, M.H.H.; Lam, T.T.Y.; Leung, G.M.; Rose, J.; Sanchez-Cid, C.; et al. Toward a universal unit for quantification of antibiotic resistance genes in environmental samples. Environ. Sci. Technol. 2023, 57, 9713–9721. [Google Scholar] [CrossRef]
- Moura, A.; Soares, M.; Pereira, C.; Leitao, N.; Henriques, I.; Correia, A. INTEGRALL: A database and search engine for integrons, integrases and gene cassettes. Bioinformatics 2009, 25, 1096–1098. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, 61–65. [Google Scholar] [CrossRef]
- Wang, M.; Liu, G.T.; Liu, M.; Tai, C.; Deng, Z.X.; Song, J.N.; Ou, H.Y. ICEberg 3.0: Functional categorization and analysis of the integrative and conjugative elements in bacteria. Nucleic Acids Res. 2023, 52, 732–737. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Li, D.H.; Liu, C.M.; Luo, R.B.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, J.; Zhang, Z.; Peng, Z.; Dai, X.; Ni, B.-J. Polyethylene terephthalate microplastic fibers increase the release of extracellular antibiotic resistance genes during sewage sludge anaerobic digestion. Water Res. 2022, 217, 118426. [Google Scholar] [CrossRef]
- Luo, T.; Dai, X.; Wei, W.; Xu, Q.; Ni, B.-J. Microplastics enhance the prevalence of antibiotic resistance genes in anaerobic sludge digestion by enriching antibiotic-resistant bacteria in surface biofilm and facilitating the vertical and horizontal gene transfer. Environ. Sci. Technol. 2023, 57, 14611–14621. [Google Scholar] [CrossRef]
- Abu Lila, A.S.; Alharby, T.N.; Alanazi, J.; Alanazi, M.; Abdallah, M.H.; Rizvi, S.M.D.; Moin, A.; Khafagy, E.; Tabrez, S.; Al Balushi, A.A.; et al. Clinical resistant strains of Enterococci and their correlation to reduced susceptibility to biocides: Phenotypic and genotypic analysis of Macrolides, Lincosamides, and Streptogramins. Antibiotics 2023, 12, 461. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, R.W.; Al Kobaisi, M.; Jones, L.A.; Vinu, A.; Bhosale, S.V. The supramolecular self-assembly of aminoglycoside antibiotics and their applications. ChemistryOpen 2019, 8, 1154–1166. [Google Scholar] [CrossRef]
- Jiang, H.; Dong, Y.Z.; Jiao, X.; Tang, B.; Feng, T.; Li, P.; Fang, J.H. In vivo fitness of sul gene-dependent sulfonamide-resistant Escherichia coli in the mammalian gut. mSystems 2024, 9, e00836-24. [Google Scholar] [CrossRef]
- Xu, X.; Li, X.; Luo, M.; Liu, P.; Su, K.W.; Qing, Y.; Chen, S.; Qiu, J.F.; Li, Y.L. Molecular characterisations of integrons in clinical isolates of Klebsiella pneumoniae in a Chinese tertiary hospital. Microb. Pathog. 2017, 104, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.N.; Gaston, J.M.; Dai, C.L.; Zhao, S.; Poyet, M.; Groussin, M.; Yin, X.; Li, L.G.; van Loosdrecht, M.C.; Topp, E. An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 2021, 12, 4765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Q.; Abbas, M.; Rehman, M.U.; Huang, Y.H.; Zhou, R.; Gong, S.Y.; Yang, H.; Chen, S.L.; Wang, M.S.; Cheng, A.C. Dissemination of antibiotic resistance genes (ARGs) via integrons in Escherichia coli: A risk to human health. Environ. Pollut. 2020, 266, 115260. [Google Scholar] [CrossRef]
- Wang, P.L.; Qiao, Z.R.; Li, X.N.; Wu, D.; Xie, B. Fate of integrons, antibiotic resistance genes and associated microbial community in food waste and its large-scale biotreatment systems. Environ. Int. 2020, 144, 106013. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Jia, W.Q.; Xu, L.B.; Zhang, M.J.; Huang, Y. The plastisphere of biodegradable and conventional microplastics from residues exhibit distinct microbial structure, network and function in plastic-mulching farmland. J. Hazard Mater. 2023, 442, 130011. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Chen, Z.Y.; Mu, Q.H.; Wu, X.Y.; Zhang, J.J.; Mao, D.Q.; Luo, Y.; Alvarez, P.J.J. Ionic liquid enriches the antibiotic resistome, especially efflux pump genes, before significantly affecting microbial community structure. Environ. Sci. Technol. 2020, 54, 4305–4315. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, P.; Rajabnia, M.; Maali, A.; Ferdosi-Shahandashti, E. Integron and its role in antimicrobial resistance: A literature review on some bacterial pathogens. Iran. J. Basic Med. Sci. 2021, 24, 136–142. [Google Scholar] [PubMed]
- Benler, S.; Faure, G.; Altae-Tran, H.; Shmakov, S.; Zheng, F.; Koonin, E. Cargo genes of Tn7-Like transposons comprise an enormous diversity of defense systems, mobile genetic elements, and antibiotic resistance genes. Mbio 2021, 12, e02938-21. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Gutiérrez, I.; Cantu, L.; Shanahan, J.; Girguis, M.; de la Cruz, M.; Mota-Bravo, L. Cryptic environmental conjugative plasmid recruits a novel hybrid transposon resulting in a new plasmid with higher dispersion potential. mSphere 2024, 9, e00252-24. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wang, J.J.; Huang, L.P.; Wang, X.L.; Zhou, J.T.; Wang, J. Effect of immobilized anthraquinone-2-sulfonate on antibiotic resistance genes and microbial community in biofilms of anaerobic reactors. J. Environ. Manag. 2021, 282, 111967. [Google Scholar] [CrossRef] [PubMed]
- Stenger, K.S.; Wikmark, O.G.; Bezuidenhout, C.C.; Molale-Tom, L.G. Microplastics pollution in the ocean: Potential carrier of resistant bacteria and resistance genes. Environ. Pollut. 2021, 291, 118130. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.J.; Zhang, L.J.; Jia, B.Y.; Yang, B.; Luo, Z.L.; Yang, J.K.; Boguta, P.; Su, X.T. Effects of enzymes on organic matter conversion in anaerobic fermentation of sludge to produce volatile fatty acids. Bioresour. Technol. 2022, 366, 128227. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.D.; Yang, H.F.; Zhao, Z.Y.; Zou, H.J.; Zhu, R.L.; Jiang, Q.; Sun, T.; Li, M.X.; Li, L.; Shi, D.Z.; et al. Comprehensively evaluating the digestive performance of sludge with different lignocellulosic components in mesophilic anaerobic digester. Bioresour. Technol. 2019, 293, 122042. [Google Scholar] [CrossRef]
- Wu, D.; Li, L.; Zhen, F.; Liu, H.L.; Xiao, F.; Sun, Y.M.; Peng, X.Y.; Li, Y.; Wang, X.M. Thermodynamics of volatile fatty acid degradation during anaerobic digestion under organic overload stress: The potential to better identify process stability. Water Res. 2022, 214, 118187. [Google Scholar] [CrossRef]
- Gou, Z.C.; Zheng, H.Y.; He, Z.Q.; Su, Y.J.; Chen, S.J.; Chen, H.; Chen, G.; Ma, N.L.; Sun, Y. The combined action of biochar and nitrogen-fixing bacteria on microbial and enzymatic activities of soil N cycling. Environ. Pollut. 2023, 317, 120790. [Google Scholar] [CrossRef]
- Liu, Y.; Ye, X.M.; Chen, K.Q.; Wu, X.Y.; Jiao, L.H.; Zhang, H.Y.; Zhu, F.; Xi, Y.L. Effect of nanobubble water on medium chain carboxylic acids production in anaerobic digestion of cow manure. Waste Manage. 2024, 184, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Das, S. Bacterial enzymatic degradation of recalcitrant organic pollutants: Catabolic pathways and genetic regulations. Environ. Sci. Pollut. R. 2023, 30, 79676–79705. [Google Scholar] [CrossRef] [PubMed]
- Fauser, P.; Vorkamp, K.; Strand, J. Residual additives in marine microplastics and their risk assessment-A critical review. Mar. Pollut. Bull. 2022, 177, 113467. [Google Scholar] [CrossRef] [PubMed]
- Cazaudehore, G.; Monlau, F.; Gassie, C.; Lallement, A.; Guyoneaud, R. Active microbial communities during biodegradation of biodegradable plastics by mesophilic and thermophilic anaerobic digestion. J. Hazard Mater. 2023, 443, 130208. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.G.; Wang, H.; Blaustein, R.A.; Guo, L.; Ye, Q.; Fu, Y.L.; Fan, J.H.; Su, X.M.; Hartmann, E.M.; Shen, C.F. Pangenomic and functional investigations for dormancy and biodegradation features of an organic pollutant-degrading bacterium Rhodococcus biphenylivorans TG9. Sci. Total Environ. 2022, 809, 151141. [Google Scholar] [CrossRef] [PubMed]
- Li, M.J.; Chen, H.B. Enhanced short-chain fatty acid production from sludge anaerobic fermentation by combined pretreatment with sodium pyrophosphate and thermal hydrolysis. Bioresour. Technol. 2024, 406, 131067. [Google Scholar] [CrossRef]
- Hu, X.J.; Waigi, M.G.; Yang, B.; Gao, Y.Z. Impact of plastic particles on the horizontal transfer of antibiotic resistance genes to bacterium: Dependent on particle sizes and antibiotic resistance gene vector replication capacities. Environ. Sci. Technol. 2022, 56, 14948–14959. [Google Scholar] [CrossRef] [PubMed]
- He, A.Y.; Dean, J.M.; Lodhi, I.J. Peroxisomes as cellular adaptors to metabolic and environmental stress. Trends Cell Biol. 2021, 31, 656–670. [Google Scholar] [CrossRef]
- Yang, G.; Cao, J.M.; Cui, H.L.; Zhan, X.M.; Duan, G.L.; Zhu, Y.G. Artificial sweetener enhances the spread of antibiotic resistance genes during anaerobic digestion. Environ. Sci. Technol. 2023, 57, 10919–10928. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Huang, Q.S.; Sun, J.; Wang, J.Y.; Wu, S.L.; Ni, B.J. Polyvinyl chloride microplastics affect methane production from the anaerobic digestion of waste activated sludge through leaching toxic Bisphenol-A. Environ. Sci. Technol. 2019, 53, 2509–2517. [Google Scholar] [CrossRef]
- Qiu, D.Y.; Ke, M.J.; Zhang, Q.; Zhang, F.; Lu, T.; Sun, L.W.; Qian, H.F. Response of microbial antibiotic resistance to pesticides: An emerging health threat. Sci. Total Environ. 2022, 850, 158057. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Y.; Zhang, G.S.; Zhang, D.W.; Zhu, N.Z.; Bo, J.P.; Meng, X.Z.; Chen, Y.; Qin, Y.; Liu, H.J.; Li, W.Y. Microplastic biofilms promote the horizontal transfer of antibiotic resistance genes in estuarine environments. Mar. Environ. Res. 2024, 202, 106777. [Google Scholar] [CrossRef] [PubMed]
- Hua, M.X.; Dai, D.F.; Du, P.C.; Chen, N.; Duan, A.; Yue, J.L.; Jia, H.B.; Rong, C.B.; Li, A.; Zeng, H.; et al. A chromosome-encoded T4SS independently contributes to horizontal gene transfer in Enterococcus faecalis. Cell Rep. 2022, 41, 14. [Google Scholar] [CrossRef]
- Ilangovan, A.; Connery, S.; Waksman, G. Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol. 2015, 23, 301–310. [Google Scholar] [CrossRef]
- Priyadarshanee, M.; Das, S.; Vandana. Bacterial extracellular polymeric substances: Biosynthesis and interaction with environmental pollutants. Chemosphere 2023, 332, 138876. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Yuan, D. Metagenomic Analysis Reveals the Effects of Microplastics on Antibiotic Resistance Genes in Sludge Anaerobic Digestion. Toxics 2024, 12, 920. https://doi.org/10.3390/toxics12120920
Li Z, Yuan D. Metagenomic Analysis Reveals the Effects of Microplastics on Antibiotic Resistance Genes in Sludge Anaerobic Digestion. Toxics. 2024; 12(12):920. https://doi.org/10.3390/toxics12120920
Chicago/Turabian StyleLi, Zhonghong, and Donghai Yuan. 2024. "Metagenomic Analysis Reveals the Effects of Microplastics on Antibiotic Resistance Genes in Sludge Anaerobic Digestion" Toxics 12, no. 12: 920. https://doi.org/10.3390/toxics12120920
APA StyleLi, Z., & Yuan, D. (2024). Metagenomic Analysis Reveals the Effects of Microplastics on Antibiotic Resistance Genes in Sludge Anaerobic Digestion. Toxics, 12(12), 920. https://doi.org/10.3390/toxics12120920





