Characteristics of the Proteolytic Enzymes Produced by Lactic Acid Bacteria
Abstract
1. Introduction
2. Characteristics of Lactic Acid Bacteria
3. Enzymatic Activities of Proteases
- -
- aminopeptidases (EC 3.4.11), dipeptidyl peptidases (EC 3.4.14), and tripeptide peptidases (EC 3.4.14), which cleave one, two, or three amino acid residues from the N-terminus;
- -
- Ep tripeptidases (EC 3.4.11), – which remove a terminal amino acid from a tripeptide;
- -
- carboxypeptidases (EC 3.4.12): serine (EC 3.4.16), metallocarboxypeptidase (EC 3.4.17), and cysteine (EC 3.4.18), which remove one amino acid residue from the C-terminus;
- -
- dipeptidases (EC 3.4.13), which hydrolyze the peptide bond in the dipeptide;
- -
- peptidyl dipeptidases (EC 3.4.15), which remove two residues from the C-terminus,
- -
4. Nitrogen Metabolism of Bacteria—Proteolysis
| Enzymes. | Microorganism | Substrate | Reference |
|---|---|---|---|
| PrtP | Lacticaseibacillus rhamnosus CGMCC11055 | β-casein | [49] |
| - | Limosilactobacillus fermentum R6 | casein | [8] |
| PrtR | Lacticaseibacillus rhamnosus OXY | azocasein | [29] |
| PrtP | Lacticaseibacillus paracasei BL312(VSL#3) | casein | [50] |
| - | Lacticaseibacillus casei PRA205 | β-, αs1-, κ-, αs2-caseins | [51] |
| PrtP | Lactiplantibacillus plantarum BGSJ3–18 | β-casein | [52] |
| PrtS | Streptococcus thermophilus 4F44 | β-casein | [53] |
| PrtS | Streptococcus thermophilus | β-, αs1- and αs2-CN | [54] |
| PrtB | Lactobacillus delbrueckii subsp. bulgaricus 92059 | β-, α-casein | [55] |
| PrtL | Lactobacillus delbrueckii subsp. lactis CRL 581 | αs1-, β-, and κ-casein | [56] |
| PrtH | Lactobacillus helveticus CNRZ32 | casein | [57] |
| PrtR | Lacticaseibacillus rhamnosus BGT10 | β-casein | [58] |
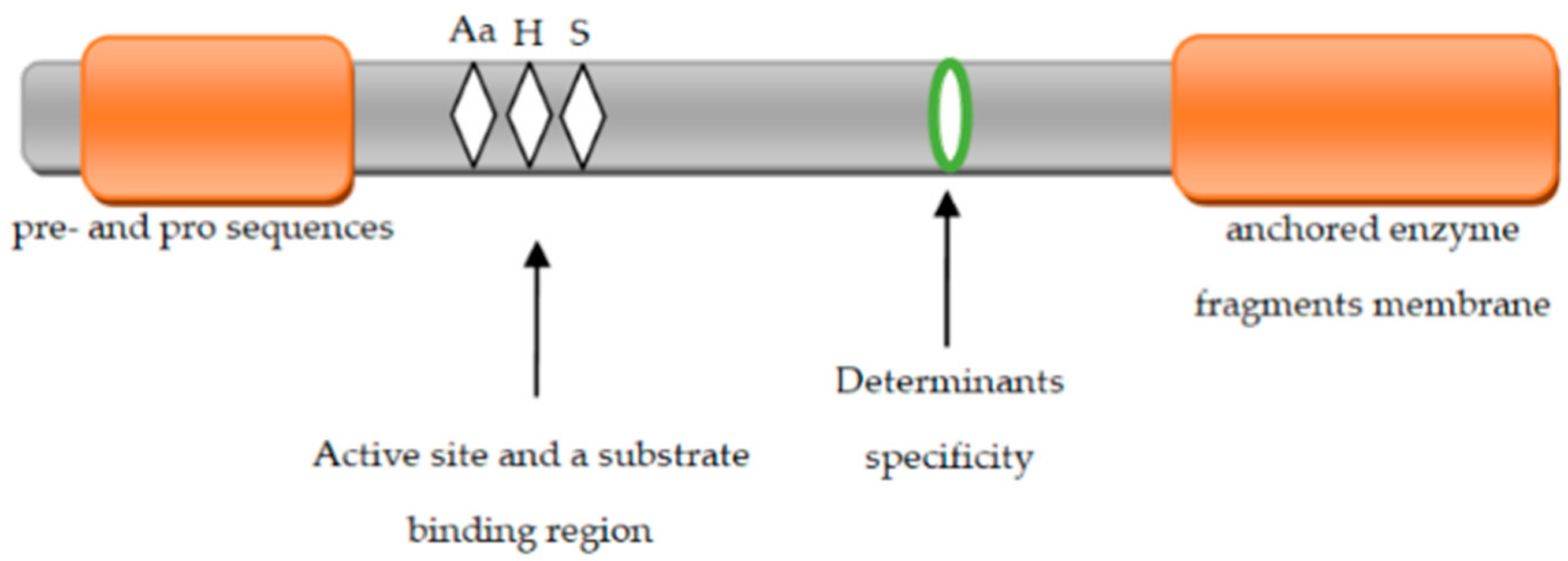
5. Structure of Extracellular Proteinases
6. Peptide Transport System
7. Regulation of the Proteolytic System
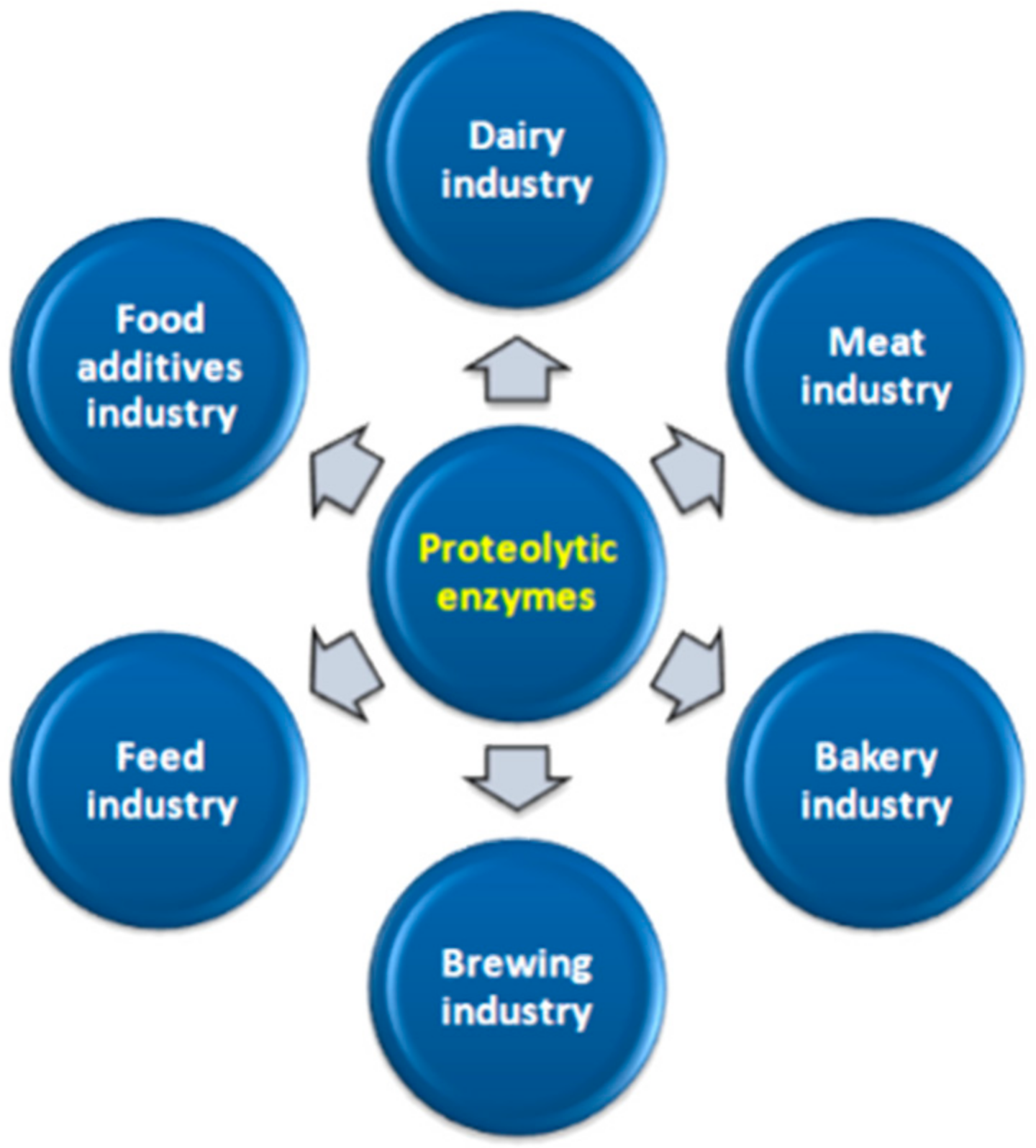
8. Industrial Use of Proteolytic Enzymes
8.1. Dairy Industry
8.2. Meat Industry
8.3. Bakery Industry
8.4. Brewing Industry
8.5. Food Additives Industry
8.6. Feed Industry
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sandoval, B.A.; Hyster, T.K. Emerging strategies for expanding the toolbox of enzymes in biocatalysis. Curr. Opin. Chem. Biol. 2020, 55, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Abd Latip, M.A.; Abdul Hamid, A.A.; Nordin, N.F.H. Microbial hydrolytic enzymes: In silico studies between polar and tropical regions. Polar Sci. 2019, 20, 9–18. [Google Scholar] [CrossRef]
- Do, T.; Page, J.E.; Walker, S. Uncovering the activities, biological roles, and regulation of bacterial cell wall hydrolases and tailoring enzymes. J. Biol. Chem. 2020, 295, 3347–3361. [Google Scholar] [CrossRef] [PubMed]
- Savijoki, K.; Ingmer, H.; Varmanen, P. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 2006, 71, 394–406. [Google Scholar] [CrossRef]
- Kenny, O.; FitzGerald, R.J.; O’Cuinn, G.; Beresford, T.; Jordan, K. Growth phase and growth medium effects on the peptidase activities of Lactobacillus helveticus. Int. Dairy J. 2003, 13, 509–516. [Google Scholar] [CrossRef]
- Razzaq, A.; Shamsi, S.; Ali, A.; Ali, Q.; Sajjad, M.; Malik, A.; Ashraf, M. Microbial proteases applications. Front. Bioeng. Biotechnol. 2019, 7, 110. [Google Scholar] [CrossRef]
- Sharma, M.; Gat, Y.; Arya, S.; Kumar, V.; Panghal, A.; Kumar, A. A review on microbial alkaline protease: An essential tool for various industrial approaches. Ind. Biotechnol. 2019, 15, 69–78. [Google Scholar] [CrossRef]
- Sun, F.; Hu, Y.; Yin, X.; Kong, B.; Qin, L. Production, purification and biochemical characterization of the microbial protease produced by Lactobacillus fermentum R6 isolated from Harbin dry sausages. Process Biochem. 2020, 89, 37–45. [Google Scholar] [CrossRef]
- García-Cano, I.; Rocha-Mendoza, D.; Ortega-Anaya, J.; Wang, K.; Kosmerl, E.; Jiménez-Flores, R. Lactic acid bacteria isolated from dairy products as potential producers of lipolytic, proteolytic and antibacterial proteins. Appl. Microbiol. Biotechnol. 2019, 103, 5243–5257. [Google Scholar] [CrossRef]
- Sun, F.; Sun, Q.; Zhang, H.; Kong, B.; Xia, X. Purification and biochemical characteristics of the microbial extracellular protease from Lactobacillus curvatus isolated from Harbin dry sausages. Int. J. Biol. Macromol. 2019, 133, 987–997. [Google Scholar] [CrossRef]
- Matti, A.; Utami, T.; Hidayat, C.S.; Rahayu, E. Isolation, Screening, and identification of proteolytic lactic acid bacteria from indigenous Chao product. J. Aquat. Food Prod. Technol. 2019, 28, 781–793. [Google Scholar] [CrossRef]
- Othman, M.; Ariff, A.B.; Rios-Solis, L.; Halim, M. Extractive Fermentation of lactic acid in lactic acid bacteria cultivation: A review. Front. Microbiol. 2017, 8, 2285. [Google Scholar] [CrossRef] [PubMed]
- Zielińska, D.; Kolozyn-Krajewska, D. Food-origin lactic acid bacteria may exhibit probiotic properties: Review. Biomed Res. Int. 2018, 2018, 5063185. [Google Scholar] [CrossRef]
- Kavitake, D.; Kandasamy, S.; Devi, P.B.; Shetty, P.H. Recent developments on encapsulation of lactic acid bacteria as potential starter culture in fermented foods–A review. Food Biosci. 2018, 21, 34–44. [Google Scholar] [CrossRef]
- Chlebowska-Smigiel, A.; Gniewosz, M.; Kieliszek, M.; Bzducha-Wrobel, A. The Effect of pullulan on the growth and acidifying activity of selected stool microflora of human. Curr. Pharm. Biotechnol. 2017, 18, 121–126. [Google Scholar] [CrossRef]
- Chlebowska-Śmigiel, A.; Kycia, K.; Neffe-Skocińska, K.; Kieliszek, M.; Gniewosz, M.; Kołożyn-Krajewska, D. Effect of pullulan on physicochemical, microbiological, and sensory quality of yogurts. Curr. Pharm. Biotechnol. 2019, 20, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Janiszewska-Turak, E.; Hornowska, Ł.; Pobiega, K.; Gniewosz, M.; Witrowa-Rajchert, D. The influence of Lactobacillus bacteria type and kind of carrier on the properties of spray-dried microencapsules of fermented beetroot powders. Int. J. Food Sci. Technol. 2020, in press. [Google Scholar] [CrossRef]
- Cirlini, M.; Ricci, A.; Galaverna, G.; Lazzi, C. Application of lactic acid fermentation to elderberry juice: Changes in acidic and glucidic fractions. LWT 2020, 118, 108779. [Google Scholar] [CrossRef]
- Wu, C.; Li, T.; Qi, J.; Jiang, T.; Xu, H.; Lei, H. Effects of lactic acid fermentation-based biotransformation on phenolic profiles, antioxidant capacity and flavor volatiles of apple juice. LWT 2020, 122, 109064. [Google Scholar] [CrossRef]
- Philippe, C.; Chaïb, A.; Jaomanjaka, F.; Cluzet, S.; Lagarde, A.; Ballestra, P.; Decendit, A.; Petrel, M.; Claisse, O.; Goulet, A.; et al. Wine phenolic compounds differently affect the host-killing activity of two lytic bacteriophages infecting the lactic acid bacterium Oenococcus oeni. Viruses 2020, 12, 1316. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W.H.; Haberer, P.; Geisen, R.; Björkroth, J.; Schillinger, U. Taxonomy and important features of probiotic microorganisms in food and nutrition. Am. J. Clin. Nutr. 2001, 73, 365S–373S. [Google Scholar] [CrossRef]
- Worsztynowicz, P.; Białas, W.; Grajek, W. Integrated approach for obtaining bioactive peptides from whey proteins hydrolysed using a new proteolytic lactic acid bacteria. Food Chem. 2020, 312, 126035. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Ma, J.; Xu, M.; Agyei, D. Cell-envelope proteinases from lactic acid bacteria: Biochemical features and biotechnological applications. Compr. Rev. Food Sci. Food Saf. 2021, 20, 369–400. [Google Scholar] [CrossRef]
- Linares-Morales, J.R.; Cuellar-Nevárez, G.E.; Rivera-Chavira, B.E.; Gutiérrez-Méndez, N.; Pérez-Vega, S.B.; Nevárez-Moorillón, G.V. Selection of lactic acid bacteria isolated from fresh fruits and vegetables based on their antimicrobial and enzymatic activities. Foods 2020, 9, 1399. [Google Scholar] [CrossRef]
- Cichosz, G.; Kornacki, M.; Giczewska, M.; Konopka, A. Aktywność peptydazowa wybranych szczepów Lactobacillus. Żywność Nauk. Technol. Jakość 2006, 46, 66–74. (In Polish) [Google Scholar]
- Saidi, Y.; del Rio, B.; Senouci, D.E.; Redruello, B.; Martinez, B.; Ladero, V.; Kihal, M.; Alvarez, M.A. Polyphasic characterisation of non-starter lactic acid bacteria from Algerian raw Camel’s milk and their technological aptitudes. Food Technol. Biotechnol. 2020, 58, 1–30. [Google Scholar] [CrossRef]
- Toe, C.J.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R.; Idrus, Z. Extracellular proteolytic activity and amino acid production by lactic acid bacteria isolated from Malaysian foods. Int. J. Mol. Sci. 2019, 20, 1777. [Google Scholar] [CrossRef]
- Lim, Y.H.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdullah, N. Comparative studies of versatile extracellular proteolytic activities of lactic acid bacteria and their potential for extracellular amino acid productions as feed supplements. J. Anim. Sci. Biotechnol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Waśko, A.; Kieliszek, M.; Targoński, Z. Purification and characterization of a proteinase from the probiotic Lactobacillus rhamnosus OXY. Prep. Biochem. Biotechnol. 2012, 42, 476–488. [Google Scholar] [CrossRef]
- Zhao, C.J.; Schieber, A.; Gänzle, M.G. Formation of taste-active amino acids, amino acid derivatives and peptides in food fermentations–A review. Food Res. Int. 2016, 89, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, R.; Altier, C.; Oikonomopoulou, K.; Hollenberg, M.D. Proteinases, their extracellular targets, and inflammatory signaling. Pharmacol. Rev. 2016, 68, 1110–1142. [Google Scholar] [CrossRef]
- Gurumallesh, P.; Alagu, K.; Ramakrishnan, B.; Muthusamy, S. A systematic reconsideration on proteases. Int. J. Biol. Macromol. 2019, 128, 254–267. [Google Scholar] [CrossRef]
- Mamo, J.; Assefa, F. The role of microbial aspartic protease enzyme in food and beverage industries. J. Food Qual. 2018, 2018, 7957269. [Google Scholar] [CrossRef]
- Sims, A.H.; Dunn-Coleman, N.S.; Robson, G.D.; Oliver, S.G. Glutamic protease distribution is limited to filamentous fungi. FEMS Microbiol. Lett. 2004, 239, 95–101. [Google Scholar] [CrossRef]
- Ewert, J.; Schlierenkamp, F.; Nesensohn, L.; Fischer, L.; Stressler, T. Improving the colloidal and sensory properties of a caseinate hydrolysate using particular exopeptidases. Food Funct. 2018, 9, 5989–5998. [Google Scholar] [CrossRef]
- Bansal, P.; Kumar, R.; Singh, J.; Dhanda, S. Next generation sequencing, biochemical characterization, metabolic pathway analysis of novel probiotic Pediococcus acidilactici NCDC 252 and it’s evolutionary relationship with other lactic acid bacteria. Mol. Biol. Rep. 2019, 46, 5883–5895. [Google Scholar] [CrossRef]
- Daliri, E.B.M.; Lee, B.H.; Park, B.J.; Kim, S.H.; Oh, D.H. Antihypertensive peptides from whey proteins fermented by lactic acid bacteria. Food Sci. Biotechnol. 2018, 27, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, R.T.; Badgujar, S.B. Biological aspects of proteolytic enzymes: A Review. J. Pharm. Res. 2010, 3, 2048–2068. [Google Scholar]
- Hernandez-Valdes, J.A.; aan de Stegge, M.; Hermans, J.; Teunis, J.; van Tatenhove-Pel, R.J.; Teusink, B.; Bachmann, H.; Kuipers, O.P. Enhancement of amino acid production and secretion by Lactococcus lactis using a droplet-based biosensing and selection system. Metab. Eng. Commun. 2020, 11, e00133. [Google Scholar] [CrossRef] [PubMed]
- Laroute, V.; Tormo, H.; Couderc, C.; Mercier-Bonin, M.; Le Bourgeois, P.; Cocaign-Bousquet, M.; Daveran-Mingot, M.-L. From genome to phenotype: An integrative approach to evaluate the biodiversity of Lactococcus lactis. Microorganisms 2017, 5, 27. [Google Scholar] [CrossRef]
- Gómez de Cadiñanos, L.P.; García-Cayuela, T.; Martínez-Cuesta, M.C.; Peláez, C.; Requena, T. Expression of amino acid converting enzymes and production of volatile compounds by Lactococcus lactis IFPL953. Int. Dairy J. 2019, 96, 29–35. [Google Scholar] [CrossRef]
- Tagliazucchi, D.; Martini, S.; Solieri, L. Bioprospecting for bioactive peptide production by lactic acid bacteria isolated from fermented dairy food. Fermentation 2019, 5, 96. [Google Scholar] [CrossRef]
- Hebert, E.M.; Raya, R.R.; De Giori, G.S. Nutritional requirements and nitrogen-dependent regulation of proteinase activity of Lactobacillus helveticus CRL 1062. Appl. Environ. Microbiol. 2000, 66, 5316–5321. [Google Scholar] [CrossRef]
- Von Schillde, M.A.; Hörmannsperger, G.; Weiher, M.; Alpert, C.A.; Hahne, H.; Bäuerl, C.; Van Huynegem, K.; Steidler, L.; Hrncir, T.; Pérez-Martínez, G.; et al. Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe 2012, 11, 387–396. [Google Scholar] [CrossRef]
- Kunji, E.R.S. The proteolytic systems of lactic acid bacteria. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 1996, 70, 187–221. [Google Scholar] [CrossRef]
- Reid, J.R.; Coolbear, T.; Moore, C.H.; Harding, D.R.; Pritchard, G.G. Involvement of enzyme-substrate charge interactions in the caseinolytic specificity of lactococcal cell envelope-associated proteinases. Appl. Environ. Microbiol. 1995, 61, 3934–3939. [Google Scholar] [CrossRef]
- Juillard, V.; Laan, H.; Kunji, E.R.S.; Jeronimus-Stratingh, C.M.; Bruins, A.P.; Konings, W.N. The extracellular P(I)-type proteinase of Lactococcus lactis hydrolyzes β-casein into more than one hundred different oligopeptides. J. Bacteriol. 1995, 177, 3472–3478. [Google Scholar] [CrossRef][Green Version]
- Pritchard, G.G.; Coolbear, T. The physiology and biochemistry of the proteolytic system in lactic acid bacteria. FEMS Microbiol. Rev. 1993, 12, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Ouyang, X.; Xin, Y.; Wang, Y.; Zhang, S.; Kong, J. Characterization of a New Cell Envelope Proteinase PrtP from Lactobacillus rhamnosus CGMCC11055. J. Agric. Food Chem. 2016, 64, 6985–6992. [Google Scholar] [CrossRef] [PubMed]
- Coll-Marqués, J.M.; Bäuerl, C.; Zúñiga, M.; Pérez-Martínez, G. Differences in the expression of cell envelope proteinases (CEP) in two Lactobacillus paracasei probiotic strains. FEMS Microbiol. Lett. 2020, 367, fnaa102. [Google Scholar] [CrossRef]
- Solieri, L.; De Vero, L.; Tagliazucchi, D. Peptidomic study of casein proteolysis in bovine milk by Lactobacillus casei PRA205 and Lactobacillus rhamnosus PRA331. Int. Dairy J. 2018, 85, 237–246. [Google Scholar] [CrossRef]
- Topisirovic, L.; Strahinic, I.; Kojic, M.; Tolinacki, M.; Fira, D. The presence of prtP proteinase gene in natural isolate Lactobacillus plantarum BGSJ3-18. Lett. Appl. Microbiol. 2010, 50, 43–49. [Google Scholar] [CrossRef]
- Chang, O.K.; Roux, É.; Awussi, A.A.; Miclo, L.; Jardin, J.; Jameh, N.; Dary, A.; Humbert, G.; Perrin, C. Use of a free form of the Streptococcus thermophilus cell envelope protease PrtS as a tool to produce bioactive peptides. Int. Dairy J. 2014, 38, 104–115. [Google Scholar] [CrossRef]
- Miclo, L.; Roux, É.; Genay, M.; Brusseaux, É.; Poirson, C.; Jameh, N.; Perrin, C.; Dary, A. Variability of hydrolysis of β-, α s1-, and α s2-caseins by 10 strains of Streptococcus thermophilus and resulting bioactive peptides. J. Agric. Food Chem. 2012, 60, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Habermann, D.; Kliche, T.; Klempt, M.; Wutkowski, A.; Clawin-Rädecker, I.; Koberg, S.; Brinks, E.; Koudelka, T.; Tholey, A.; et al. Soluble Lactobacillus delbrueckii subsp. bulgaricus 92059 PrtB proteinase derivatives for production of bioactive peptide hydrolysates from casein. Appl. Microbiol. Biotechnol. 2019, 103, 2731–2743. [Google Scholar] [CrossRef]
- Villegas, J.M.; Brown, L.; Savoy de Giori, G.; Hebert, E.M. Characterization of the mature cell surface proteinase of Lactobacillus delbrueckii subsp. lactis CRL 581. Appl. Microbiol. Biotechnol. 2015, 99, 4277–4286. [Google Scholar] [CrossRef]
- Genay, M.; Sadat, L.; Gagnaire, V.; Lortal, S. PrtH2, Not prtH, Is the ubiquitous cell wall proteinase gene in Lactobacillus helveticus. Appl. Environ. Microbiol. 2009, 75, 3238–3249. [Google Scholar] [CrossRef] [PubMed]
- Lozo, J.; Strahinic, I.; Dalgalarrondo, M.; Chobert, J.M.; Haertlé, T.; Topisirovic, L. Comparative analysis of β-casein proteolysis by PrtP proteinase from Lactobacillus paracasei subsp. paracasei BGHN14, PrtR proteinase from Lactobacillus rhamnosus BGT10 and PrtH proteinase from Lactobacillus helveticus BGRA43. Int. Dairy J. 2011, 21, 863–868. [Google Scholar] [CrossRef]
- Navarre, W.W.; Schneewind, O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol. Microbiol. 1994, 14, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Espla, M.D.; Garault, P.; Monnet, V.; Rul, F. Streptococcus thermophilus cell wall-anchored proteinase: Release, purification, and biochemical and genetic characterization. Appl. Environ. Microbiol. 2000, 66, 4772–4778. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie van Leeuwenhoek 1999, 76, 139–155. [Google Scholar] [CrossRef]
- Haandrikman, A.J.; Kok, J.; Venema, G. Lactococcal proteinase maturation protein PrtM is a lipoprotein. J. Bacteriol. 1991, 173, 4517–4525. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haandrikman, A.J.; Kok, J.; Laan, H.; Soemitro, S.; Ledeboer, A.M.; Konings, W.N.; Venema, G. Identification of a gene required for maturation of an extracellular lactococcal serine proteinase. J. Bacteriol. 1989, 171, 2789–2794. [Google Scholar] [CrossRef]
- Kok, J. Genetics of proteolytic enzymes of lactococci and their role in cheese flavor development. J. Dairy Sci. 1993, 76, 2056–2064. [Google Scholar] [CrossRef]
- Holck, A.; Naes, H. Cloning, sequencing and expression of the gene encoding the cell-envelope-associated proteinase from Lactobacillus paracasei subsp. paracasei NCDO 151. J. Gen. Microbiol. 1992, 138, 1353–1364. [Google Scholar] [CrossRef]
- Brown, L.; Pingitore, E.V.; Mozzi, F.; Saavedra, L.M.; Villegas, J.M.; Hebert, E. Lactic acid bacteria as cell factories for the generation of bioactive peptides. Protein Pept. Lett. 2017, 24, 146–155. [Google Scholar] [CrossRef]
- Vos, P.; Van Asseldonk, M.; Van Jeveren, F.; Siezen, R.; Simons, G.; De Vos, W.M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J. Bacteriol. 1989, 171, 2795–2802. [Google Scholar] [CrossRef] [PubMed]
- Bruinenberg, P.G.; Vos, P.; De Vos, W.M. Proteinase overproduction in Lactococcus lactis strains: Regulation and effect on growth and acidification in milk. Appl. Environ. Microbiol. 1992, 58, 78–84. [Google Scholar] [CrossRef]
- Wu, Q.; Shah, N.P. High γ-aminobutyric acid production from lactic acid bacteria: Emphasis on Lactobacillus brevis as a functional dairy starter. Crit. Rev. Food Sci. Nutr. 2017, 57, 3661–3672. [Google Scholar] [CrossRef]
- Pastar, I.; Tonic, I.; Golic, N.; Kojic, M.; Van Kranenburg, R.; Kleerebezem, M.; Topisirovic, L.; Jovanovic, G. Identification and genetic characterization of a novel proteinase, PrtR, from the human isolate Lactobacillus rhamnosus BGT10. Appl. Environ. Microbiol. 2003, 69, 5802–5811. [Google Scholar] [CrossRef] [PubMed]
- Paštar, I.; Fira, D.; Strahinić, I.; Krstić, K.; Begović, J.; Topisirović, L.; Jovanović, G. Analysis of the presence of prtR proteinase gene in natural isolates of Lactobacillus rhamnosus. Folia Microbiol. 2006, 51, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Venegas-Ortega, M.G.; Flores-Gallegos, A.C.; Martínez-Hernández, J.L.; Aguilar, C.N.; Nevárez-Moorillón, G.V. Production of bioactive peptides from lactic acid bacteria: A sustainable approach for healthier foods. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1039–1051. [Google Scholar] [CrossRef]
- Kok, J.; Venema, G. Genetic manipulation of the peptidolytic system in lactic acid bacteria. Int. Dairy J. 1995, 5, 737–755. [Google Scholar] [CrossRef][Green Version]
- Vermeulen, N.; Pavlovic, M.; Ehrmann, M.A.; Gänzle, M.G.; Vogel, R.F. Functional characterization of the proteolytic system of Lactobacillus sanfranciscensis DSM 20451T during growth in sourdough. Appl. Environ. Microbiol. 2005, 71, 6260–6266. [Google Scholar] [CrossRef] [PubMed]
- Martín, C.; Fernández-Vega, I.; Suárez, J.E.; Quirós, L.M. Adherence of Lactobacillus salivarius to HeLa cells promotes changes in the expression of the genes involved in biosynthesis of their ligands. Front. Immunol. 2020, 10, 3019. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, G.; Zhai, Z.; Zhou, P.; Hao, Y. Global transcriptomic analysis and function identification of malolactic enzyme pathway of Lactobacillus paracasei L9 in response to bile stress. Front. Microbiol. 2018, 9, 1978. [Google Scholar] [CrossRef]
- Zeng, Z.; Zuo, F.; Yu, R.; Zhang, B.; Ma, H.; Chen, S. Characterization of a lactose-responsive promoter of ATP-binding cassette (ABC) transporter gene from Lactobacillus acidophilus 05–172. FEMS Microbiol. Lett. 2017, 364, 167. [Google Scholar] [CrossRef]
- Detmers, F.J.M.; Lanfermeijer, F.C.; Abele, R.; Jack, R.W.; Tampé, R.; Konings, W.N.; Poolman, B. Combinatorial peptide libraries reveal the ligand-binding mechanism of the oligopeptide receptor OppA of Lactococcus lactis. Proc. Natl. Acad. Sci. USA 2000, 97, 12487–12492. [Google Scholar] [CrossRef] [PubMed]
- Detmers, F.J.M.; Kunji, E.R.S.; Lanfermeijer, F.C.; Poolman, B.; Konings, W.N. Kinetics and specificity of peptide uptake by the oligopeptide transport system of Lactococcus lactis. Biochemistry 1998, 37, 16671–16679. [Google Scholar] [CrossRef]
- Foucaud, C.; Kunji, E.R.S.; Hagting, A.; Richard, J.; Konings, W.N.; Desmazeaud, M.; Poolman, B. Specificity of peptide transport systems in Lactococcus lactis: Evidence for a third system which transports hydrophobic di- and tripeptides. J. Bacteriol. 1995, 177, 4652–4657. [Google Scholar] [CrossRef][Green Version]
- Nakajima, H.; Hagting, A.; Kunji, E.R.S.; Poolman, B.; Konings, W.N. Cloning and functional expression in Escherichia coli of the gene encoding the Di- and tripeptide transport protein of Lactobacillus helveticus. Appl. Environ. Microbiol. 1997, 63, 2213–2217. [Google Scholar] [CrossRef]
- Gitton, C.; Meyrand, M.; Wang, J.; Caron, C.; Trubuil, A.; Guillot, A.; Mistou, M.Y. Proteomic signature of Lactococcus lactis NCDO763 cultivated in milk. Appl. Environ. Microbiol. 2005, 71, 7152–7163. [Google Scholar] [CrossRef][Green Version]
- Jensen, M.P.; Ardö, Y. Variation in aminopeptidase and aminotransferase activities of six cheese related Lactobacillus helveticus strains. Int. Dairy J. 2010, 20, 149–155. [Google Scholar] [CrossRef]
- Griffiths, M.W.; Tellez, A.M. Lactobacillus helveticus: The proteolytic system. Front. Microbiol. 2013, 4, 30. [Google Scholar] [CrossRef]
- Guan, N.; Li, J.; Shin, H.D.; Du, G.; Chen, J.; Liu, L. Microbial response to environmental stresses: From fundamental mechanisms to practical applications. Appl. Microbiol. Biotechnol. 2017, 101, 3991–4008. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, J.; Wang, L.; Ma, Z.; Zhao, G.; Ge, Z.; Zhu, H.; Qiao, J. Improving nitrogen source utilization from defatted soybean meal for nisin production by enhancing proteolytic function of Lactococcus lactis F44. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Brown, L.; Villegas, J.M.; Elean, M.; Fadda, S.; Mozzi, F.; Saavedra, L.; Hebert, E.M. YebC, a putative transcriptional factor involved in the regulation of the proteolytic system of Lactobacillus. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kunji, E.R.S.; Hagting, A.; De Vries, C.J.; Juillard, V.; Haandrikman, A.J.; Poolman, B.; Konings, W.N. Transport of β-casein-derived peptides by the oligopeptide transport system is a crucial step in the proteolytic pathway of Lactococcus lactis. J. Biol. Chem. 1995, 270, 1569–1574. [Google Scholar] [CrossRef] [PubMed]
- Guédon, E.; Serror, P.; Ehrlich, S.D.; Renault, P.; Delorme, C. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 2001, 40, 1227–1239. [Google Scholar] [CrossRef]
- Petranovic, D.; Guédon, E.; Sperandio, B.; Delorme, C.; Ehrlich, D.; Renault, P. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transaction regulator. Mol. Microbiol. 2004, 53, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Den Hengst, C.D.; Van Hijum, S.A.F.T.; Geurts, J.M.W.; Nauta, A.; Kok, J.; Kuipers, O.P. The Lactococcus lactis CodY regulon: Identification of a conserved cis-regulatory element. J. Biol. Chem. 2005, 280, 34332–34342. [Google Scholar] [CrossRef] [PubMed]
- Galia, W.; Jameh, N.; Perrin, C.; Genay, M.; Dary-Mourot, A. Acquisition of PrtS in Streptococcus thermophilus is not enough in certain strains to achieve rapid milk acidification. Dairy Sci. Technol. 2016, 96, 623–636. [Google Scholar] [CrossRef]
- Yamamoto, N.; Wakai, T. Genome-wide motif predictions of BCARR-box in the amino-acid repressed genes of Lactobacillus helveticus CM4. BMC Microbiol. 2017, 17, 224. [Google Scholar] [CrossRef] [PubMed]
- Vido, K.; Le Bars, D.; Mistou, M.Y.; Anglade, P.; Gruss, A.; Gaudu, P. Proteome analyses of heme-dependent respiration in Lactococcus lactis: Involvement of the proteolytic system. J. Bacteriol. 2004, 186, 1648–1657. [Google Scholar] [CrossRef]
- Guédon, E.; Renault, P.; Ehrlich, S.D.; Delorme, C. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 2001, 183, 3614–3622. [Google Scholar] [CrossRef] [PubMed]
- Morel, F.; Lamarque, M.; Bissardon, I.; Atlan, D.; Galinier, A. Autoregulation of the biosynthesis of the CcpA-like protein, PepR1, in Lactobacillus delbrueckii subsp. bulgaricus. J. Mol. Microbiol. Biotechnol. 2001, 3, 63–66. [Google Scholar]
- Morel, F.; Frot-Coutaz, J.; Aubel, D.; Portalier, R.; Atlan, D. Characterization of a prolidase from Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397 with an unusual regulation of biosynthesis. Microbiology 1999, 145, 437–446. [Google Scholar] [CrossRef]
- Abada, E.A. Application of microbial enzymes in the dairy industry. In Enzymes in Food Biotechnology: Production, Applications, and Future Prospects; Academic Press: Cambridge, MA, USA, 2018; pp. 61–72. ISBN 9780128132807. [Google Scholar]
- Worsztynowicz, P.; Olejnik-Schmidt, A.; Białas, W.; Grajek, W. Identification and partial characterization of proteolytic activity of Enterococcus faecalis relevant to their application in the dairy industry. Acta Biochim. Pol. 2019, 66, 61–69. [Google Scholar] [CrossRef]
- Munir, M.; Nadeem, M.; Qureshi, T.M.; Leong, T.S.H.; Gamlath, C.J.; Martin, G.J.O.; Ashokkumar, M. Effects of high pressure, microwave and ultrasound processing on proteins and enzyme activity in dairy systems—A review. Innov. Food Sci. Emerg. Technol. 2019, 57, 102192. [Google Scholar] [CrossRef]
- Ozturkoglu-Budak, S.; Wiebenga, A.; Bron, P.A.; de Vries, R.P. Protease and lipase activities of fungal and bacterial strains derived from an artisanal raw ewe’s milk cheese. Int. J. Food Microbiol. 2016, 237, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Aguilar, J.G.; Sato, H.H. Microbial proteases: Production and application in obtaining protein hydrolysates. Food Res. Int. 2018, 103, 253–262. [Google Scholar] [CrossRef]
- Biscola, V.; Choiset, Y.; Rabesona, H.; Chobert, J.M.; Haertlé, T.; Franco, B.D.G.M. Brazilian artisanal ripened cheeses as sources of proteolytic lactic acid bacteria capable of reducing cow milk allergy. J. Appl. Microbiol. 2018, 125, 564–574. [Google Scholar] [CrossRef]
- Ahmad, M.N.; Mat Noh, N.A.; Abdullah, E.N.; Yarmo, M.A.; Mat Piah, M.B.; Ku Bulat, K.H. Optimization of a protease extraction using a statistical approach for the production of an alternative meat tenderizer from Spondias cytherea roots. J. Food Process. Preserv. 2019, 43, e14192. [Google Scholar] [CrossRef]
- Da Silva, R.R. Bacterial and fungal proteolytic enzymes: Production, catalysis and potential applications. Appl. Biochem. Biotechnol. 2017, 183, 1–19. [Google Scholar] [CrossRef]
- Singh, P.K.; Shrivastava, N.; Ojha, B.K. Enzymes in the meat industry. In Enzymes in Food Biotechnology: Production, Applications, and Future Prospects; Academic Press: Cambridge, MA, USA, 2019; pp. 111–128. ISBN 9780128132807. [Google Scholar]
- Arshad, M.S.; Kwon, J.-H.; Imran, M.; Sohaib, M.; Aslam, A.; Nawaz, I.; Amjad, Z.; Khan, U.; Javed, M. Plant and bacterial proteases: A key towards improving meat tenderization, a mini review. Cogent Food Agric. 2016, 2, 1261780. [Google Scholar] [CrossRef]
- Krasnowska, G. Próba wykorzystania enzymów pochodzenia mikrobiologicznego do degradacji surowców zwierzęcych bogatych w tkankę łączną. ŻYWNOŚĆ. Nauk. Technol. Jakość 2004, 1, 84–93. (In Polish) [Google Scholar]
- Lorenzo, J.M.; Munekata, P.E.S.; Gómez, B.; Barba, F.J.; Mora, L.; Pérez-Santaescolástica, C.; Toldrá, F. Bioactive peptides as natural antioxidants in food products–A review. Trends Food Sci. Technol. 2018, 79, 136–147. [Google Scholar] [CrossRef]
- Zotta, T.; Ricciardi, A.; Parente, E. Enzymatic activities of lactic acid bacteria isolated from Cornetto di Matera sourdoughs. Int. J. Food Microbiol. 2007, 115, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Martorana, A.; Giuffrè, A.M.; Capocasale, M.; Zappia, C.; Sidari, R. Sourdoughs as a source of lactic acid bacteria and yeasts with technological characteristics useful for improved bakery products. Eur. Food Res. Technol. 2018, 244, 1873–1885. [Google Scholar] [CrossRef]
- Saa, D.L.T.; Nissen, L.; Gianotti, A. Metabolomic approach to study the impact of flour type and fermentation process on volatile profile of bakery products. Food Res. Int. 2019, 119, 510–516. [Google Scholar] [CrossRef]
- Scarnato, L.; Montanari, C.; Serrazanetti, D.I.; Aloisi, I.; Balestra, F.; Del Duca, S.; Lanciotti, R. New bread formulation with improved rheological properties and longer shelf-life by the combined use of transglutaminase and sourdough. LWT Food Sci. Technol. 2017, 81, 101–110. [Google Scholar] [CrossRef]
- Mohan Kumar, B.V.; Sarabhai, S.; Prabhasankar, P. Targeted degradation of gluten proteins in wheat flour by prolyl endoprotease and its utilization in low immunogenic pasta for gluten sensitivity population. J. Cereal Sci. 2019, 87, 59–67. [Google Scholar] [CrossRef]
- Moghaddam, M.F.T.; Jalali, H.; Nafchi, A.M.; Nouri, L. Evaluating the effects of lactic acid bacteria and olive leaf extract on the quality of gluten-free bread. Gene Rep. 2020, 21, 100771. [Google Scholar] [CrossRef]
- Sarabhai, S.; Tamilselvan, T.; Prabhasankar, P. Role of enzymes for improvement in gluten-free foxtail millet bread: It’s effect on quality, textural, rheological and pasting properties. LWT 2020, 137, 110365. [Google Scholar] [CrossRef]
- Puligundla, P.; Smogrovicova, D.; Mok, C.; Obulam, V.S.R. Recent developments in high gravity beer-brewing. Innov. Food Sci. Emerg. Technol. 2020, 64, 102399. [Google Scholar] [CrossRef]
- San Martin, D.; Orive, M.; Iñarra, B.; Castelo, J.; Estévez, A.; Nazzaro, J.; Iloro, I.; Elortza, F.; Zufía, J. Brewers’ spent yeast and grain protein hydrolysates as second-generation feedstuff for aquaculture feed. Waste Biomass Valoriz. 2020, 11, 5307–5320. [Google Scholar] [CrossRef]
- Kok, Y.J.; Ye, L.; Muller, J.; Ow, D.S.W.; Bi, X. Brewing with malted barley or raw barley: What makes the difference in the processes? Appl. Microbiol. Biotechnol. 2019, 103, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Mathias, T.R.; de Aguiar, P.F.; Silva, J.B.A.; de Mello, P.P.M.; Sérvulo, E.F.C. Brewery wastes reuse for protease production by lactic acid bacteria fermentation. Food Technol. Biotechnol. 2017, 55, 218–224. [Google Scholar] [CrossRef]
- Connolly, A.; Cermeño, M.; Crowley, D.; O’Callaghan, Y.; O’Brien, N.M.; FitzGerald, R.J. Characterisation of the in vitro bioactive properties of alkaline and enzyme extracted brewers’ spent grain protein hydrolysates. Food Res. Int. 2019, 121, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Marson, G.V.; de Castro, R.J.S.; da Costa Machado, M.T.; da Silva Zandonadi, F.; Barros, H.D.d.F.Q.; Maróstica Júnior, M.R.; Sussulini, A.; Hubinger, M.D. Proteolytic enzymes positively modulated the physicochemical and antioxidant properties of spent yeast protein hydrolysates. Process Biochem. 2020, 91, 34–45. [Google Scholar] [CrossRef]
- Haard, N.F. A review of proteotlytic enzymes from marine organisms and their application in the food industry. J. Aquat. Food Prod. Technol. 1992, 1, 17–35. [Google Scholar] [CrossRef]
- Hou, Y.; Wu, Z.; Dai, Z.; Wang, G.; Wu, G. Protein hydrolysates in animal nutrition: Industrial production, bioactive peptides, and functional significance. J. Anim. Sci. Biotechnol. 2017, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Merenstein, D.; Merrifield, C.A.; Hutkins, R. Probiotics for human use. Nutr. Bull. 2018, 43, 212–225. [Google Scholar] [CrossRef]
- Pokora, M.; Eckert, E.; Zambrowicz, A.; Bobak, Ł.; Szołtysik, M.; Dązbrowska, A.; Chrzanowska, J.; Polanowski, A.; Trziszka, T. Biological and functional properties of proteolytic enzyme-modified egg protein by-products. Food Sci. Nutr. 2013, 1, 184–195. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kieliszek, M.; Pobiega, K.; Piwowarek, K.; Kot, A.M. Characteristics of the Proteolytic Enzymes Produced by Lactic Acid Bacteria. Molecules 2021, 26, 1858. https://doi.org/10.3390/molecules26071858
Kieliszek M, Pobiega K, Piwowarek K, Kot AM. Characteristics of the Proteolytic Enzymes Produced by Lactic Acid Bacteria. Molecules. 2021; 26(7):1858. https://doi.org/10.3390/molecules26071858
Chicago/Turabian StyleKieliszek, Marek, Katarzyna Pobiega, Kamil Piwowarek, and Anna M. Kot. 2021. "Characteristics of the Proteolytic Enzymes Produced by Lactic Acid Bacteria" Molecules 26, no. 7: 1858. https://doi.org/10.3390/molecules26071858
APA StyleKieliszek, M., Pobiega, K., Piwowarek, K., & Kot, A. M. (2021). Characteristics of the Proteolytic Enzymes Produced by Lactic Acid Bacteria. Molecules, 26(7), 1858. https://doi.org/10.3390/molecules26071858









