ATP Analogues for Structural Investigations: Case Studies of a DnaB Helicase and an ABC Transporter
Abstract
1. Introduction
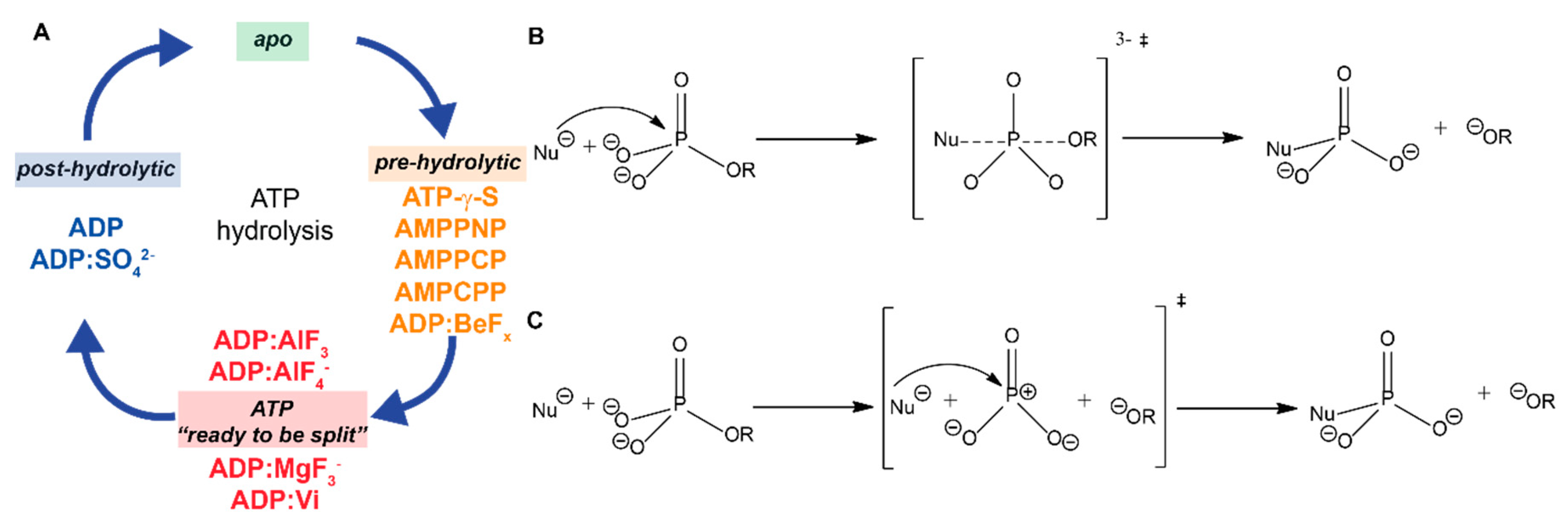
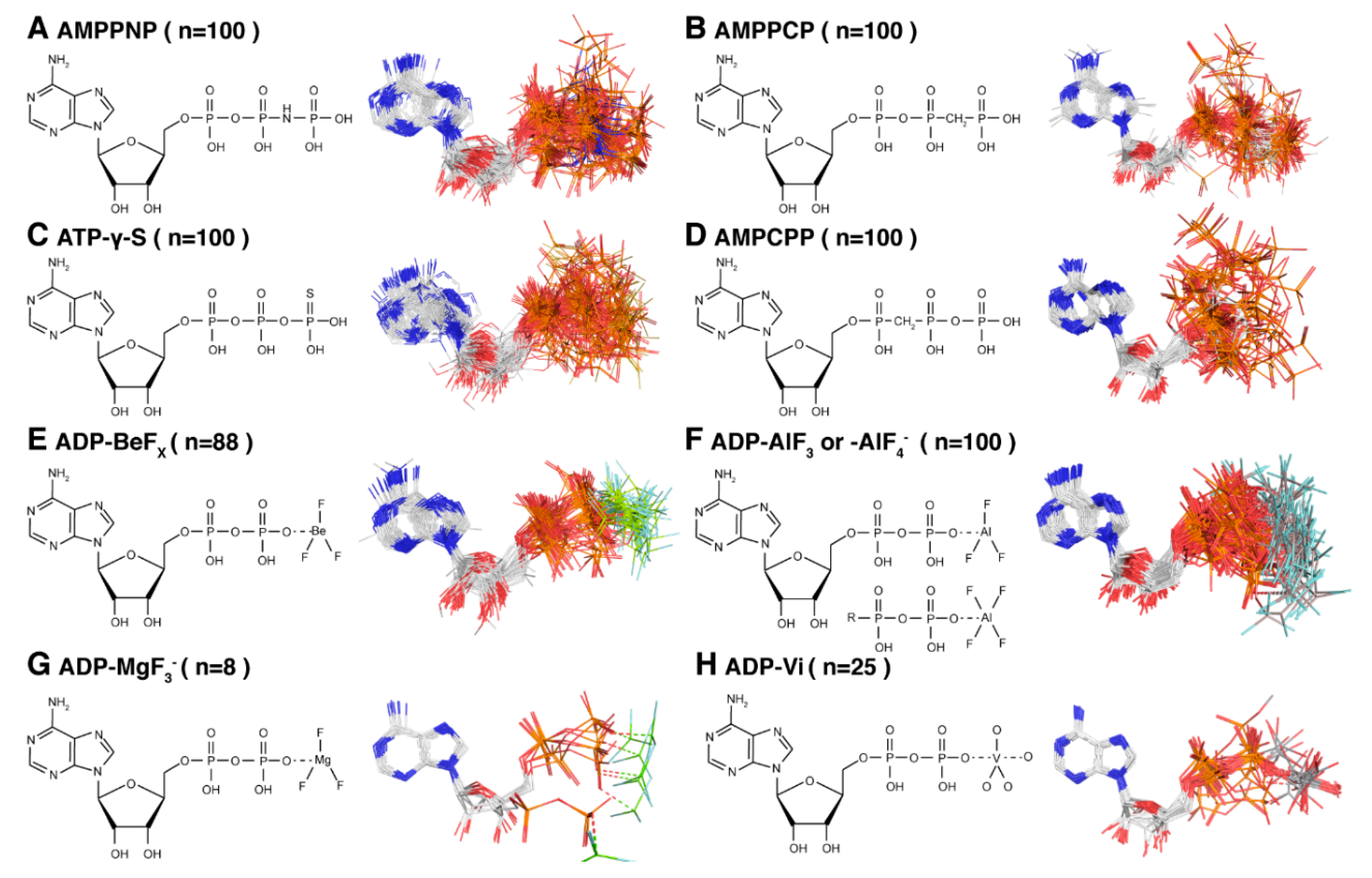
2. The Different Hydrolysis States and the ATP-Mimics Used to Induce Them
3. The DnaB Helicase and the ABC Transporter BmrA
4. Results and Discussion
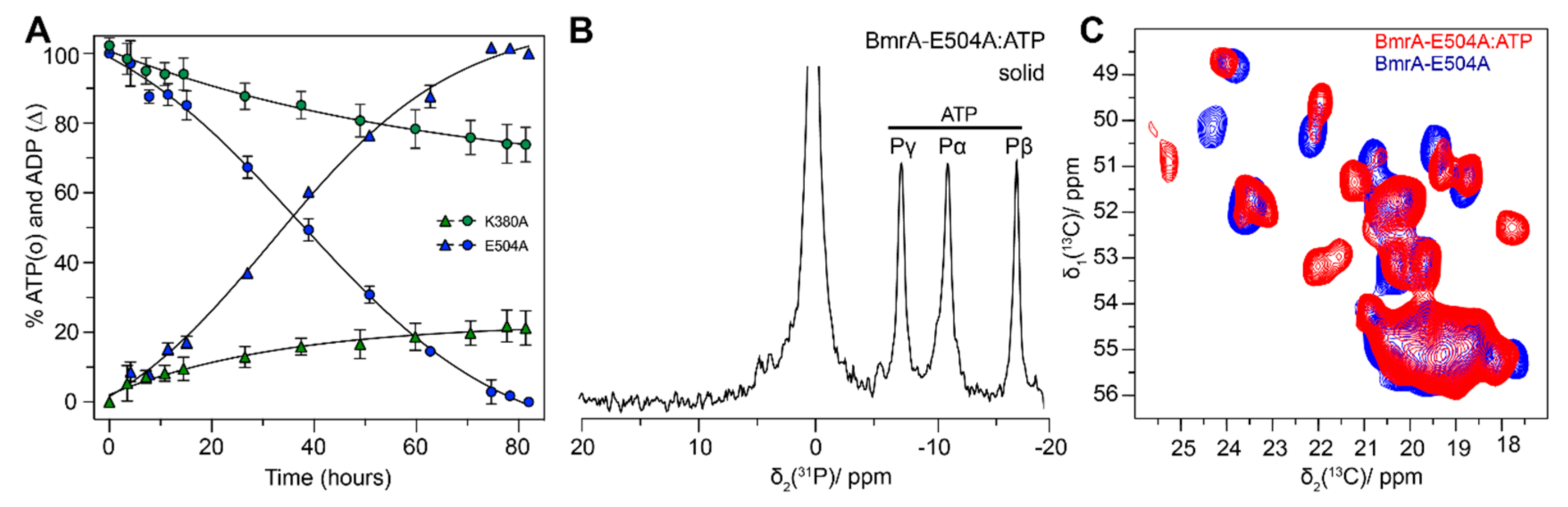
4.1. The Pre-Hydrolytic State Mimicked by AMPPCP, AMPPNP and ATP-γ-S
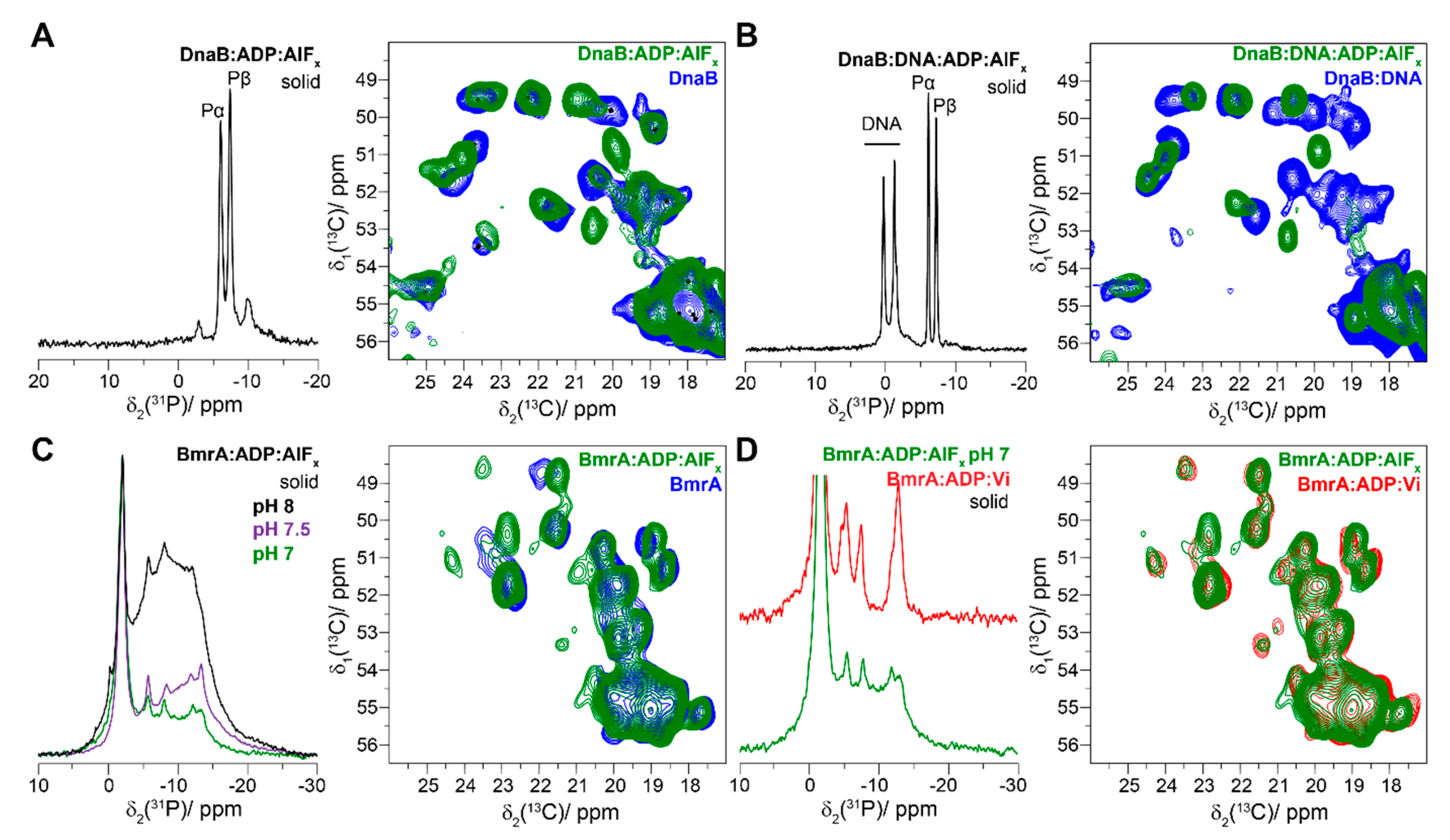
4.2. The Transition-State Analogues ATP/ADP:Vi and Aluminium Fluorides (ADP:AlFx)
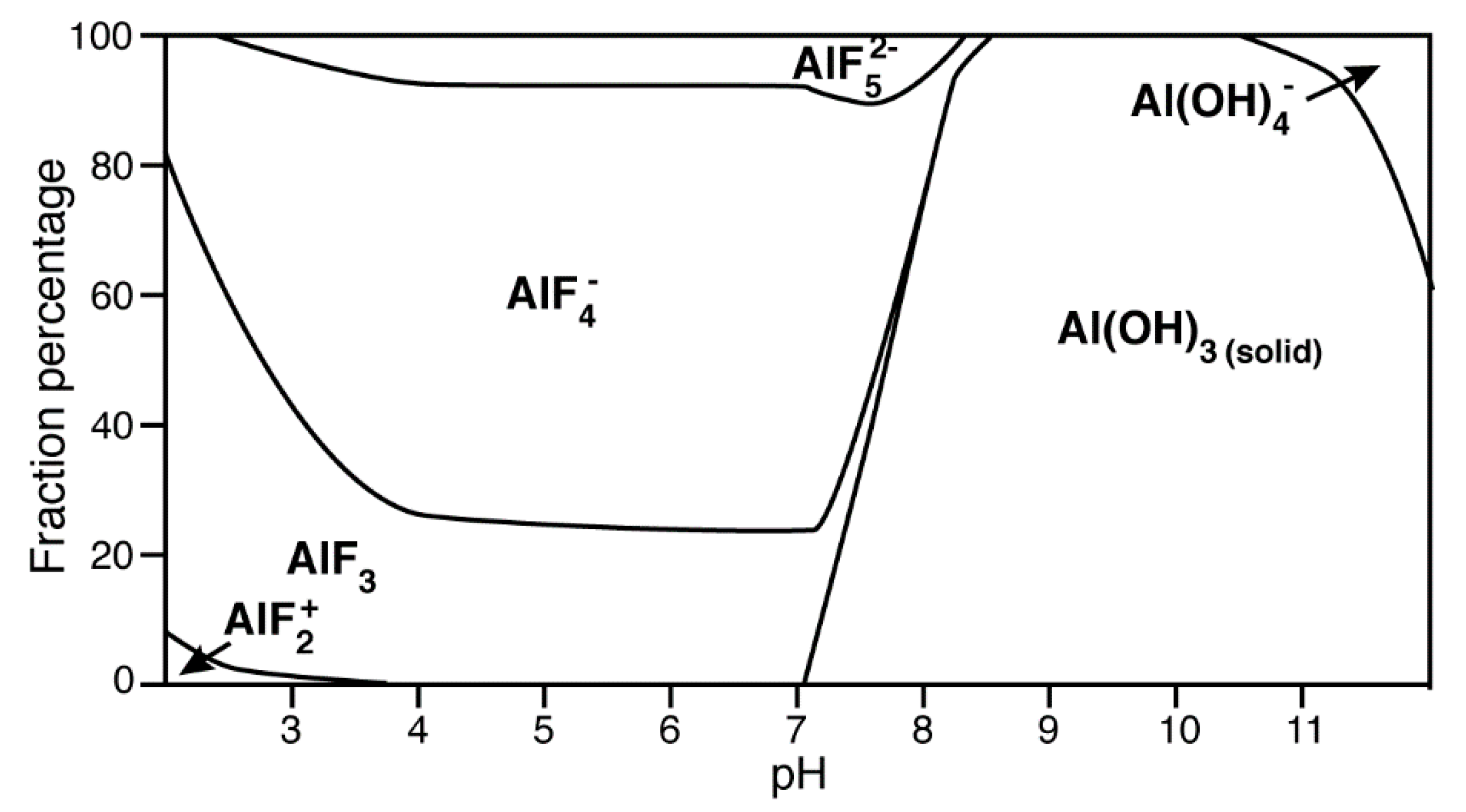
4.3. Aluminium Fluorides (AlFx) as Transition-State Mimic
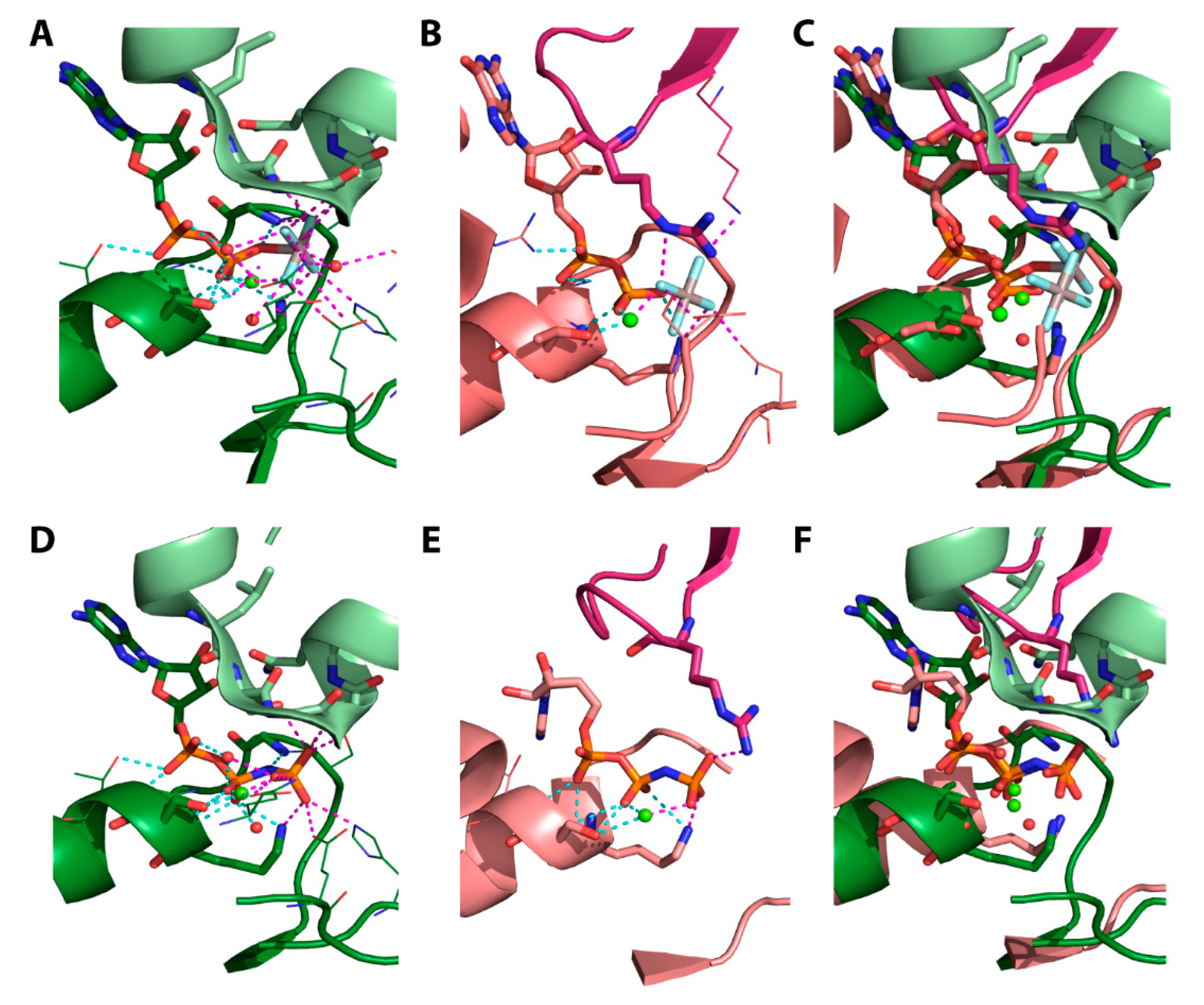
4.4. The Post-Hydrolytic State Induced by ADP
4.5. Structural Considerations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Manna, R.N.; Dutta, M.; Jana, B. Mechanistic study of the ATP hydrolysis reaction in dynein motor protein. Phys. Chem. Chem. Phys. 2020, 22, 1534–1542. [Google Scholar] [CrossRef] [PubMed]
- Mächtel, R.; Narducci, A.; Griffith, D.A.; Cordes, T.; Orelle, C. An integrated transport mechanism of the maltose ABC importer. Res. Microbiol. 2019, 170, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Prieß, M.; Göddeke, H.; Groenhof, G.; Schäfer, L.V. Molecular mechanism of ATP hydrolysis in an ABC transporter. ACS Central Sci. 2018, 4, 1334–1343. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M.; Matson, S.W. History of DNA Helicases. Genes 2020, 11, 255. [Google Scholar] [CrossRef]
- Ma, W.; Schulten, K. Mechanism of substrate translocation by a ring-shaped ATPase motor at millisecond resolution. J. Am. Chem. Soc. 2015, 137, 3031–3040. [Google Scholar] [CrossRef]
- Dittrich, M.; Schulten, K. PcrA Helicase, a prototype ATP-driven molecular motor. Structure 2006, 14, 1345–1353. [Google Scholar] [CrossRef]
- Grigorenko, B.L.; Rogov, A.V.; Topol, I.A.; Burt, S.K.; Martinez, H.M.; Nemukhin, A.V. Mechanism of the myosin catalyzed hydrolysis of ATP as rationalized by molecular modeling. Proc. Natl. Acad. Sci. USA 2007, 104, 7057–7061. [Google Scholar] [CrossRef]
- Davidson, R.B.; Hendrix, J.; Geiss, B.J.; Mccullagh, M. Allostery in the dengue virus NS3 helicase: Insights into the NTPase cycle from molecular simulations. PLoS Comput. Biol. 2018, 14, e1006103. [Google Scholar] [CrossRef]
- Geourjon, C.; Orelle, C.; Steinfels, E.; Blanchet, C.; Deléage, G.; Di Pietro, A.; Jault, J.M. A common mechanism for ATP hydrolysis in ABC transporter and helicase superfamilies. Trends Biochem. Sci. 2001, 26, 539–544. [Google Scholar] [CrossRef]
- Gardiennet, C.; Schütz, A.K.; Hunkeler, A.; Kunert, B.; Terradot, L.; Böckmann, A.; Meier, B.H. A sedimented sample of a 59 kDa dodecameric helicase yields high-resolution solid-state NMR spectra. Angew. Chem. Int. Ed. 2012, 51, 7855–7858. [Google Scholar] [CrossRef]
- Wiegand, T.; Lacabanne, D.; Torosyan, A.; Boudet, J.; Cadalbert, R.; Allain, F.H.-T.; Meier, B.H.; Böckmann, A. Sedimentation yields long-term stable protein samples as shown by solid-state NMR. Front. Mol. Biosci. 2020, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, T. A solid-state NMR tool box for the investigation of ATP-fueled protein engines. Prog. Nucl. Magn. Reson. Spectrosc. 2020, 117, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Takegoshi, K.; Nakamura, S.; Terao, T. 13C-13C polarization transfer by resonant interference recoupling under magic-angle spinning in solid-state NMR. Chem. Phys. Lett. 1999, 307, 295–302. [Google Scholar] [CrossRef]
- Takegoshi, K.; Nakamura, S.; Terao, T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001, 344, 631–637. [Google Scholar] [CrossRef]
- Böckmann, A.; Gardiennet, C.; Verel, R.; Hunkeler, A.; Loquet, A.; Pintacuda, G.; Emsley, L.; Meier, B.H.; Lesage, A. Characterization of different water pools in solid-state NMR protein samples. J. Biomol. NMR 2009, 45, 319–327. [Google Scholar] [CrossRef]
- Cox, N.; Lubitz, W.; Savitsky, A. W-band ELDOR-detected NMR (EDNMR) spectroscopy as a versatile technique for the characterisation of transition metal-ligand interactions. Mol. Phys. 2013, 111, 2788–2808. [Google Scholar] [CrossRef]
- Cox, N.; Nalepa, A.; Lubitz, W.; Savitsky, A. ELDOR-detected NMR: A general and robust method for electron-nuclear hyperfine spectroscopy? J. Magn. Reson. 2017, 280, 63–78. [Google Scholar] [CrossRef]
- Goldfarb, D. ELDOR-Detected NMR. In eMagRes; Wiley: Hoboken, NJ, USA, 2017; Volume 563, pp. 101–114. [Google Scholar]
- Schosseler, P.; Wacker, T.; Schweiger, A. Pulsed ELDOR detected NMR. Chem. Phys. Lett. 1994, 224, 319–324. [Google Scholar] [CrossRef]
- Giannoulis, A.; Feintuch, A.; Barak, Y.; Mazal, H.; Albeck, S.; Unger, T.; Yang, F.; Su, X.-C.; Goldfarb, D. Two closed ATP- and ADP-dependent conformations in yeast Hsp90 chaperone detected by Mn(II) EPR spectroscopic techniques. Proc. Natl. Acad. Sci. USA 2019, 117, 395–404. [Google Scholar] [CrossRef]
- Flowers, S.; Biswas, E.E.; Biswas, S.B. Conformational dynamics of DnaB helicase upon DNA and nucleotide-binding: Analysis by intrinsic tryptophan fluorescence quenching. Biochemistry 2003, 42, 1910–1921. [Google Scholar] [CrossRef]
- Robson, A.; Booth, A.E.; Gold, V.A.; Clarke, A.R.; Collinson, I. A Large conformational change couples the ATP binding site of SecA to the SecY protein channel. J. Mol. Biol. 2007, 374, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Orelle, C.; Gubellini, F.; Durand, A.; Marco, S.; Levy, D.; Gros, P.; Di Pietro, A.; Jault, J.-M. Conformational change induced by ATP binding in the multidrug ATP-binding cassette transporter BmrA. Biochemistry 2008, 47, 2404–2412. [Google Scholar] [CrossRef] [PubMed]
- Kühner, S.; Fischer, S. Structural mechanism of the ATP-induced dissociation of rigor myosin from actin. Proc. Natl. Acad. Sci. USA 2011, 108, 7793–7798. [Google Scholar] [CrossRef] [PubMed]
- Wikström, M. Biophysical and structural aspects of bioenergetics. R. Soc. Chem. 2007, 1, 414. [Google Scholar]
- Chen, B.; Doucleff, M.; Wemmer, D.E.; De Carlo, S.; Huang, H.H.; Nogales, E.; Hoover, T.R.; Kondrashkina, E.; Guo, L.; Nixon, B.T. ATP Ground-and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, σ54. Structures 2007, 15, 429–440. [Google Scholar] [CrossRef][Green Version]
- Chabre, M. Aluminofluoride and beryllofluoride complexes: A new phosphate analogs in enzymology. Trends Biochem. Sci. 1990, 15, 6–10. [Google Scholar] [CrossRef]
- Yount, R.G.; Babcock, D.; Ballantyne, W.; Ojala, D. Adenylyl imidodiphosphate, an adenosine triphosphate analog containing a P-N-P linkage. Biochemistry 1971, 10, 2484–2489. [Google Scholar] [CrossRef]
- Myers, T.C.; Nakamura, K.; Flesher, J.W. Phosphonic acid analogs of nucleoside phosphates. I. The synthesis of 5″-adenylyl methylenediphosphonate, a phosphonic acid analog of ATP. J. Am. Chem. Soc. 2002, 85, 3292–3295. [Google Scholar] [CrossRef]
- Goody, R.S.; Eckstein, F. Thiophosphate analogs of nucleoside di- and triphosphates. J. Am. Chem. Soc. 1971, 93, 6252–6257. [Google Scholar] [CrossRef]
- Mannherz, H.G.; Brehme, H.; Lamp, U. Depolymerisation of F-actin to G-actin and its repolymerisation in the presence of analogs of adenosine triphosphate. JBIC J. Biol. Inorg. Chem. 1975, 60, 109–116. [Google Scholar] [CrossRef]
- Watanabe, F.; Hashimoto, T.; Tagawa, K. Energy-independent protection of the oxidative phosphorylation capacity of mitochondria against anoxic damage by ATP and its nonmetabolizable analogs1. J. Biochem. 1985, 97, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Shimizu, T.; Morii, H.; Tanokura, M. Hydrolysis of AMPPNP by the motor domain of ncd, a kinesin-related protein. FEBS Lett. 1997, 409, 29–32. [Google Scholar] [CrossRef]
- Olesen, C.; Picard, M.; Winther, A.-M.L.; Gyrup, C.; Morth, J.P.; Oxvig, C.; Møller, J.V.; Nissen, P. The structural basis of calcium transport by the calcium pump. Nat. Cell Biol. 2007, 450, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Siarheyeva, A.; Liu, R.; Sharom, F.J. Characterization of an asymmetric occluded state of P-glycoprotein with two bound nucleotides. J. Biol. Chem. 2010, 285, 7575–7586. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, A.D.; Sheth, P.R.; Basso, A.D.; Paliwal, S.; Gray, K.; Fischmann, T.O.; Le, H.V. Structural basis of CX-4945 binding to human protein kinase CK2. FEBS Lett. 2011, 585, 104–110. [Google Scholar] [CrossRef]
- Timachi, M.H.; Hutter, C.A.; Hohl, M.; Assafa, T.; Böhm, S.; Mittal, A.; Seeger, M.A.; Bordignon, E. Exploring conformational equilibria of a heterodimeric ABC transporter. eLife 2017, 6, 257. [Google Scholar] [CrossRef]
- Wiegand, T.; Cadalbert, R.; Gardiennet, C.; Timmins, J.; Terradot, L.; Böckmann, A.; Meier, B.H. Monitoring ssDNA binding to the DnaB helicase from helicobacter pylori by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. 2016, 55, 14164–14168. [Google Scholar] [CrossRef]
- Kung, G.; Runquist, J.A.; Miziorko, H.M.; Harrison, D.H.T. Identification of the allosteric regulatory site in bacterial phosphoribulokinase. Biochemistry 1999, 38, 15157–15165. [Google Scholar] [CrossRef]
- Nitta, R.; Kikkawa, M.; Okada, Y.; Hirokawa, N. KIF1A Alternately uses two loops to bind microtubules. Science 2004, 305, 678–683. [Google Scholar] [CrossRef]
- Reddy, M.C.M.; Palaninathan, S.K.; Shetty, N.D.; Owen, J.L.; Watson, M.D.; Sacchettini, J.C. High resolution crystal structures of Mycobacterium tuberculosis Adenosine Kinase. J. Biol. Chem. 2007, 282, 27334–27342. [Google Scholar] [CrossRef]
- Shintre, C.A.; Pike, A.C.W.; Li, Q.; Kim, J.-I.; Barr, A.J.; Goubin, S.; Shrestha, L.; Yang, J.; Berridge, G.; Ross, J.; et al. Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc. Natl. Acad. Sci. USA 2013, 110, 9710–9715. [Google Scholar] [CrossRef] [PubMed]
- Hassett, A.; Blaettler, W.; Knowles, J.R. Pyruvate kinase: Is the mechanism of phospho transfer associative or dissociative? Biochemistry 1982, 21, 6335–6340. [Google Scholar] [CrossRef]
- Admiraal, S.J.; Herschlag, D. Mapping the transition state for ATP hydrolysis: Implications for enzymatic catalysis. Chem. Biol. 1995, 2, 729–739. [Google Scholar] [CrossRef]
- Kamerlin, S.C.L.; Florián, J.; Warshel, A. Associative versus dissociative mechanisms of phosphate monoester hydrolysis: On the interpretation of activation entropies. ChemPhysChem 2008, 9, 1767–1773. [Google Scholar] [CrossRef]
- Yang, Y.; Cui, Q. The hydrolysis activity of adenosine triphosphate in myosin: A Theoretical analysis of anomeric effects and the nature of the transition state. J. Phys. Chem. A 2009, 113, 12439–12446. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.R.; Plotnikov, N.V.; Warshel, A. Addressing open questions about phosphate hydrolysis pathways by careful free energy mapping. J. Phys. Chem. B 2012, 117, 153–163. [Google Scholar] [CrossRef]
- Knowles, J.R. Enzyme-catalyzed phosphoryl transfer reactions. Annu. Rev. Biochem. 1980, 49, 877–919. [Google Scholar] [CrossRef]
- Sondek, J.; Lambright, D.G.; Noel, J.P.; Hamm, H.E.; Sigler, P.B. GTPase mechanism of G proteins from the 1.7-Å crystal structure of transducin α-GDP AIF−4. Nat. Cell Biol. 1994, 372, 276–279. [Google Scholar] [CrossRef]
- Wittinghofer, A. Signaling mechanistics: Aluminum fluoride for molecule of the year. Curr. Biol. 1997, 7, R682–R685. [Google Scholar] [CrossRef]
- Graham, D.L.; Lowe, P.N.; Grime, G.W.; Marsh, M.; Rittinger, K.; Smerdon, S.J.; Gamblin, S.J.; Eccleston, J.F. MgF3- as a transition state analog of phosphoryl transfer. Chem. Biol. 2002, 9, 375–381. [Google Scholar] [CrossRef]
- Davies, D.R.; Hol, W.G. The power of vanadate in crystallographic investigations of phosphoryl transfer enzymes. FEBS Lett. 2004, 577, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Davidson, A.L. Vanadate-induced trapping of nucleotides by purified maltose transport complex requires ATP hydrolysis. J. Bacteriol. 2000, 182, 6570–6576. [Google Scholar] [CrossRef] [PubMed]
- Loo, T.W.; Clarke, D.M. Vanadate trapping of nucleotide at the ATP-binding sites of human multidrug resistance P-glycoprotein exposes different residues to the drug-binding site. Proc. Natl. Acad. Sci. USA 2002, 99, 3511–3516. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Richards, N.G.; Waltho, J.P.; Blackburn, G.M. Metal fluorides as analogues for studies on phosphoryl transfer enzymes. Angew. Chem. Int. Ed. 2017, 56, 4110–4128. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Cliff, M.J.; Baxter, N.J.; Dannatt, H.R.W.; Hounslow, A.M.; Bowler, M.W.; Blackburn, G.M.; Waltho, J.P. Charge-balanced metal fluoride complexes for protein kinase a with adenosine diphosphate and substrate peptide SP20. Angew. Chem. Int. Ed. 2012, 51, 12242–12245. [Google Scholar] [CrossRef]
- Baxter, N.J.; Blackburn, G.M.; Marston, J.P.; Hounslow, A.M.; Cliff, M.J.; Bermel, W.; Williams, N.H.; Hollfelder, F.; Wemmer, A.D.E.; Waltho, J.P. Anionic charge is prioritized over geometry in aluminum and magnesium fluoride transition state analogs of phosphoryl transfer enzymes. J. Am. Chem. Soc. 2008, 130, 3952–3958. [Google Scholar] [CrossRef]
- Jin, Y.; Molt, R.W.; Blackburn, G.M. Metal fluorides: Tools for structural and computational analysis of phosphoryl transfer enzymes. Top Curr Chem (Cham) 2017, 375, 1–31. [Google Scholar] [CrossRef]
- Akabayov, S.R.; Akabayov, B. Vanadate in structural biology. Inorganica Chim. Acta 2014, 420, 16–23. [Google Scholar] [CrossRef]
- Chen, J.; Sharma, S.; Quiocho, F.A.; Davidson, A.L. Trapping the transition state of an ATP-binding cassette transporter: Evidence for a concerted mechanism of maltose transport. Proc. Natl. Acad. Sci. USA 2001, 98, 1525–1530. [Google Scholar] [CrossRef]
- Münnich, S.; Taft, M.H.; Manstein, D.J. Crystal structure of human myosin 1c-The Motor in GLUT4 exocytosis: Implications for Ca2+ regulation and 14-3-3 binding. J. Mol. Biol. 2014, 426, 2070–2081. [Google Scholar] [CrossRef]
- Chinthalapudi, K.; Heissler, S.M.; Preller, M.; Sellers, J.R.; Manstein, D.J. Mechanistic insights into the active site and allosteric communication pathways in human nonmuscle myosin-2C. eLife 2017, 6, e32742. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Zalyte, R.; Urnavicius, L.; Carter, A.P. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nat. Cell Biol. 2015, 518, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Oldham, M.L.; Chen, J. Snapshots of the maltose transporter during ATP hydrolysis. Proc. Natl. Acad. Sci. USA 2011, 108, 15152–15156. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Januliene, D.; Mehdipour, A.R.; Thomas, C.; Stefan, E.; Brüchert, S.; Kuhn, B.T.; Geertsma, E.R.; Hummer, G.; Tampé, R.; et al. Conformation space of a heterodimeric ABC exporter under turnover conditions. Nat. Cell Biol. 2019, 571, 580–583. [Google Scholar] [CrossRef]
- Jiang, J.; Maes, E.G.; Taylor, A.B.; Wang, L.; Hinck, A.P.; Lafer, E.M.; Sousa, R. Structural basis of J Cochaperone binding and regulation of Hsp70. Mol. Cell 2007, 28, 422–433. [Google Scholar] [CrossRef]
- Luo, D.; Xu, T.; Watson, R.P.; Scherer-Becker, D.; Sampath, A.; Jahnke, W.; Yeong, S.S.; Wang, C.H.; Lim, S.P.; Strongin, A.; et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008, 27, 3209–3219. [Google Scholar] [CrossRef]
- Dumais, M.; Davies, D.R.; Lin, T.; Staker, B.L.; Myler, P.J.; Van Voorhis, W.C. Structure and analysis of nucleoside diphosphate kinase from Borrelia burgdorferi prepared in a transition-state complex with ADP and vanadate moieties. Acta Crystallogr. F Struct. Biol. Commun. 2018, 74, 373–384. [Google Scholar] [CrossRef]
- Chen, C.; Saxena, A.K.; Simcoke, W.N.; Garboczi, D.N.; Pedersen, P.L.; Ko, Y.H. Mitochondrial ATP synthase. Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism. J Biol Chem 2006, 281, 13777–13783. [Google Scholar] [CrossRef]
- Luo, D.; Nakazawa, M.; Yoshida, Y.; Cai, J.; Imai, S. Effects of three different Ca(2+) pump ATPase inhibitors on evoked contractions in rabbit aorta and activities of Ca(2+) pump ATPases in porcine aorta. Gen. Pharmacol. Vasc. Syst. 2000, 34, 211–220. [Google Scholar] [CrossRef]
- Drakou, C.E.; Malekkou, A.; Hayes, J.M.; Lederer, C.W.; Leonidas, D.; Oikonomakos, N.G.; Lamond, A.I.; Santama, N.; Zographos, S. hCINAP is an atypical mammalian nuclear adenylate kinase with an ATPase motif: Structural and functional studies. Proteins: Struct. Funct. Bioinform. 2011, 80, 206–220. [Google Scholar] [CrossRef]
- He, C.; Chen, J.; Wang, H.; Wan, Y.; Zhou, J.; Dan, Z.; Zeng, Y.; Xu, W.; Zhu, Y.; Huang, W.; et al. Crystal structures of rice hexokinase 6 with a series of substrates shed light on its enzymatic mechanism. Biochem. Biophys. Res. Commun. 2019, 515, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.-C.; Shi, W.; Rinaldo-Matthis, A.; Tyler, P.C.; Evans, G.B.; Clinch, K.; Almo, S.C.; Schramm, V.L. Four generations of transition-state analogues for human purine nucleoside phosphorylase. Proc. Natl. Acad. Sci. USA 2010, 107, 4805–4812. [Google Scholar] [CrossRef] [PubMed]
- Menz, R.; Walker, J.E.; Leslie, A.G. Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites. Cell 2001, 106, 331–341. [Google Scholar] [CrossRef]
- Dinescu, A.; Bhansali, V.S.; Cundari, T.R.; Luo, J.-L.; Anderson, M.E. Function of conserved residues of human glutathione synthetase. J. Biol. Chem. 2004, 279, 22412–22421. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. Why nature chose phosphate to modify proteins. Philos. Trans. R. Soc. B: Biol. Sci. 2012, 367, 2513–2516. [Google Scholar] [CrossRef]
- Bagshaw, C. ATP analogues at a glance. J. Cell Sci. 2001, 114, 459–460. [Google Scholar]
- Elphick, L.M.; Lee, S.E.; Gouverneur, V.; Mann, D.J. Using chemical genetics and ATP analogues to dissect protein kinase function. ACS Chem. Biol. 2007, 2, 299–314. [Google Scholar] [CrossRef]
- Wiberg, K. Application of the pople-santry-segal CNDO method to the cyclopropylcarbinyl and cyclobutyl cation and to bicyclobutane. Tetrahedron 1968, 24, 1083–1096. [Google Scholar] [CrossRef]
- Gardiennet, C.; Wiegand, T.; Bazin, A.; Cadalbert, R.; Kunert, B.; Lacabanne, D.; Gutsche, I.; Terradot, L.; Meier, B.H.; Böckmann, A. Solid-state NMR chemical-shift perturbations indicate domain reorientation of the DnaG primase in the primosome of Helicobacter pylori. J. Biomol. NMR 2016, 64, 189–195. [Google Scholar] [CrossRef]
- Keller, K.; Wiegand, T.; Cadalbert, R.; Meier, B.H.; Böckmann, A.; Jeschke, G.; Yulikov, M. High-spin Metal Centres in Dipolar EPR Spectroscopy. Chim. Int. J. Chem. 2018, 72, 216–220. [Google Scholar] [CrossRef]
- Wiegand, T.; Cadalbert, R.; Lacabanne, D.; Timmins, J.; Terradot, L.; Böckmann, A.; Meier, B.H. The conformational changes coupling ATP hydrolysis and translocation in a bacterial DnaB helicase. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, T.; Cadalbert, R.; von Schroetter, C.; Allain, F.H.T.; Meier, B.H. Segmental isotope labelling and solid-state NMR of a 12 × 59 kDa motor protein: Identification of structural variability. J. Biomol. NMR 2018, 71, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, T.; Gardiennet, C.; Cadalbert, R.; Lacabanne, D.; Kunert, B.; Terradot, L.; Böckmann, A.; Meier, B.H. Variability and conservation of structural domains in divide-and-conquer approaches. J. Biomol. NMR 2016, 65, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, T.; Gardiennet, C.; Ravotti, F.; Bazin, A.; Kunert, B.; Lacabanne, D.; Cadalbert, R.; Güntert, P.; Terradot, L.; Böckmann, A.; et al. Solid-state NMR sequential assignments of the N-terminal domain of HpDnaB helicase. Biomol. NMR Assign. 2015, 10, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, T.; Lacabanne, D.; Keller, K.; Cadalbert, R.; Lecoq, L.; Yulikov, M.; Terradot, L.; Jeschke, G.; Meier, B.H.; Böckmann, A. Solid-state NMR and EPR Spectroscopy of Mn2+-substituted ATP-fueled protein engines. Angew. Chem. Int. Ed. 2017, 56, 3369–3373. [Google Scholar] [CrossRef]
- Wiegand, T.; Liao, W.-C.; Ong, T.-C.; Däpp, A.; Cadalbert, R.; Copéret, C.; Böckmann, A.; Meier, B.H. Protein-nucleotide contacts in motor proteins detected by DNP-enhanced solid-state NMR. J. Biomol. NMR 2017, 69, 157–164. [Google Scholar] [CrossRef]
- Wiegand, T.; Schledorn, M.; Malär, A.A.; Cadalbert, R.; Däpp, A.; Terradot, L.; Meier, B.H.; Böckmann, A. Nucleotide binding modes in a motor protein revealed by 31P-and 1H-detected MAS solid-state NMR spectroscopy. ChemBioChem 2019, 21, 324–330. [Google Scholar] [CrossRef]
- Kunert, B.; Gardiennet, C.; Lacabanne, D.; Calles-Garcia, D.; Falson, P.; Jault, J.-M.; Meier, B.H.; Penin, F.; Bã¶ckmann, A.; Böckmann, A. Efficient and stable reconstitution of the ABC transporter BmrA for solid-state NMR studies. Front. Mol. Biosci. 2014, 1, 5. [Google Scholar] [CrossRef][Green Version]
- Lacabanne, D.; Kunert, B.; Gardiennet, C.; Meier, B.H.; Böckmann, A. Sample preparation for membrane protein structural studies by solid-state NMR. Adv. Struct. Saf. Stud. 2017, 1635, 345–358. [Google Scholar] [CrossRef]
- Lacabanne, D.; Lends, A.; Danis, C.; Kunert, B.; Fogeron, M.-L.; Jirasko, V.; Chuilon, C.; Lecoq, L.; Orelle, C.; Chaptal, V.; et al. Gradient reconstitution of membrane proteins for solid-state NMR studies. J. Biomol. NMR 2017, 69, 81–91. [Google Scholar] [CrossRef]
- Lacabanne, D.; Orelle, C.; Lecoq, L.; Kunert, B.; Chuilon, C.; Wiegand, T.; Ravaud, S.; Jault, J.-M.; Meier, B.H.; Böckmann, A. Flexible-to-rigid transition is central for substrate transport in the ABC transporter BmrA from Bacillus subtilis. Commun. Biol. 2019, 2, 149. [Google Scholar] [CrossRef]
- Aravind, L.; Iyer, L.M.; Leipe, D.D.; Koonin, E.V. A novel family of P-loop NTPases with an unusual phyletic distribution and transmembrane segments inserted within the NTPase domain. Genome Biol. 2004, 5, R30. [Google Scholar] [CrossRef] [PubMed]
- Leipe, D.D.; Aravind, L.; Grishin, N.V.; Koonin, E.V. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000, 10, 5–16. [Google Scholar] [PubMed]
- Leipe, D.D.; Wolf, Y.I.; Koonin, E.V.; Aravind, L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002, 317, 41–72. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.; Saraste, M.; Runswick, M.; Gay, N. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982, 1, 945–951. [Google Scholar] [CrossRef]
- Saraste, M.; Sibbald, P.R.; Wittinghofer, A. The P-loop-A common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990, 15, 430–434. [Google Scholar] [CrossRef]
- Iyer, L.M.; Leipe, D.D.; Koonin, E.V.; Aravind, L. Evolutionary history and higher order classification of AAA + ATPases. J. Struct. Biol. 2004, 146, 11–31. [Google Scholar] [CrossRef]
- Orelle, C.; Dalmas, O.; Gros, P.; Di Pietro, A.; Jault, J.-M. The conserved glutamate residue adjacent to the walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J. Biol. Chem. 2003, 278, 47002–47008. [Google Scholar] [CrossRef]
- Feniouk, B.A.; Yoshida, M. Regulatory mechanisms of proton-translocating F(O)F (1)-ATP synthase. Results Probl. in cell Differ. 2008, 45, 279–308. [Google Scholar] [CrossRef]
- Wittinghofer, A. Phosphoryl transfer in Ras proteins, conclusive or elusive? Trends Biochem. Sci. 2006, 31, 20–23. [Google Scholar] [CrossRef]
- Wittinghofer, A.; Vetter, I.R. Structure-function relationships of the G Domain, a canonical switch motif. Annu. Rev. Biochem. 2011, 80, 943–971. [Google Scholar] [CrossRef] [PubMed]
- Gerwert, K.; Mann, D.; Kötting, C. Common mechanisms of catalysis in small and heterotrimeric GTPases and their respective GAPs. Biol. Chem. 2017, 398, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Shalaeva, D.N.; Cherepanov, D.A.; Galperin, M.Y.; Golovin, A.V.; Mulkidjanian, A.Y. Evolution of cation binding in the active sites of P-loop nucleoside triphosphatases in relation to the basic catalytic mechanism. Elife 2018, 7, e37373e. [Google Scholar] [CrossRef]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Ford, R.C.; Beis, K. Learning the ABCs one at a time: Structure and mechanism of ABC transporters. Biochem. Soc. Trans. 2019, 47, 23–36. [Google Scholar] [CrossRef]
- Ogawa, T.; Saijo, S.; Shimizu, N.; Jiang, X.; Hirokawa, N. Mechanism of catalytic microtubule depolymerization via KIF2-tubulin transitional conformation. Cell Rep. 2017, 20, 2626–2638. [Google Scholar] [CrossRef]
- Steinfels, E.; Orelle, C.; Fantino, J.-R.; Dalmas, O.; Rigaud, J.-L.; Denizot, F.; Di Pietro, A.; Jault, J.-M. Characterization of YvcC (BmrA), a multidrug ABC transporter constitutively expressed in Bacillus subtilis. Biochemistry 2004, 43, 7491–7502. [Google Scholar] [CrossRef]
- Tombline, G.; Senior, A.E. The occluded nucleotide conformation of P-glycoprotein. J. Bioenerg. Biomembr. 2005, 37, 497–500. [Google Scholar] [CrossRef]
- Van Veen, H.W.; Margolles, A.; Müller, M.; Higgins, C.F.; Konings, W.N. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J. 2000, 19, 2503–2514. [Google Scholar] [CrossRef]
- Orelle, C.; Jault, J.-M. Structures and transport mechanisms of the ABC efflux pumps. In Efflux-Mediated Antimicrobial Resistance in Bacteria; Springer Science and Business Media LLC: Basel, Switzerland, 2016; pp. 73–98. [Google Scholar]
- Collauto, A.; Mishra, S.; Litvinov, A.; Mchaourab, H.S.; Goldfarb, D. Direct spectroscopic detection of ATP turnover reveals mechanistic divergence of ABC exporters. Structure 2017, 25, 1264–1274.e3. [Google Scholar] [CrossRef]
- Soni, R.K.; Mehra, P.; Choudhury, N.R.; Mukhopadhyay, G.; Dhar, S.K. Functional characterization of Helicobacter pylori DnaB helicase. Nucleic Acids Research 2003, 31, 6828–6840. [Google Scholar] [CrossRef] [PubMed]
- Lapina, O.; Shubin, A.; Khabibulin, D.; Terskikh, V.V.; Bodart, P.; Amoureux, J.-P. Solid-state NMR for characterization of vanadium-containing systems. Catal. Today 2003, 78, 91–104. [Google Scholar] [CrossRef]
- Fernandez, C.; Bodart, P.; Amoureux, J.-P. Determination of 51V quadrupole and chemical shift tensor orientations in V2O5 by analysis of magic-angle spinning nuclear magnetic resonance spectra. Solid State Nucl. Magn. Reson. 1994, 3, 79–91. [Google Scholar] [CrossRef]
- Rehder, D.; Polenova, T.; Buhl, M. Vanadium-51 NMR. In Annual Reports on NMR Spectroscopy; Elsevier BV: Amsterdam, The Netherlands, 2007; Volume 62, pp. 49–114. [Google Scholar]
- Aureliano, M.; Tiago, T.; Gândara, R.M.; Sousa, A.; Moderno, A.; Kaliva, M.; Salifoglou, A.; Duarte, R.O.; Moura, J.J.G. Interactions of vanadium(V)-citrate complexes with the sarcoplasmic reticulum calcium pump. J. Inorg. Biochem. 2005, 99, 2355–2361. [Google Scholar] [CrossRef]
- Fenn, A.; Wächtler, M.; Gutmann, T.; Breitzke, H.; Buchholz, A.; Lippold, I.; Plass, W.; Buntkowsky, G. Correlations between 51V solid-state NMR parameters and chemical structure of vanadium (V) complexes as models for related metalloproteins and catalysts. Solid State Nucl. Magn. Reson. 2009, 36, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Pooransingh-Margolis, N.; Renirie, R.; Hasan, Z.; Wever, R.; Vega, A.J.; Polenova, T. 51V solid-state magic angle spinning NMR spectroscopy of vanadium chloroperoxidase. J. Am. Chem. Soc. 2006, 128, 5190–5208. [Google Scholar] [CrossRef]
- McCann, N.; Wagner, M.; Hasse, H. A thermodynamic model for vanadate in aqueous solution-equilibria and reaction enthalpies. Dalton Trans. 2013, 42, 2622–2628. [Google Scholar] [CrossRef] [PubMed]
- Iannuzzi, M.; Young, T.; Frankel, G.S. Aluminum alloy corrosion inhibition by vanadates. J. Electrochem. Soc. 2006, 153, B533–B541. [Google Scholar] [CrossRef]
- Lewinson, O.; Orelle, C.; Seeger, M.A. Structures of ABC transporters: Handle with care. FEBS Letters 2020. [Google Scholar] [CrossRef]
- Müller, B. ChemEQL Version 3.2; Swiss Federal Institute of Aquatic Science and Technology: Kastanienbaum, Switzerland, 2015. [Google Scholar]
- Bao, H.; Dalal, K.; Cytrynbaum, E.N.; Duong, F. Sequential action of MalE and maltose allows coupling ATP hydrolysis to translocation in the MalFGK2 transporter. J. Biol. Chem. 2015, 290, 25452–25460. [Google Scholar] [CrossRef]
- Mehmood, S.; Domene, C.; Forest, E.; Jault, J.-M. Dynamics of a bacterial multidrug ABC transporter in the inward-and outward-facing conformations. Proc. Natl. Acad. Sci. USA 2012, 109, 10832–10836. [Google Scholar] [CrossRef] [PubMed]
- Singleton, M.R.; Sawaya, M.R.; Ellenberger, T.; Wigley, D.B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 2000, 101, 589–600. [Google Scholar] [CrossRef]
- Bailey, S.; Eliason, W.K.; Steitz, T.A. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 2007, 318, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef] [PubMed]
- Enemark, E.J.; Joshua-Tor, L. On helicases and other motor proteins. Curr. Opin. Struct. Biol. 2008, 18, 243–257. [Google Scholar] [CrossRef]
- Wang, G.; Klein, M.G.; Tokonzaba, E.; Zhang, Y.; Holden, L.G.; Chen, X.S. The structure of a DnaB-family replicative helicase and its interactions with primase. Nat. Struct. Mol. Biol. 2007, 15, 94–100. [Google Scholar] [CrossRef]
- Itsathitphaisarn, O.; Wing, R.A.; Eliason, W.K.; Wang, J.; Steitz, T.A. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 2012, 151, 267–277. [Google Scholar] [CrossRef]
- Gao, Y.; Cui, Y.; Fox, T.; Lin, S.; Wang, H.; De Val, N.; Zhou, Z.H.; Yang, W. Structures and operating principles of the replisome. Science 2019, 363, eaav7003. [Google Scholar] [CrossRef]
- Gordon, J.A. Use of vanadate as protein-phosphotyrosine phosphatase inhibitor. In Methods in Enzymology; Academic Press: San Diego, CA, USA, 1991; Volume 201, pp. 477–482. [Google Scholar]
- Gor’kov, P.L.; Witter, R.; Chekmenev, E.Y.; Nozirov, F.; Fu, R.; Brey, W.W. Low-E probe for (19)F-(1)H NMR of dilute biological solids. J. Magn. Reson. 2007, 189, 182–189. [Google Scholar] [CrossRef]
- Fogh, R.; Ionides, J.; Ulrich, E.; Boucher, W.; Vranken, W.; Linge, J.P.; Habeck, M.; Rieping, W.; Bhat, T.N.; Westbrook, J.; et al. The CCPN project: An interim report on a data model for the NMR community. Nat. Struct. Biol. 2002, 9, 416–418. [Google Scholar] [CrossRef]
- Stevens, T.J.; Fogh, R.H.; Boucher, W.; Higman, V.A.; Eisenmenger, F.; Bardiaux, B.; van Rossum, B.J.; Oschkinat, H.; Laue, E.D. A software framework for analysing solid-state MAS NMR data. J. Biomol. NMR 2011, 51, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
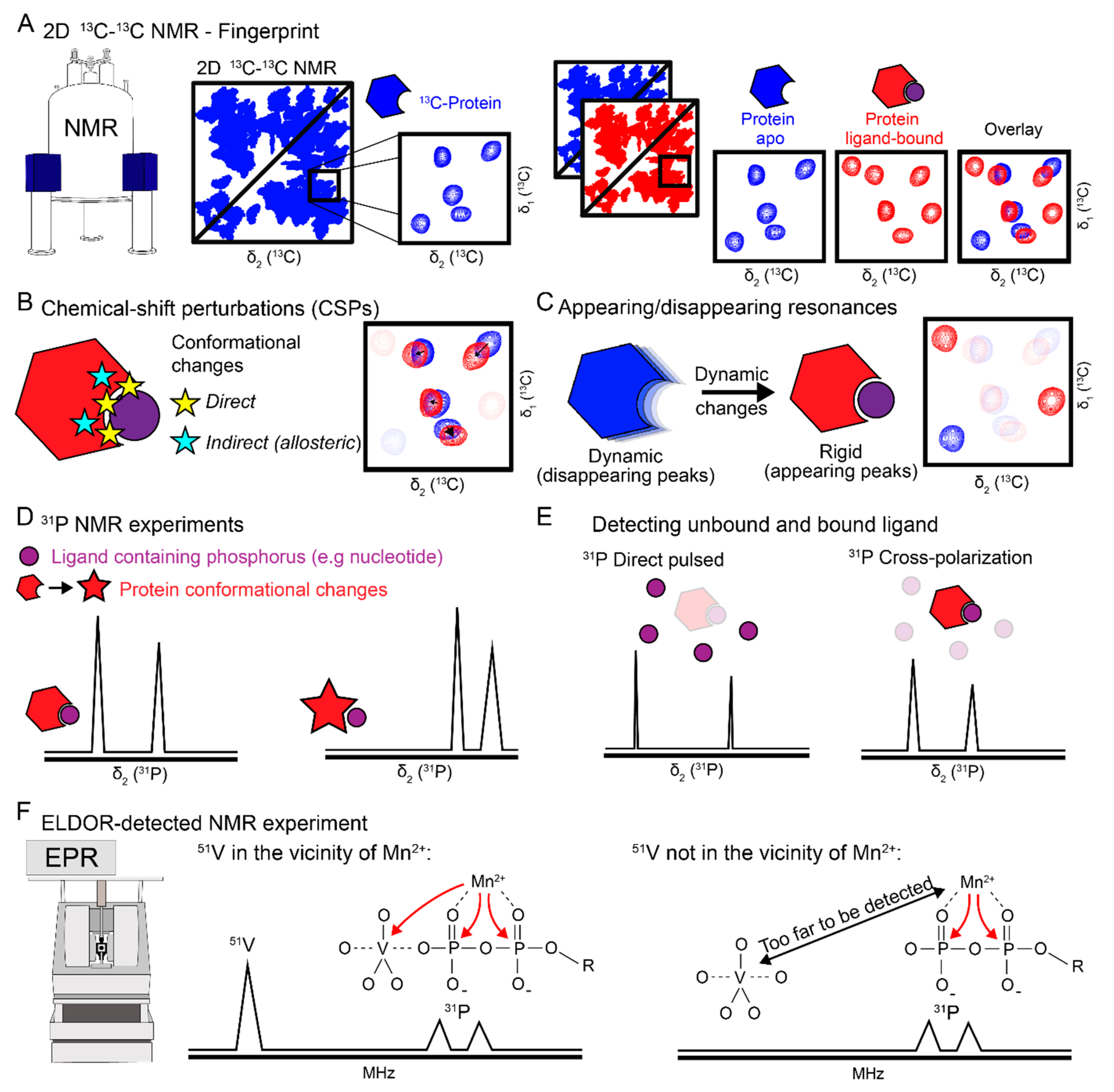



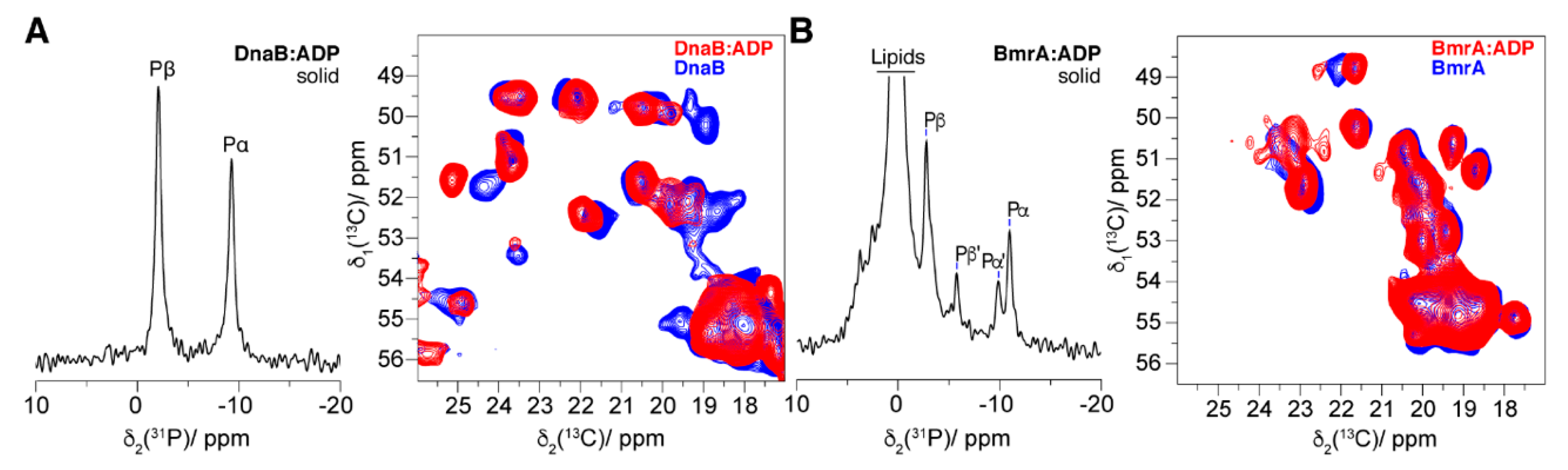
| Magnetic-Resonance Technique | Protein Conformational Changes | Protein Dynamic Changes | Nucleotide Binding | Nucleotide Conformation | Vanadate Binding | Metal Co-Factor Binding |
|---|---|---|---|---|---|---|
| NMR 13C/15N fingerprints | x | x | x | |||
| CSPs | x | x | ||||
| Disappearing/appearing resonances | x | x | ||||
| 31P CP-MAS | x | |||||
| 31P chemical-shift values | x | x | x | |||
| 51V NMR | x | |||||
| EDNMR | x | x |
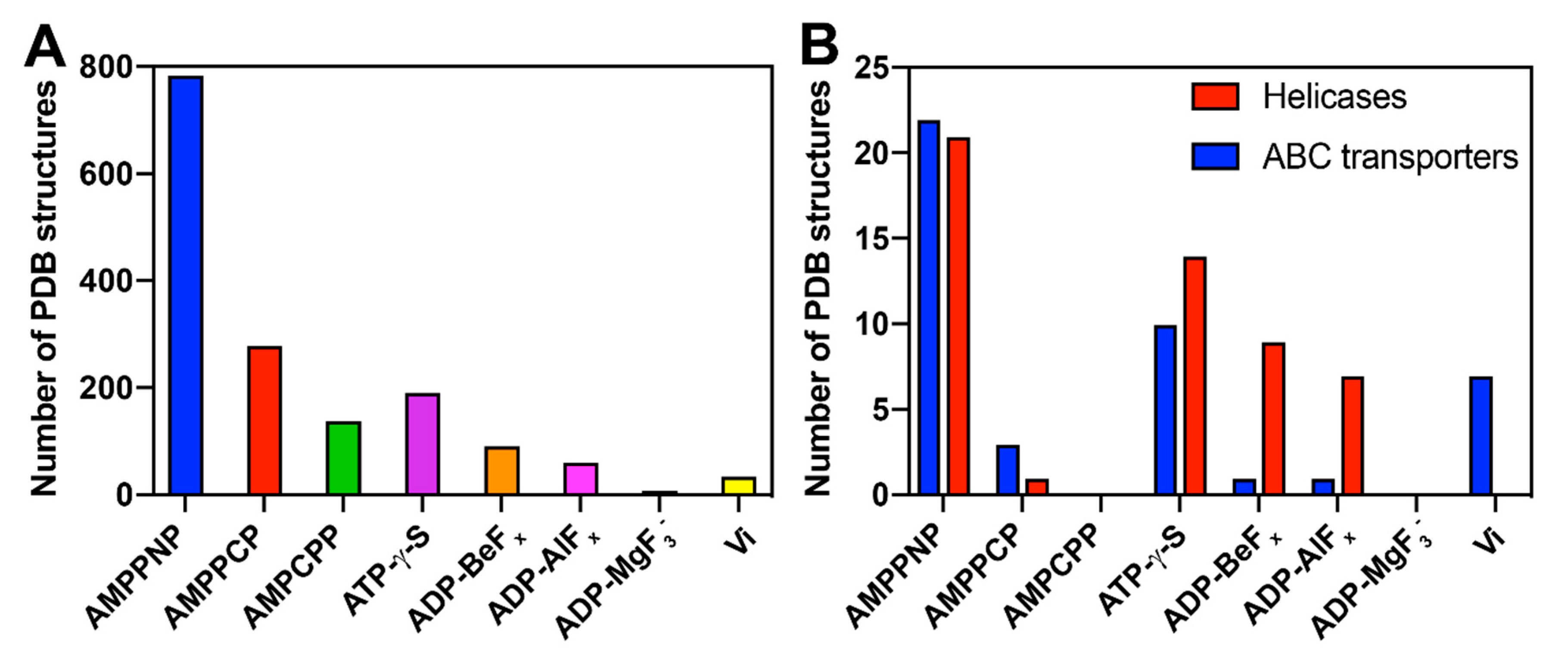
| Analogue | State Being Mimicked | Suitability | Comments | |
|---|---|---|---|---|
| DnaB | BmrA | |||
| AMPPNP | Pre-hydrolytic | - | -- | Hydrolysed by DnaB in the presence of DNA Hydrolysed by BmrA |
| AMPPCP | Pre-hydrolytic | ++ | -- | Rigidifies DnaB in the presence of DNA Hydrolysis observed |
| ATP-γ-S | Pre-hydrolytic | -- | -- | Hydrolysed by BmrA and DnaB |
| Vi | Transition-state | -- | ++ | Inhibits the DNA binding in DnaB Provides the transition state for BmrA |
| AlFx | Transition-state | ++ | + | Provides the transition state for DnaB and BmrA Starts to precipitate at pH ≥7 |
| ADP | Post-hydrolytic | ++ | ++ | Provides the post-hydrolysis state in both systems |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lacabanne, D.; Wiegand, T.; Wili, N.; Kozlova, M.I.; Cadalbert, R.; Klose, D.; Mulkidjanian, A.Y.; Meier, B.H.; Böckmann, A. ATP Analogues for Structural Investigations: Case Studies of a DnaB Helicase and an ABC Transporter. Molecules 2020, 25, 5268. https://doi.org/10.3390/molecules25225268
Lacabanne D, Wiegand T, Wili N, Kozlova MI, Cadalbert R, Klose D, Mulkidjanian AY, Meier BH, Böckmann A. ATP Analogues for Structural Investigations: Case Studies of a DnaB Helicase and an ABC Transporter. Molecules. 2020; 25(22):5268. https://doi.org/10.3390/molecules25225268
Chicago/Turabian StyleLacabanne, Denis, Thomas Wiegand, Nino Wili, Maria I. Kozlova, Riccardo Cadalbert, Daniel Klose, Armen Y. Mulkidjanian, Beat H. Meier, and Anja Böckmann. 2020. "ATP Analogues for Structural Investigations: Case Studies of a DnaB Helicase and an ABC Transporter" Molecules 25, no. 22: 5268. https://doi.org/10.3390/molecules25225268
APA StyleLacabanne, D., Wiegand, T., Wili, N., Kozlova, M. I., Cadalbert, R., Klose, D., Mulkidjanian, A. Y., Meier, B. H., & Böckmann, A. (2020). ATP Analogues for Structural Investigations: Case Studies of a DnaB Helicase and an ABC Transporter. Molecules, 25(22), 5268. https://doi.org/10.3390/molecules25225268