The Role of miR-155 in Nutrition: Modulating Cancer-Associated Inflammation
Abstract
:1. Introduction
2. MiRNAs and Nutrition
3. The Role(s) of MiR-155 in Intra- and Intercellular Signaling
4. The Role of MiR-155 in Regulation of the Immune Response
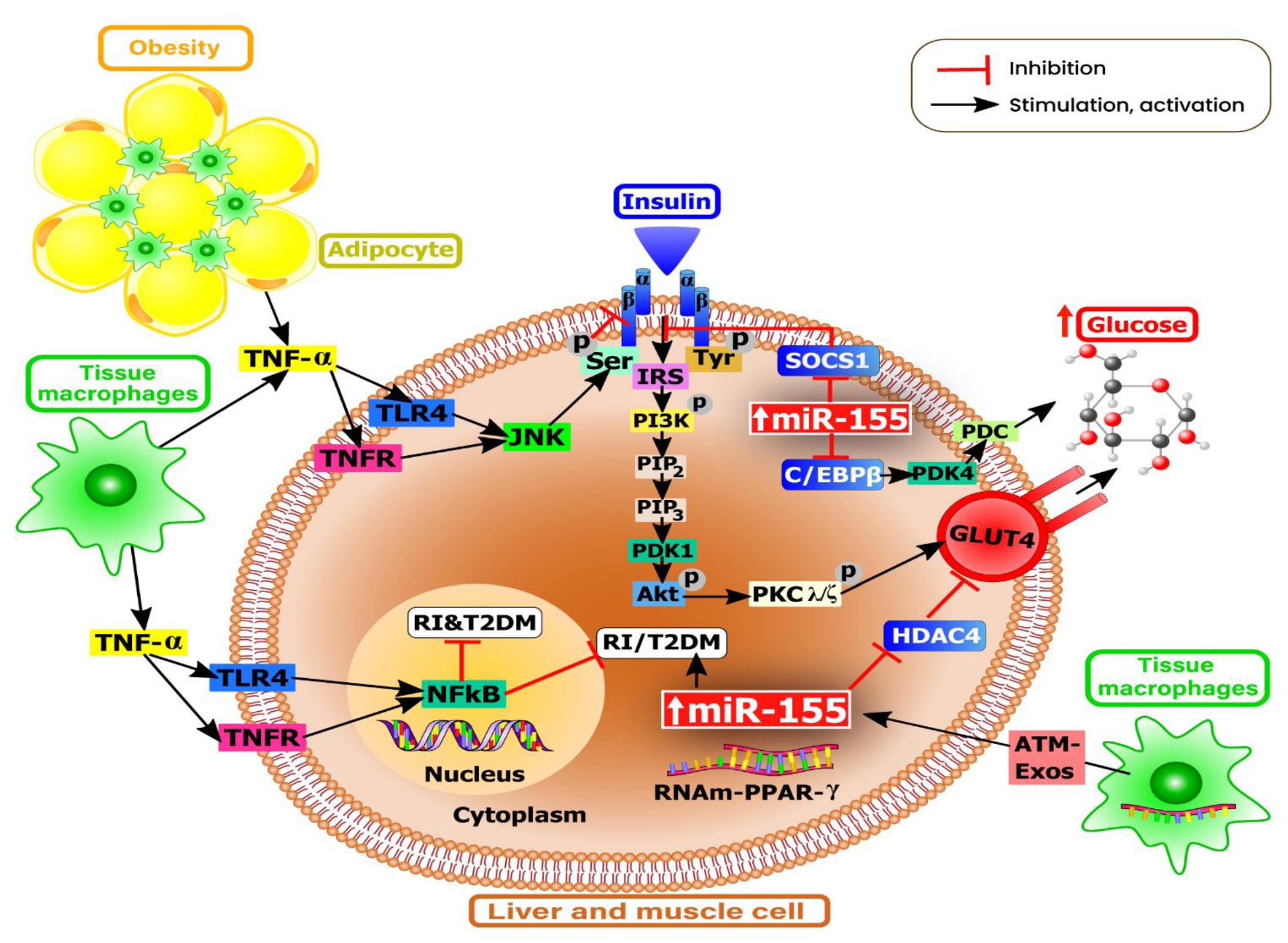
5. The Role of MiR-155 in Metabolism
6. The Role of MiR-155 in Inflammation
7. The Role of MiR-155 in Response to Various Dietary Nutrients
8. MiR-155 in Carcinogenesis and MiRNA-Based Monitoring of Disease Progression
9. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- García-Segura, L.; Pérez-Andrade, M.; Miranda-Ríos, J. The emerging role of MicroRNAs in the regulation of gene expression by nutrients. J. Nutr. Nutr. 2013, 6, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Khoshbin, K.; Camilleri, M. Effects of dietary components on intestinal permeability in health and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G589–G608. [Google Scholar] [CrossRef] [PubMed]
- Mullins, V.A.; Bresette, W.; Johnstone, L.; Hallmark, B.; Chilton, F.H. Genomics in personalized nutrition: Can you “eat for your genes”? Nutrients 2020, 12, 3118. [Google Scholar] [CrossRef] [PubMed]
- Bara, T., Jr.; Gurzu, S.; Sugimura, H.; Bara, T.; Beleaua, M.A.; Jung, I. A systematic review of the possible carcinogenic role of the aristolochic acid. Rom. J. Morphol. Embryol. 2017, 58, 41–44. [Google Scholar] [PubMed]
- Arias-Del-Val, J.; Santo-Domingo, J.; García-Casas, P.; Alvarez-Illera, P.; Núñez Galindo, A.; Wiederkehr, A.; Fonteriz, R.I.; Montero, M.; Alvarez, J. Regulation of inositol 1,4,5-trisphosphate-induced Ca(2+) release from the endoplasmic reticulum by AMP-activated kinase modulators. Cell Calcium 2019, 77, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.Q.; Wei, H.Y.; Huang, G.R.; Xu, L.Y.; Chen, Y.L.; Qi, J.; Xian, W.; Qin, Y.C.; Wei, L.D.; Zhao, L.J.; et al. Molecular mechanisms of apoptosis in hepatocellular carcinoma cells induced by ethanol extracts of Solanum lyratum Thumb through the mitochondrial pathway. World J. Gastroenterol. 2017, 23, 1010–1017. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; An, N.; Xu, W.; Zhang, W.; Meng, X.; Chen, G.; Liu, W. Functional characterization of the upstream components of the Hog1-like kinase cascade in hyperosmotic and carbon sensing in Trichoderma reesei. Biotechnol. Biofuels 2018, 11, 97. [Google Scholar] [CrossRef] [Green Version]
- Tao, H.; Cui, L.; Li, Y.; Zhou, X.; Ma, G.; Yao, L.; Fu, J.; Li, W.; Cai, Y.; Zhou, H.; et al. Association of tag SNPs and rare CNVs of the MIR155HG/miR-155 gene with epilepsy in the Chinese Han population. BioMed Res. Int. 2015, 837213. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; He, H.; Liu, A.; Liu, H.; Huang, H.; Zhao, C.; Jing, L.; Ni, J.; Yin, L.; Hu, S.; et al. Natural functional SNPs in miR-155 alter its expression level, blood cell counts, and immune responses. Front. Immunol. 2016, 7, 295. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Xu, M.; Tu, J.; Zhao, Z.; Gao, J.; Chen, M.; Song, J.; Zhu, H.; Cheng, X.; Hui, J.; et al. MiR-155 and its functional variant rs767649 contribute to the susceptibility and survival of hepatocellular carcinoma. Oncotarget 2016, 7, 60303–60309. [Google Scholar] [CrossRef]
- Due, H.; Svendsen, P.; Bødker, J.S.; Schmitz, A.; Bøgsted, M.; Johnsen, H.E.; El-Galaly, T.C.; Roug, A.S.; Dybkær, K. miR-155 as a Biomarker in B-Cell Malignancies. Biomed. Res. Int. 2016, 2016, 9513037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, C.; Yang, F.; Qin, Z.; Jing, X.; Shu, Y.; Shen, H. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: A systematic review with meta-analysis. BMC Cancer 2019, 19, 1103. [Google Scholar] [CrossRef] [Green Version]
- Kudela, E.; Samec, M.; Koklesova, L.; Liskova, A.; Kubatka, P.; Kozubik, E.; Rokos, T.; Pribulova, T.; Gabonova, E.; Smolar, M.; et al. miRNA expression profiles in luminal A breast cancer-implications in biology, prognosis, and prediction of response to hormonal treatment. Int. J. Mol. Sci. 2020, 21, 7691. [Google Scholar] [CrossRef]
- Camp, K.M.; Trujillo, E. Position of the Academy of Nutrition and Dietetics: Nutritional genomics. J. Acad. Nutr. Diet. 2014, 114, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Petrie, J.R.; Guzik, T.J.; Touyz, R.M. Diabetes, hypertension, and cardiovascular disease: Clinical insights and vascular mechanisms. Can. J. Cardiol. 2018, 34, 575–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasper, P.; Martin, A.; Lang, S.; Kütting, F.; Goeser, T.; Demir, M.; Steffen, H.M. NAFLD and cardiovascular diseases: A clinical review. Clin. Res. Cardiol. 2020. [Google Scholar] [CrossRef]
- Deng, T.; Lyon, C.J.; Bergin, S.; Caligiuri, M.A.; Hsueh, W.A. Obesity, inflammation, and cancer. Annu. Rev. Pathol. 2016, 11, 421–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanoaga, O.; Jurj, A.; Raduly, L.; Cojocneanu-Petric, R.; Fuentes-Mattei, E.; Wu, O.; Braicu, C.; Gherman, C.D.; Berindan-Neagoe, I. Implications of dietary ω-3 and ω-6 polyunsaturated fatty acids in breast cancer. Exp. Ther. Med. 2018, 15, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Giugliano, D.; Ceriello, A.; Esposito, K. The effects of diet on inflammation: Emphasis on the metabolic syndrome. J. Am. Coll. Cardiol. 2006, 48, 677–685. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Lewis, E.D.; Pae, M.; Meydani, S.N. Nutritional modulation of immune function: Analysis of evidence, mechanisms, and clinical relevance. Front. Immunol. 2018, 9, 3160. [Google Scholar] [CrossRef]
- Iddir, M.; Brito, A.; Dingeo, G.; Fernandez Del Campo, S.S.; Samouda, H.; La Frano, M.R.; Bohn, T. Strengthening the immune system and reducing inflammation and oxidative stress through diet and nutrition: Considerations during the COVID-19 crisis. Nutrients 2020, 12, 1562. [Google Scholar] [CrossRef] [PubMed]
- Esser, N.; Legrand-Poels, S.; Piette, J.; Scheen, A.J.; Paquot, N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res. Clin. Pract. 2014, 105, 141–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braicu, C.; Zanoaga, O.; Zimta, A.A.; Tigu, A.B.; Kilpatrick, K.L.; Bishayee, A.; Nabavi, S.M.; Berindan-Neagoe, I. Natural compounds modulate the crosstalk between apoptosis- and autophagy-regulated signaling pathways: Controlling the uncontrolled expansion of tumor cells. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [Green Version]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [Green Version]
- Dykes, I.M.; Emanueli, C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genom. Proteom. Bioinform 2017, 15, 177–186. [Google Scholar] [CrossRef]
- Rane, S.; Sayed, D.; Abdellatif, M. MicroRNA with a macroFunction. Cell Cycle 2007, 6, 1850–1855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, H.W.; Mendell, J.T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 2006, 94, 776–780. [Google Scholar] [CrossRef]
- Yao, S. MicroRNA biogenesis and their functions in regulating stem cell potency and differentiation. Biol. Proced. Online 2016, 18, 8. [Google Scholar] [CrossRef] [Green Version]
- Leichter, A.L.; Sullivan, M.J.; Eccles, M.R.; Chatterjee, A. MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Mol. Cancer 2017, 16, 15. [Google Scholar] [CrossRef] [Green Version]
- Blenkiron, C.; Miska, E.A. miRNAs in cancer: Approaches, aetiology, diagnostics and therapy. Hum. Mol. Genet. 2007, 16, R106–R113. [Google Scholar] [CrossRef]
- Zhang, T.; Ji, C.; Shi, R. miR-142-3p promotes pancreatic β cell survival through targeting FOXO1 in gestational diabetes mellitus. Int. J. Clin. Exp. Pathol. 2019, 12, 1529–1538. [Google Scholar]
- Okamoto, A.; Sehouli, J.; Yanaihara, N.; Hirata, Y.; Braicu, I.; Kim, B.G.; Takakura, S.; Saito, M.; Yanagida, S.; Takenaka, M.; et al. Somatic copy number alterations associated with Japanese or endometriosis in ovarian clear cell adenocarcinoma. PLoS ONE 2015, 10, e0116977. [Google Scholar] [CrossRef] [PubMed]
- Strubberg, A.M.; Madison, B.B. MicroRNAs in the etiology of colorectal cancer: Pathways and clinical implications. Dis. Model. Mech. 2017, 10, 197–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braicu, C.; Gulei, D.; Raduly, L.; Harangus, A.; Rusu, A.; Berindan-Neagoe, I. Altered expression of miR-181 affects cell fate and targets drug resistance-related mechanisms. Mol. Asp. Med. 2019, 70, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Braicu, C.; Buiga, R.; Cojocneanu, R.; Buse, M.; Raduly, L.; Pop, L.A.; Chira, S.; Budisan, L.; Jurj, A.; Ciocan, C.; et al. Connecting the dots between different networks: MiRNAs associated with bladder cancer risk and progression. J. Exp. Clin. Cancer Res. 2019, 38, 433. [Google Scholar] [CrossRef] [PubMed]
- Cojocneanu, R.; Braicu, C.; Raduly, L.; Jurj, A.; Zanoaga, O.; Magdo, L.; Irimie, A.; Muresan, M.S.; Ionescu, C.; Grigorescu, M.; et al. Plasma and tissue specific miRNA expression pattern and functional analysis associated to colorectal cancer patients. Cancers 2020, 12, 843. [Google Scholar] [CrossRef] [Green Version]
- Ranjbar, R.; Karimian, A.; Aghaie Fard, A.; Tourani, M.; Majidinia, M.; Jadidi-Niaragh, F.; Yousefi, B. The importance of miRNAs and epigenetics in acute lymphoblastic leukemia prognosis. J. Cell Physiol. 2019, 234, 3216–3230. [Google Scholar] [CrossRef]
- Castell-Auví, A.; Cedó, L.; Movassat, J.; Portha, B.; Sánchez-Cabo, F.; Pallarès, V.; Blay, M.; Pinent, M.; Ardévol, A. Procyanidins modulate microRNA expression in pancreatic islets. J. Agric. Food Chem. 2013, 61, 355–363. [Google Scholar] [CrossRef]
- Ross, S.A.; Davis, C.D. The emerging role of microRNAs and nutrition in modulating health and disease. Annu. Rev. Nutr. 2014, 34, 305–336. [Google Scholar] [CrossRef] [PubMed]
- Beckett, E.L.; Yates, Z.; Veysey, M.; Duesing, K.; Lucock, M. The role of vitamins and minerals in modulating the expression of microRNA. Nutr. Res. Rev. 2014, 27, 94–106. [Google Scholar] [CrossRef] [Green Version]
- Michell, D.L.; Vickers, K.C. Lipoprotein carriers of microRNAs. Biochim. Biophys. Acta 2016, 1861, 2069–2074. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.M.; Lim, S.M.; Yoo, J.A.; Woo, M.J.; Cho, K.H. Consumption of high-dose vitamin C (1250 mg per day) enhances functional and structural properties of serum lipoprotein to improve anti-oxidant, anti-atherosclerotic, and anti-aging effects via regulation of anti-inflammatory microRNA. Food Funct. 2015, 6, 3604–3612. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ye, J.; Guo, R.; Liang, X.; Yang, L. TRIF regulates BIC/miR-155 via the ERK signaling pathway to control the ox-LDL-induced macrophage inflammatory response. J. Immunol. Res. 2018, 2018, 6249085. [Google Scholar] [CrossRef]
- Rustum, Y.M.; Chintala, S.; Durrani, F.A.; Bhattacharya, A. Non-Coding micro RNAs and hypoxia-inducible factors are selenium targets for development of a mechanism-based combination strategy in clear-cell renal cell Carcinoma-Bench-to-Bedside therapy. Int. J. Mol. Sci. 2018, 19, 3378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Sun, Z.; Xu, Z.; Liu, T.; Pan, T.; Li, S. Down-regulation of microRNA-155 promotes selenium deficiency-induced apoptosis by tumor necrosis factor receptor superfamily member 1B in the broiler spleen. Oncotarget 2017, 8, 58513–58525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurj, A.; Zanoaga, O.; Braicu, C.; Lazar, V.; Tomuleasa, C.; Irimie, A.; Berindan-Neagoe, I. A comprehensive picture of extracellular vesicles and their contents. Molecular transfer to cancer cells. Cancers 2020, 12, 298. [Google Scholar] [CrossRef] [Green Version]
- Braicu, C.; Tomuleasa, C.; Monroig, P.; Cucuianu, A.; Berindan-Neagoe, I.; Calin, G.A. Exosomes as divine messengers: Are they the Hermes of modern molecular oncology? Cell Death Differ. 2015, 22, 34–45. [Google Scholar] [CrossRef]
- Braicu, C.; Cojocneanu-Petric, R.; Chira, S.; Truta, A.; Floares, A.; Petrut, B.; Achimas-Cadariu, P.; Berindan-Neagoe, I. Clinical and pathological implications of miRNA in bladder cancer. Int. J. Nanomed. 2015, 10, 791–800. [Google Scholar] [CrossRef] [Green Version]
- Braicu, C.; Calin, G.A.; Berindan-Neagoe, I. MicroRNAs and cancer therapy—From bystanders to major players. Curr. Med. Chem. 2013, 20, 3561–3573. [Google Scholar] [CrossRef] [PubMed]
- Park, H.K.; Jo, W.; Choi, H.J.; Jang, S.; Ryu, J.E.; Lee, H.J.; Lee, H.; Kim, H.; Yu, E.S.; Son, W.C. Time-course changes in the expression levels of miR-122, -155, and -21 as markers of liver cell damage, inflammation, and regeneration in acetaminophen-induced liver injury in rats. J. Vet. Sci. 2016, 17, 45–51. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. miR-155 gene: A typical multifunctional microRNA. Biochim. Biophys. Acta 2009, 1792, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Calame, K. MicroRNA-155 function in B Cells. Immunity 2007, 27, 825–827. [Google Scholar] [CrossRef] [Green Version]
- Eissa, M.G.; Artlett, C.M. The microRNA miR-155 is essential in fibrosis. Non-coding RNA 2019, 5, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchiya, M.; Kalurupalle, S.; Kumar, P.; Ghoshal, S.; Zhang, Y.; Lehrmann, E.; Becker, K.G.; Gorospe, M.; Biswas, R. RPTOR, a novel target of miR-155, elicits a fibrotic phenotype of cystic fibrosis lung epithelium by upregulating CTGF. RNA Biol. 2016, 13, 837–847. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Shan, S.; Huo, Y.; Xie, Z.; Fang, Y.; Qi, Z.; Chen, F.; Li, Y.; Sun, B. MiR-155-5p inhibits PDK1 and promotes autophagy via the mTOR pathway in cervical cancer. Int. J. Biochem. Cell Biol. 2018, 99, 91–99. [Google Scholar] [CrossRef]
- Chen, H.; Liu Gao, M.Y.; Zhang, L.; He, F.L.; Shi, Y.K.; Pan, X.H.; Wang, H. MicroRNA-155 affects oxidative damage through regulating autophagy in endothelial cells. Oncol. Lett. 2019, 17, 2237–2243. [Google Scholar] [CrossRef]
- Wan, J.; Yang, X.; Ren, Y.; Li, X.; Zhu, Y.; Haddock, A.N.; Ji, B.; Xia, L.; Lu, N. Inhibition of miR-155 reduces impaired autophagy and improves prognosis in an experimental pancreatitis mouse model. Cell Death Dis. 2019, 10, 303. [Google Scholar] [CrossRef] [Green Version]
- Valeri, N.; Gasparini, P.; Fabbri, M.; Braconi, C.; Veronese, A.; Lovat, F.; Adair, B.; Vannini, I.; Fanini, F.; Bottoni, A.; et al. Modulation of mismatch repair and genomic stability by miR-155. Proc. Natl. Acad. Sci. USA 2010, 107, 6982–6987. [Google Scholar] [CrossRef] [Green Version]
- Lánczky, A.; Nagy, Á.; Bottai, G.; Munkácsy, G.; Szabó, A.; Santarpia, L.; Győrffy, B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016, 160, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Loeb, G.B.; Khan, A.A.; Canner, D.; Hiatt, J.B.; Shendure, J.; Darnell, R.B.; Leslie, C.S.; Rudensky, A.Y. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 2012, 48, 760–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Rhee, J.K.; Yoo, H.J.; Lee, H.J.; Lee, E.J.; Lee, J.W.; Yu, J.H.; Son, B.H.; Gong, G.; Kim, S.B.; et al. Bioinformatic and metabolomic analysis reveals miR-155 regulates thiamine level in breast cancer. Cancer Lett. 2015, 357, 488–497. [Google Scholar] [CrossRef]
- Cui, W.; Meng, W.; Zhao, L.; Cao, H.; Chi, W.; Wang, B. TGF-β-induced long non-coding RNA MIR155HG promotes the progression and EMT of laryngeal squamous cell carcinoma by regulating the miR-155-5p/SOX10 axis. Int. J. Oncol. 2019, 54, 2005–2018. [Google Scholar] [CrossRef] [Green Version]
- Kong, J.; Li, L.; Lu, Z.; Song, J.; Yan, J.; Yang, J.; Gu, Z.; Da, Z. MicroRNA-155 suppresses mesangial cell proliferation and TGF-β1 production via inhibiting CXCR5-ERK signaling pathway in Lupus nephritis. Inflammation 2019, 42, 255–263. [Google Scholar] [CrossRef] [Green Version]
- Tarassishin, L.; Loudig, O.; Bauman, A.; Shafit-Zagardo, B.; Suh, H.S.; Lee, S.C. Interferon regulatory factor 3 inhibits astrocyte inflammatory gene expression through suppression of the proinflammatory miR-155 and miR-155*. Glia 2011, 59, 1911–1922. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.R.; Zhu, H.F.; Zhu, Y. Knockout of microRNA-155 ameliorates the Th17/Th9 immune response and promotes wound healing. Curr. Med. Sci. 2019, 39, 954–964. [Google Scholar] [CrossRef]
- Quinn, S.R.; Mangan, N.E.; Caffrey, B.E.; Gantier, M.P.; Williams, B.R.; Hertzog, P.J.; McCoy, C.E.; O’Neill, L.A. The role of Ets2 transcription factor in the induction of microRNA-155 (miR-155) by lipopolysaccharide and its targeting by interleukin-10. J. Biol. Chem. 2014, 289, 4316–4325. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Xiong, S.; Jiang, P.; Liu, R.; Liu, X.; Qian, J.; Zheng, X.; Chu, Y. Glucocorticoids inhibit lipopolysaccharide-mediated inflammatory response by downregulating microRNA-155: A novel anti-inflammation mechanism. Free Radic. Biol. Med. 2012, 52, 1307–1317. [Google Scholar] [CrossRef]
- Mahesh, G.; Biswas, R. MicroRNA-155: A master regulator of inflammation. J. Interf. Cytokine Res. 2019, 39, 321–330. [Google Scholar] [CrossRef]
- Shantikumar, S.; Caporali, A.; Emanueli, C. Role of microRNAs in diabetes and its cardiovascular complications. Cardiovasc. Res. 2012, 93, 583–593. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Mei, H.; Chang, X.; Chen, F.; Zhu, Y.; Han, X. Adipocyte-derived microvesicles from obese mice induce M1 macrophage phenotype through secreted miR-155. J. Mol. Cell Biol. 2016, 8, 505–517. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.M.; Gilchrist, D.S.; Nijjar, J.; Araldi, E.; Ramirez, C.M.; Lavery, C.A.; Fernández-Hernando, C.; McInnes, I.B.; Kurowska-Stolarska, M. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS ONE 2013, 8, e72324. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, A.D.; Fonken, L.K.; Gushchina, L.V.; Aubrecht, T.G.; Maurya, S.K.; Periasamy, M.; Nelson, R.J.; Popovich, P.G. miR-155 deletion in female mice prevents diet-induced obesity. Sci. Rep. 2016, 6, 22862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Lee, E.; Jung, J.; Lee, J.W.; Kim, H.J.; Kim, J.; Yoo, H.J.; Lee, H.J.; Chae, S.Y.; Jeon, S.M.; et al. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene 2018, 37, 2982–2991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, M.; Mehta, A.; Zhao, J.L.; Lee, K.; Marinov, G.K.; Garcia-Flores, Y.; Lu, L.-F.; Rudensky, A.Y.; Baltimore, D. An NF-κB-microRNA regulatory network tunes macrophage inflammatory responses. Nat. Commun. 2017, 8, 851. [Google Scholar] [CrossRef] [Green Version]
- Testa, U.; Pelosi, E.; Castelli, G.; Labbaye, C. miR-146 and miR-155: Two key modulators of immune response and tumor development. Non-Coding RNA 2017, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Han, S.R.; Kang, Y.H.; Jeon, H.; Lee, S.; Park, S.J.; Song, D.Y.; Min, S.S.; Yoo, S.M.; Lee, M.S.; Lee, S.H. Differential expression of miRNAs and behavioral change in the cuprizone-induced demyelination mouse model. Int. J. Mol. Sci. 2020, 21, 646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, A.E.; Boesch-Saadatmandi, C.; Dose, J.; Schultheiss, G.; Rimbach, G. Anti-inflammatory potential of allyl-isothiocyanate--role of Nrf2, NF-(κ) B and microRNA-155. J. Cell Mol. Med. 2012, 16, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Wei, Y. Modulators of microRNA function in the immune system. Int. J. Mol. Sci. 2020, 21, 2357. [Google Scholar] [CrossRef]
- Alivernini, S.; Gremese, E.; McSharry, C.; Tolusso, B.; Ferraccioli, G.; McInnes, I.B.; Kurowska-Stolarska, M. MicroRNA-155-at the critical interface of innate and adaptive immunity in arthritis. Front. Immunol. 2017, 8, 1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bala, S.; Tilahun, Y.; Taha, O.; Alao, H.; Kodys, K.; Catalano, D.; Szabo, G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J. Transl. Med. 2012, 10, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and their potential for inflammatory diseases treatment. Front Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef] [Green Version]
- Ceppi, M.; Pereira, P.M.; Dunand-Sauthier, I.; Barras, E.; Reith, W.; Santos, M.A.; Pierre, P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2735–2740. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef] [Green Version]
- Thai, T.H.; Calado, D.P.; Casola, S.; Ansel, K.M.; Xiao, C.; Xue, Y.; Murphy, A.; Frendewey, D.; Valenzuela, D.; Kutok, J.L.; et al. Regulation of the germinal center response by microRNA-155. Science 2007, 316, 604–608. [Google Scholar] [CrossRef]
- Yin, Q.; Wang, X.; McBride, J.; Fewell, C.; Flemington, E. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J. Biol. Chem. 2008, 283, 2654–2662. [Google Scholar] [CrossRef] [Green Version]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, A.; Vigorito, E.; Clare, S.; Warren, M.V.; Couttet, P.; Soond, D.R.; van Dongen, S.; Grocock, R.J.; Das, P.P.; Miska, E.A.; et al. Requirement of bic/microRNA-155 for normal immune function. Science 2007, 316, 608–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigorito, E.; Perks, K.L.; Abreu-Goodger, C.; Bunting, S.; Xiang, Z.; Kohlhaas, S.; Das, P.P.; Miska, E.A.; Rodriguez, A.; Bradley, A.; et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007, 27, 847–859. [Google Scholar] [CrossRef] [Green Version]
- Vigorito, E.; Kohlhaas, S.; Lu, D.; Leyland, R. miR-155: An ancient regulator of the immune system. Immunol. Rev. 2013, 253, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Schambach, F.; DeJong, C.S.; Hammond, S.M.; Reiner, S.L. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur. J. Immunol. 2010, 40, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Agbu, P.; Carthew, R.W. MicroRNA-mediated regulation of glucose and lipid metabolism. Nat. Rev. Mol. Cell Biol. 2021, 22, 425–438. [Google Scholar] [CrossRef] [PubMed]
- Klöting, N.; Berthold, S.; Kovacs, P.; Schön, M.R.; Fasshauer, M.; Ruschke, K.; Stumvoll, M.; Blüher, M. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS ONE 2009, 4, e4699. [Google Scholar] [CrossRef] [Green Version]
- Padmanaban, V.; Prakash, S.S. Strain, diet, and gender influence the role of miR155 in Diabetes mellitus. Indian J. Endocrinol. Metab. 2018, 22, 570–572. [Google Scholar]
- Zhang, B.H.; Shen, C.A.; Zhu, B.W.; An, H.Y.; Zheng, B.; Xu, S.B.; Sun, J.C.; Sun, P.C.; Zhang, W.; Wang, J.; et al. Insight into miRNAs related with glucometabolic disorder. Biomed. Pharm. 2019, 111, 657–665. [Google Scholar] [CrossRef]
- Lin, X.; Qin, Y.; Jia, J.; Lin, T.; Lin, X.; Chen, L.; Zeng, H.; Han, Y.; Wu, L.; Huang, S.; et al. MiR-155 enhances insulin sensitivity by coordinated regulation of multiple genes in mice. PLoS Genet. 2016, 12, e1006308. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Jia, J.; Du, T.; Li, W.; Wang, X.; Wei, J.; Lin, X.; Zeng, H.; Yao, L.; Chen, X.; et al. Overexpression of miR-155 in the liver of transgenic mice alters the expression profiling of hepatic genes associated with lipid metabolism. PLoS ONE 2015, 10, e0118417. [Google Scholar] [CrossRef] [Green Version]
- Tili, E.; Michaille, J.J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007, 179, 5082–5089. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, Y.; Li, L.; Su, B.; Yang, L.; Fan, W.; Yin, Q.; Chen, L.; Cui, T.; Zhang, J.; et al. Involvement of inflammation-related miR-155 and miR-146a in diabetic nephropathy: Implications for glomerular endothelial injury. BMC Nephrol. 2014, 15, 142. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Cui, Y.; Li, B.; Luo, X.; Li, B.; Tang, Y. miR-155 regulates high glucose-induced cardiac fibrosis via the TGF-β signaling pathway. Mol. Biosyst. 2016, 13, 215–224. [Google Scholar] [CrossRef]
- Johnson, C.; Drummer, C., 4th; Virtue, A.; Gao, T.; Wu, S.; Hernandez, M.; Singh, L.; Wang, H.; Yang, X.F. Increased expression of resistin in microRNA-155-deficient white adipose tissues may be a possible driver of metabolically healthy obesity transition to classical obesity. Front. Physiol. 2018, 9, 1297. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Zou, M.; Li, P.; Liu, H. MicroRNA regulation of energy metabolism to induce chemoresistance in cancers. Technol. Cancer Res. Treat. 2018, 17, 1533033818805997. [Google Scholar] [CrossRef] [PubMed]
- Nejad, C.; Stunden, H.J.; Gantier, M.P. A guide to miRNAs in inflammation and innate immune responses. FEBS J. 2018, 285, 3695–3716. [Google Scholar] [CrossRef]
- Salimi, V.; Ramezani, A.; Mirzaei, H.; Tahamtan, A.; Faghihloo, E.; Rezaei, F.; Naseri, M.; Bont, L.; Mokhtari-Azad, T.; Tavakoli-Yaraki, M. Evaluation of the expression level of 12/15 lipoxygenase and the related inflammatory factors (CCL5, CCL3) in respiratory syncytial virus infection in mice model. Microb. Pathog. 2017, 109, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, X.; Wu, M. miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct. Target. Ther. 2018, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Q.; Wu, R.; Zhao, M.; Garcia-Gomez, A.; Ballestar, E. miRNAs as therapeutic targets in inflammatory disease. Trends Pharm. Sci. 2019, 40, 853–865. [Google Scholar] [CrossRef]
- Nazari-Jahantigh, M.; Wei, Y.; Noels, H.; Akhtar, S.; Zhou, Z.; Koenen, R.R.; Heyll, K.; Gremse, F.; Kiessling, F.; Grommes, J.; et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J. Clin. Investig. 2012, 122, 4190–4202. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Guo, R.; Shi, Y.; Qi, F.; Guo, C.; Yang, L. miR-155 regulated inflammation response by the SOCS1-STAT3-PDCD4 axis in atherogenesis. Mediat. Inflamm. 2016, 2016, 8060182. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, Z.; Xiao, q.; Hu, H.; Tian, S.Y.; Lu, Z.J.; Zhang, T.Z.; Bai, Y.L. Down-regulation of microRNA-155 attenuates retinal neovascularization via the PI3K/Akt pathway. Mol. Vis. 2015, 21, 1173–1184. [Google Scholar] [PubMed]
- Huang, R.S.; Hu, G.Q.; Lin, B.; Lin, Z.Y.; Sun, C.C. MicroRNA-155 silencing enhances inflammatory response and lipid uptake in oxidized low-density lipoprotein-stimulated human THP-1 macrophages. J. Investig. Med. 2010, 58, 961–967. [Google Scholar] [CrossRef]
- Du, F.; Yu, F.; Wang, Y.; Hui, Y.; Carnevale, K.; Fu, M.; Lu, H.; Fan, D. MicroRNA-155 deficiency results in decreased macrophage inflammation and attenuated atherogenesis in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Arango, D.; Diosa-Toro, M.; Rojas-Hernandez, L.S.; Cooperstone, J.L.; Schwartz, S.J.; Mo, X.; Jiang, J.; Schmittgen, T.D.; Doseff, A.I. Dietary apigenin reduces LPS-induced expression of miR-155 restoring immune balance during inflammation. Mol. Nutr. Food Res. 2015, 59, 763–772. [Google Scholar] [CrossRef]
- Louafi, F.; Martinez-Nunez, R.T.; Sanchez-Elsner, T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta}. J. Biol. Chem. 2010, 285, 41328–41336. [Google Scholar] [CrossRef] [Green Version]
- Min, M.; Peng, L.; Yang, Y.; Guo, M.; Wang, W.; Sun, G. MicroRNA-155 is involved in the pathogenesis of ulcerative colitis by targeting FOXO3a. Inflamm. Bowel Dis. 2014, 20, 652–659. [Google Scholar] [CrossRef]
- Zhu, M.; Wei, Y.; Geißler, C.; Abschlag, K.; Corbalán Campos, J.; Hristov, M.; Möllmann, J.; Lehrke, M.; Karshovska, E.; Schober, A. Hyperlipidemia-induced microRNA-155-5p improves β-cell function by targeting Mafb. Diabetes 2017, 66, 3072–3084. [Google Scholar] [CrossRef] [Green Version]
- Lv, X.; Yao, L.; Zhang, J.; Han, P.; Li, C. Inhibition of microRNA-155 sensitizes lung cancer cells to irradiation via suppression of HK2-modulated glucose metabolism. Mol. Med. Rep. 2016, 14, 1332–1338. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, Y.; Kakoi, K.; Kimura, A.; Takada, I.; Kashiwagi, I.; Wakabayashi, Y.; Morita, R.; Nomura, M.; Yoshimura, A. Smad2 and Smad3 are redundantly essential for the suppression of iNOS synthesis in macrophages by regulating IRF3 and STAT1 pathways. Int. Immunol. 2012, 24, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Boesch-Saadatmandi, C.; Loboda, A.; Wagner, A.E.; Stachurska, A.; Jozkowicz, A.; Dulak, J.; Döring, F.; Wolffram, S.; Rimbach, G. Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: Role of miR-155. J. Nutr. Biochem. 2011, 22, 293–299. [Google Scholar] [CrossRef]
- Kocic, H.; Damiani, G.; Stamenkovic, B.; Tirant, M.; Jovic, A.; Tiodorovic, D.; Peris, K. Dietary compounds as potential modulators of microRNA expression in psoriasis. Ther. Adv. Chronic Dis. 2019, 10, 2040622319864805. [Google Scholar] [CrossRef] [Green Version]
- Tomé-Carneiro, J.; Larrosa, M.; Yáñez-Gascón, M.J.; Dávalos, A.; Gil-Zamorano, J.; Gonzálvez, M.; García-Almagro, F.J.; Ruiz Ros, J.A.; Tomás-Barberán, F.A.; Espín, J.C.; et al. One-year supplementation with a grape extract containing resveratrol modulates inflammatory-related microRNAs and cytokines expression in peripheral blood mononuclear cells of type 2 diabetes and hypertensive patients with coronary artery disease. Pharm. Res. 2013, 72, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Scalbert, A.; Morand, C.; Rémésy, C.; Jiménez, L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moens, B.; Decanine, D.; Menezes, S.M.; Khouri, R.; Silva-Santos, G.; Lopez, G.; Alvarez, C.; Talledo, M.; Gotuzzo, E.; de Almeida Kruschewsky, R.; et al. Ascorbic acid has superior ex vivo antiproliferative, cell death-inducing and immunomodulatory effects over IFN-α in HTLV-1-associated myelopathy. PLoS Negl. Trop. Dis. 2012, 6, e1729. [Google Scholar] [CrossRef] [Green Version]
- Cianciulli, A.; Calvello, R.; Porro, C.; Trotta, T.; Salvatore, R.; Panaro, M.A. PI3k/Akt signalling pathway plays a crucial role in the anti-inflammatory effects of curcumin in LPS-activated microglia. Int. Immunopharmacol. 2016, 36, 282–290. [Google Scholar] [CrossRef]
- Shojania, H.R.; Momeni-Moghaddam, M.; Hossini, S.E.; Armin, M.; Omrani Bidi, J. MicroRNA 155 downregulation by vitamin C-loaded human serum albumin nanoparticles during cutaneous wound healing in mice. Int. J. Low. Extrem. Wounds 2019, 18, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, N.; Talcott, S.; Safe, S.; Mertens-Talcott, S.U. Cytotoxicity of pomegranate polyphenolics in breast cancer cells in vitro and vivo: Potential role of miRNA-27a and miRNA-155 in cell survival and inflammation. Breast Cancer Res. Treat. 2012, 136, 21–34. [Google Scholar] [CrossRef]
- Ma, K.; Xu, W.; Wang, C.; Li, B.; Su, K.; Li, W. Vitamin D deficiency is associated with a poor prognosis in advanced non-small cell lung cancer patients treated with platinum-based first-line chemotherapy. Cancer Biomark. Sect. A 2017, 18, 297–303. [Google Scholar] [CrossRef]
- Karkeni, E.; Bonnet, L.; Marcotorchino, J.; Tourniaire, F.; Astier, J.; Ye, J.; Landrier, J.F. Vitamin D limits inflammation-linked microRNA expression in adipocytes in vitro and in vivo: A new mechanism for the regulation of inflammation by vitamin D. Epigenetics 2018, 13, 156–162. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Liu, W.; Sun, T.; Huang, Y.; Wang, Y.; Deb, D.K.; Yoon, D.; Kong, J.; Thadhani, R.; Li, Y.C. 1,25-Dihydroxyvitamin D promotes negative feedback regulation of TLR signaling via targeting microRNA-155-SOCS1 in macrophages. J. Immunol. 2013, 190, 3687–3695. [Google Scholar] [CrossRef] [Green Version]
- Arboleda, J.F.; Fernandez, G.J.; Urcuqui-Inchima, S. Vitamin D-mediated attenuation of miR-155 in human macrophages infected with dengue virus: Implications for the cytokine response. Infect. Genet. Evol. 2019, 69, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Roessler, C.; Kuhlmann, K.; Hellwing, C.; Leimert, A.; Schumann, J. Impact of polyunsaturated fatty acids on miRNA profiles of monocytes/macrophages and endothelial cells—A pilot study. Int. J. Mol. Sci. 2017, 18, 284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogero, M.M.; Calder, P.C. Obesity, inflammation, toll-like receptor 4 and fatty acids. Nutrients 2018, 10, 432. [Google Scholar] [CrossRef] [Green Version]
- Marques-Rocha, J.L.; Garcia-Lacarte, M.; Samblas, M.; Bressan, J.; Martínez, J.A.; Milagro, F.I. Regulatory roles of miR-155 and let-7b on the expression of inflammation-related genes in THP-1 cells: Effects of fatty acids. J. Physiol. Biochem. 2018, 74, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Kim, S.; Patterson, N.; Rooney, K.; Searles, C.D. MiRNA-155 targets myosin light chain kinase and modulates actin cytoskeleton organization in endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H1192–H1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, T.S.; Chang, C.M.; Ho, H.C.; Su, Y.C.; Chen, L.F.; Chou, P.; Lee, C.C. Impact of young age on the prognosis for oral cancer: A population-based study in Taiwan. PLoS ONE 2013, 8, e75855. [Google Scholar] [CrossRef]
- De Santis, R.; Liepelt, A.; Mossanen, J.C.; Dueck, A.; Simons, N.; Mohs, A.; Trautwein, C.; Meister, G.; Marx, G.; Ostareck-Lederer, A.; et al. miR-155 targets Caspase-3 mRNA in activated macrophages. RNA Biol. 2016, 13, 43–58. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Xie, M.; Wang, X.; Jiang, X.; Li, J.; Huang, H. miR-155 modulates TNF-α-inhibited osteogenic differentiation by targeting SOCS1 expression. Bone 2012, 51, 498–505. [Google Scholar] [CrossRef]
- Liu, H.; Zhong, L.; Yuan, T.; Chen, S.; Zhou, Y.; An, L.; Guo, Y.; Fan, M.; Li, Y.; Sun, Y.; et al. MicroRNA-155 inhibits the osteogenic differentiation of mesenchymal stem cells induced by BMP9 via downregulation of BMP signaling pathway. Int. J. Mol. Med. 2018, 41, 3379–3393. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, W.; Li, X.; He, S.; Yao, J.; Wang, X.; Zhang, D.; Sun, X. MicroRNA-155 promotes tumor growth of human hepatocellular carcinoma by targeting ARID2. Int. J. Oncol. 2016, 48, 2425–2434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, L.; Chen, J.; Zheng, Y.; Wu, C. Critical role of miR-155/FoxO1/ROS axis in the regulation of non-small cell lung carcinomas. Tumour Biol. 2016, 37, 5185–5192. [Google Scholar] [CrossRef]
- Baba, O.; Hasegawa, S.; Nagai, H.; Uchida, F.; Yamatoji, M.; Kanno, N.I.; Yamagata, K.; Sakai, S.; Yanagawa, T.; Bukawa, H. MicroRNA-155-5p is associated with oral squamous cell carcinoma metastasis and poor prognosis. J. Oral Pathol. Med. 2016, 45, 248–255. [Google Scholar] [CrossRef] [Green Version]
- Slezak-Prochazka, I.; Kluiver, J.; de Jong, D.; Smigielska-Czepiel, K.; Kortman, G.; Winkle, M.; Rutgers, B.; Koerts, J.; Visser, L.; Diepstra, A.; et al. Inhibition of the miR-155 target NIAM phenocopies the growth promoting effect of miR-155 in B-cell lymphoma. Oncotarget 2016, 7, 2391–2400. [Google Scholar] [CrossRef] [Green Version]
- Zang, Y.S.; Zhong, Y.F.; Fang, Z.; Li, B.; An, J. MiR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf-1 expression. Cancer Gene Ther. 2012, 19, 773–778. [Google Scholar] [CrossRef] [Green Version]
- Shen, R.; Wang, Y.; Wang, C.X.; Yin, M.; Liu, H.L.; Chen, J.P.; Han, J.Q.; Wang, W.B. MiRNA-155 mediates TAM resistance by modulating SOCS6-STAT3 signalling pathway in breast cancer. Am. J. Transl. Res. 2015, 7, 2115–2126. [Google Scholar] [PubMed]
- Katagiri, M.; Karasawa, H.; Takagi, K.; Nakayama, S.; Yabuuchi, S.; Fujishima, F.; Naitoh, T.; Watanabe, M.; Suzuki, T.; Unno, M.; et al. Hexokinase 2 in colorectal cancer: A potent prognostic factor associated with glycolysis, proliferation and migration. Histol. Histopathol. 2017, 32, 351–360. [Google Scholar] [PubMed]
- Chen, J.; Zhang, S.; Li, Y.; Tang, Z.; Kong, W. Hexokinase 2 overexpression promotes the proliferation and survival of laryngeal squamous cell carcinoma. Tumour Biol. 2014, 35, 3743–3753. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, L.F.; Zhang, H.W.; Hu, S.; Lu, M.H.; Liang, S.; Li, B.; Li, Y.; Li, D.; Wang, E.D.; et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012, 31, 1985–1998. [Google Scholar] [CrossRef] [Green Version]
- Natsuka, S.; Akira, S.; Nishio, Y.; Hashimoto, S.; Sugita, T.; Isshiki, H.; Kishimoto, T. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood 1992, 79, 460–466. [Google Scholar] [CrossRef] [Green Version]
- Davydov, I.V.; Krammer, P.H.; Li-Weber, M. Nuclear factor-IL6 activates the human IL-4 promoter in T cells. J. Immunol. 1995, 155, 5273–5279. [Google Scholar]
- van Dijk, T.B.; Baltus, B.; Raaijmakers, J.A.; Lammers, J.W.; Koenderman, L.; de Groot, R.P. A composite C/EBP binding site is essential for the activity of the promoter of the IL-3/IL-5/granulocyte-macrophage colony-stimulating factor receptor beta c gene. J. Immunol. 1999, 163, 2674–2680. [Google Scholar] [PubMed]
- Rostami, Z.; Khorashadizadeh, M.; Ghoncheh, M.; Naseri, M. Effect of pomegranate extract in mesenchymal stem cells by modulation of microRNA-155, microRNA-21, microRNA-23b, microRNA-126a, and PI3K\AKT1\NFĸ-B expression. DNA Cell Biol. 2020, 39, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- de la Parra, C.; Castillo-Pichardo, L.; Cruz-Collazo, A.; Cubano, L.; Redis, R.; Calin, G.A.; Dharmawardhane, S. Soy isoflavone genistein-mediated downregulation of miR-155 contributes to the anticancer effects of genistein. Nutr. Cancer 2016, 68, 154–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rokah, O.H.; Granot, G.; Ovcharenko, A.; Modai, S.; Pasmanik-Chor, M.; Toren, A.; Shomron, N.; Shpilberg, O. Downregulation of miR-31, miR-155, and miR-564 in chronic myeloid leukemia cells. PLoS ONE 2012, 7, e35501. [Google Scholar]
- Wei, R.J.; Zhang, C.H.; Yang, W.Z. MiR-155 affects renal carcinoma cell proliferation, invasion and apoptosis through regulating GSK-3β/β-catenin signaling pathway. Eur. Rev. Med. Pharm. Sci. 2017, 21, 5034–5041. [Google Scholar]
- Liu, J.; Chen, Z.; Xiang, J.; Gu, X. MicroRNA-155 acts as a tumor suppressor in colorectal cancer by targeting CTHRC1 in vitro. Oncol. Lett. 2018, 15, 5561–5568. [Google Scholar] [CrossRef]
- Rawat, M.; Kadian, K.; Gupta, Y.; Kumar, A.; Chain, P.S.G.; Kovbasnjuk, O.; Kumar, S.; Parasher, G. MicroRNA in pancreatic cancer: From biology to therapeutic potential. Genes 2019, 10, 752. [Google Scholar] [CrossRef] [Green Version]
- Vargova, K.; Pesta, M.; Obrtlikova, P.; Dusilkova, N.; Minarik, L.; Vargova, J.; Berkova, A.; Zemanova, Z.; Michalova, K.; Spacek, M.; et al. MiR-155/miR-150 network regulates progression through the disease phases of chronic lymphocytic leukemia. Blood Cancer J. 2017, 7, e585. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.; Xu, Y.; Shen, L.; Huang, H.; Yu, X.; Lu, C.; Zhong, C. MiR-155 promotes proliferation of human non-small cell lung cancer H460 cells via targeting TP53INP1. Int. J. Clin. Exp. Med. 2017, 10, 11953–11960. [Google Scholar]
- Liu, F.; Song, D.; Wu, Y.; Liu, X.; Zhu, J.; Tang, Y. MiR-155 inhibits proliferation and invasion by directly targeting PDCD4 in non-small cell lung cancer. Thorac. Cancer 2017, 8, 613–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, X.; Liu, Y.; Wang, Y.; Meng, M.; Wang, K.; Zang, X.; Zhao, S.; Sun, X.; Cui, L.; Pan, L.; et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget 2016, 7, 84508–84519. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Dietary Compound | Study Type | Dose | MiR-155 Expression Level | MiR-155 Targets | Biological Effects | Reference |
|---|---|---|---|---|---|---|
| Allyl-isothiocyanate | In vitro: RAW264.7 macrophages | 1–10 μM | ↓ | ↓Nrf2, HO, p65 | ↓Inflammation | [81] |
| In vivo: C57BL/6 mice | 15 mg/kg | ↓ | ↓Nrf2, HO, p65 | ↓Inflammation | [81] | |
| Quercetin | In vitro: RAW264.7 cells | 25–100 μM | ↓ | ↓TNF-α | ↓Inflammation | [121] |
| Resveratrol | Clinical study | 8 mg/day | ↓ | ↓TNF-α | ↓Inflammation | [123] |
| Curcumin | In vitro/BV2 microglial cells | 50 μM | ↓ | ↓PI3K, p85a, and AKT | ↓Inflammation | [126] |
| Apigenin | In vivo/Male C57BL/6J mice | 50 mg/kg | ↓ | ↓TNF-α | ↓Inflammation | [115] |
| Pomegranate polyphenolics | In vivo/Female athymic BALB/c nude mice | 0.8 mg gallic acid equivalent (GAE)/kg/day | ↓ | ↓NF-kB | ↓Inflammation | [128] |
| Vitamin C | Clinical | 1250 mg/day | ↓ | ↓ROS | ↓Inflammation ↑Antioxidant activity | [45] |
| In vitro/MT-2 cells | 100 µg/mL | ↓ | ↑IFN-γ | ↑Antiproliferative and immunomodulatory anti-HTLV-1 effects | [125] | |
| In vivo | 20 µg/mL | ↓ | ↑TGF-β1 and SMAD 1,2 | ↑Wound healing rate | [127] | |
| 1,25-dihydroxy-vitamin D | In vivo/mice | 20 nM | ↓ | ↓NF-κB | ↓Inflammation, innate immunity | [131] |
| In vivo/C57BL/6J mice | 3000 IU/kg of body weight | ↓ | ↓NF-κB | ↓Inflammation | [130] | |
| In vitro/MDM cells | 0.1 nM | ↓ | ↑SOCS-1 | ↓Inflammation | [24] | |
| PUFAs | In vitro/RAW264.7 cells | 15 µmol/L | ↓ | ↓TLR | ↓Inflammation | [133] |
| Oleic acid | In vitro/THP-1 cells | 100 μM | ↓ | ↓TLR4 | ↓ Inflammation | [135] |
| Type of Cancer | MiR-155 Expression Levels in Cancer | Targets | Reference |
|---|---|---|---|
| Colon cancer | ↑ | ↓CTHRC1 | [157] |
| Pancreatic cancer | ↑ | ↓SOCS1 | [158] |
| Breast cancer | ↑ | ↓PIK3R1 and FOXO3a | [77] |
| Chronic lymphocytic leukemia | ↑ | ↓PU.1 | [159] |
| Lung cancer | ↑ | ↓TP53INP1 | [160] |
| ↑ | ↓PDCD4 | [161] | |
| ↑ | ↓SOCS1, SOCS6, and PTEN | [162] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanoaga, O.; Braicu, C.; Chiroi, P.; Andreea, N.; Hajjar, N.A.; Mărgărit, S.; Korban, S.S.; Berindan-Neagoe, I. The Role of miR-155 in Nutrition: Modulating Cancer-Associated Inflammation. Nutrients 2021, 13, 2245. https://doi.org/10.3390/nu13072245
Zanoaga O, Braicu C, Chiroi P, Andreea N, Hajjar NA, Mărgărit S, Korban SS, Berindan-Neagoe I. The Role of miR-155 in Nutrition: Modulating Cancer-Associated Inflammation. Nutrients. 2021; 13(7):2245. https://doi.org/10.3390/nu13072245
Chicago/Turabian StyleZanoaga, Oana, Cornelia Braicu, Paul Chiroi, Nutu Andreea, Nadim Al Hajjar, Simona Mărgărit, Schuyler S. Korban, and Ioana Berindan-Neagoe. 2021. "The Role of miR-155 in Nutrition: Modulating Cancer-Associated Inflammation" Nutrients 13, no. 7: 2245. https://doi.org/10.3390/nu13072245








