Genetic Correlates of Synergy Mechanisms of Daptomycin Plus Fosfomycin in Daptomycin-Susceptible and -Resistant Methicillin-Resistant Staphylococcus aureus (MRSA)
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Minimum Inhibitory Concentrations (MICs)
2.3. Serial In Vitro Passage Experiments
2.4. CM Phospholipid (PL) Composition
2.5. Genetic Profiling
2.6. In Vitro qRT-PCR Validation of the Selected Genes
2.7. Rabbit IE Model and RNA Extraction from In Vivo Rabbit Vegetations
2.8. Statistical Analysis
3. Result and Discussion
3.1. Whole Genome Sequencing (WGS)
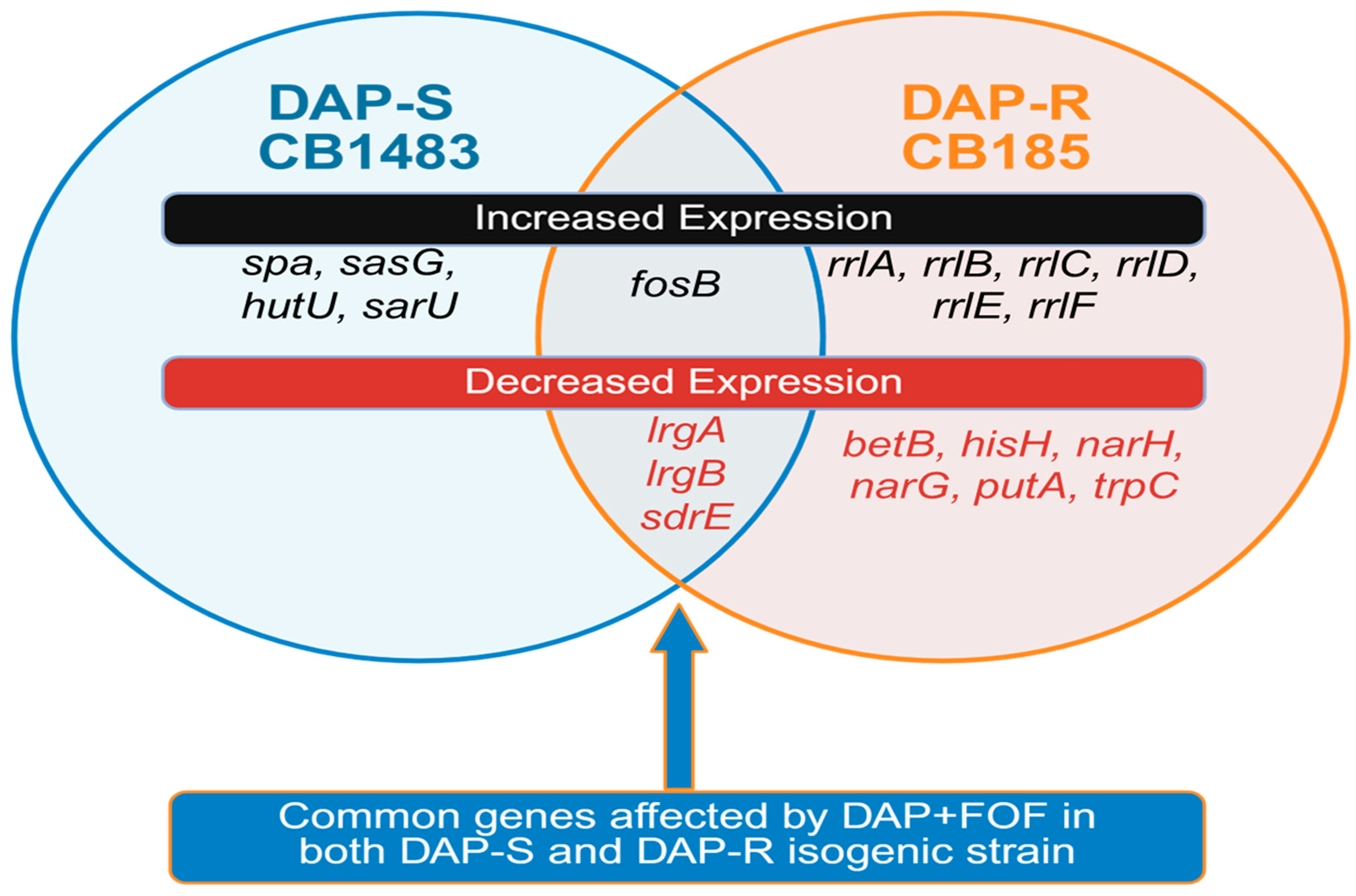
3.2. RNA Sequencing (RNA-Seq)-Based Transcriptomics
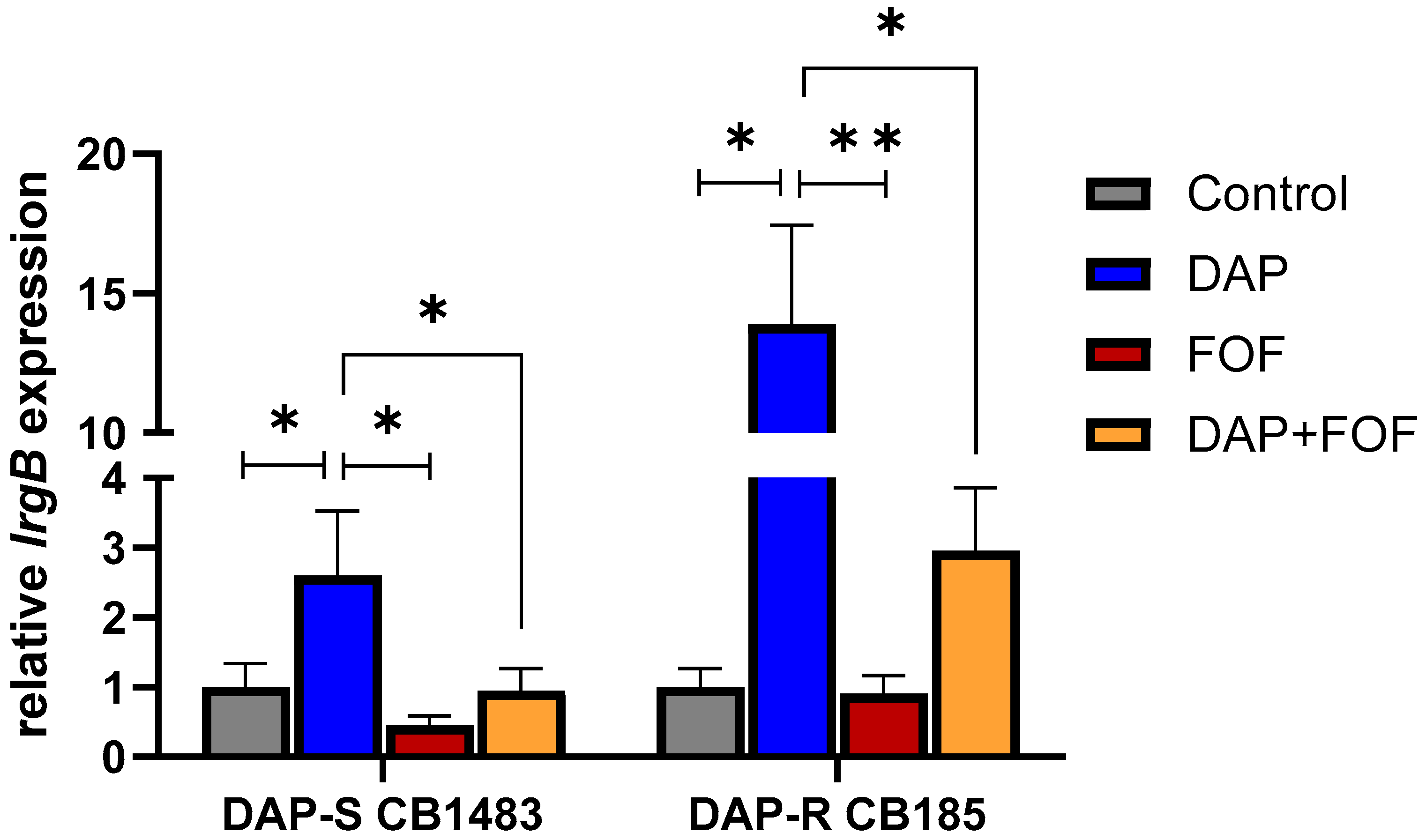
3.3. qRT-PCR Validation of the lrgB Gene
3.4. Impacts of DAP-FOF Therapy upon Expression of lrgB Genes in Rabbit IE Vegetations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roch, M.; Gagetti, P.; Davis, J.; Ceriana, P.; Errecalde, L.; Corso, A.; Rosato, A.E. Daptomycin resistance in clinical MRSA strains is associated with a high biological fitness cost. Front. Microbiol. 2017, 8, 2303. [Google Scholar] [CrossRef] [PubMed]
- Holland, T.L.; Cosgrove, S.E.; Doernberg, S.B.; Jenkins, T.C.; Turner, N.A.; Boucher, H.W.; Pavlov, O.; Titov, I.; Kosulnykov, S.; Atanasov, B.; et al. ERADICATE Study Group. Ceftobiprole for treatment of complicated staphylococcus aureus bacteremia. N. Engl. J. Med. 2023, 389, 1390–1401. [Google Scholar] [CrossRef] [PubMed]
- Fowler, V.G., Jr.; Boucher, H.W.; Corey, G.R.; Abrutyn, E.; Karchmer, A.W.; Rupp, M.E.; Levine, D.P.; Chambers, H.F.; Tally, F.P.; Vigliani, G.A.; et al. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 2006, 355, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Rose, W.E.; Fallon, M.; Moran, J.J.; Vanderloo, J.P. Vancomycin tolerance in methicillin-resistant Staphylococcus aureus: Influence of vancomycin, daptomycin, and telavancin on differential resistance gene expression. Antimicrob. Agents Chemother. 2012, 56, 4422–4427. [Google Scholar] [CrossRef]
- Marty, F.M.; Yeh, W.W.; Wennersten, C.B.; Venkataraman, L.; Albano, E.; Alyea, E.P.; Gold, H.S.; Baden, L.R.; Pillai, S.K. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J. Clin. Microbiol. 2006, 44, 595–597. [Google Scholar] [CrossRef]
- Oku, Y.; Kurokawa, K.; Ichihashi, N.; Sekimizu, K. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 2004, 150, 45–51. [Google Scholar] [CrossRef]
- Andrä, J.; Goldmann, T.; Ernst, C.M.; Peschel, A.; Gutsmann, T. Multiple peptide resistance factor (MprF)-mediated resistance of Staphylococcus aureus against antimicrobial peptides coincides with a modulated peptide interaction with artificial membranes comprising lysyl-phosphatidylglycerol. J. Biol. Chem. 2011, 286, 18692–18700. [Google Scholar] [CrossRef]
- Ernst, C.M.; Peschel, A. MprF-mediated daptomycin resistance. Int. J. Med. Microbiol. 2019, 309, 359–363. [Google Scholar] [CrossRef]
- Ernst, C.M.; Staubitz, P.; Mishra, N.N.; Yang, S.J.; Hornig, G.; Kalbacher, H.; Bayer, A.S.; Kraus, D.; Peschel, A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009, 5, e1000660. [Google Scholar] [CrossRef]
- Bayer, A.S.; Schneider, T.; Sahl, H.G. Mechanisms of daptomycin resistance in Staphylococcus aureus: Role of the cell membrane and cell wall. Ann. N. Y. Acad. Sci. 2013, 1277, 139–158. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R.; Taglialegna, A.; Rosato, A.E. Impact of PrsA on membrane lipid composition during daptomycin-resistance-mediated β-lactam sensitization in clinical MRSA strains. J. Antimicrob. Chemother. 2021, 77, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Kawada, H.; Uchida, H.; Takagi, Y.; Obata, S.; Eda, R.; Hanaki, H.; Kitasato, H. Single nucleotide polymorphism leads to daptomycin resistance causing amino acid substitution-T345I in MprF of clinically isolated MRSA strains. PLoS ONE 2021, 16, e0245732. [Google Scholar] [CrossRef] [PubMed]
- Vásquez, A.; Leidy, C.; Manrique-Moreno, M. Lysyl-Phosphatidylglycerol: A Lipid Involved in the resistance of Staphylococcus aureus to antimicrobial peptide activity. Antibiotics 2025, 14, 349. [Google Scholar] [CrossRef]
- Chen, F.J.; Lauderdale, T.L.; Lee, C.H.; Hsu, Y.C.; Huang, I.W.; Hsu, P.C.; Yang, C.S. Effect of a point mutation in mprF on susceptibility to daptomycin, vancomycin, and oxacillin in an MRSA clinical strain. Front. Microbiol. 2018, 9, 1086. [Google Scholar] [CrossRef] [PubMed]
- Camargo, I.L.; Neoh, H.M.; Cui, L.; Hiramatsu, K. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob. Agents Chemother. 2008, 52, 4289–4299. [Google Scholar] [CrossRef]
- Nguyen, A.H.; Hood, K.S.; Mileykovskaya, E.; Miller, W.R.; Tran, T.T. Bacterial cell membranes and their role in daptomycin resistance: A review. Front. Mol. Biosci. 2022, 9, 1035574. [Google Scholar] [CrossRef]
- Gaupp, R.; Lei, S.; Reed, J.M.; Peisker, H.; Boyle-Vavra, S.; Bayer, A.S.; Bischoff, M.; Herrmann, M.; Daum, R.S.; Powers, R.; et al. Staphylococcus aureus metabolic adaptations during the transition from a daptomycin susceptibility phenotype to a daptomycin nonsusceptibility phenotype. Antimicrob. Agents Chemother. 2015, 59, 4226–4238. [Google Scholar] [CrossRef]
- Rio-Marques, L.; Hartke, A.; Bizzini, A. The effect of inoculum size on selection of in vitro resistance to vancomycin, daptomycin, and linezolid in methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 2014, 20, 539–543. [Google Scholar] [CrossRef]
- Jorgensen, S.C.J.; Zasowski, E.J.; Trinh, T.D.; Lagnf, A.M.; Bhatia, S.; Sabagha, N.; Abdul-Mutakabbir, J.C.; Alosaimy, S.; Mynatt, R.P.; Davis, S.L.; et al. Daptomycin plus β-Lactam combination therapy for methicillin-resistant Staphylococcus aureus bloodstream infections: A Retrospective, comparative cohort study. Clin. Infect. Dis. 2020, 71, 1–10. [Google Scholar] [CrossRef]
- Pujol, M.; Miro, J.M.; Shaw, E.; Aguado, J.M.; San-Juan, R.; Puig-Asensio, M.; Pigrau, C.; Calbo, E.; Montejo, M.; Rodriguez-Alvarez, R.; et al. Daptomycin plus fosfomycin versus daptomycin alone for methicillin-resistant Staphylococcus aureus bacteremia and endocarditis: A randomized clinical trial. Clin. Infect. Dis. 2020, 72, 1517–1525. [Google Scholar] [CrossRef]
- Geriak, M.; Haddad, F.; Rizvi, K.; Rose, W.; Kullar, R.; LaPlante, K.; Yu, M.; Vasina, L.; Ouellette, K.; Zervos, M.; et al. Clinical data on daptomycin plus ceftaroline versus standard of care monotherapy in the treatment of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 2019, 63, e02483-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ye, C.; Liao, L.; Wang, Z.; Hu, Y.; Deng, C.; Liu, L. Adjuvant β-lactam therapy combined with vancomycin or daptomycin for methicillin-resistant Staphylococcus aureus bacteremia: A systematic review and meta-analysis. Antimicrob. Agents Chemother. 2020, 64, e01377-20. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.N.; Lew, C.; Abdelhady, W.; Lapitan, C.K.; Proctor, R.A.; Rose, W.E.; Bayer, A.S. Synergy mechanisms of daptomycin-fosfomycin combinations in daptomycin-susceptible and -resistant methicillin-resistant Staphylococcus aureus: In vitro, ex vivo, and in vivo metrics. Antimicrob. Agents Chemother. 2022, 66, e0164921. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.M.; Gardner, S.G.; Mishra, N.N.; Bayer, A.S.; Somerville, G.A. Metabolic interventions for the prevention and treatment of daptomycin non-susceptibility in Staphylococcus aureus. J. Antimicrob. Chemother. 2019, 74, 2274–2283. [Google Scholar] [CrossRef]
- Falagas, M.E.; Vouloumanou, E.K.; Samonis, G.; Vardakas, K.Z. Fosfomycin. Clin. Microbiol. Rev. 2016, 29, 321–347. [Google Scholar] [CrossRef]
- Bayer, A.S.; Mishra, N.N.; Chen, L.; Kreiswirth, B.N.; Rubio, A.; Yang, S.J. Frequency and distribution of single-nucleotide polymorphisms within mprf in methicillin-resistant Staphylococcus aureus clinical isolates and their role in cross-resistance to daptomycin and host defense antimicrobial peptides. Antimicrob. Agents Chemother. 2015, 59, 4930–4937. [Google Scholar] [CrossRef]
- Mishra, N.N.; McKinnell, J.; Yeaman, M.R.; Rubio, A.; Nast, C.C.; Chen, L.; Kreiswirth, B.N.; Bayer, A.S. In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 2011, 55, 4012–4018. [Google Scholar] [CrossRef]
- Mishra, N.N.; Bayer, A.S.; Baines, S.L.; Hayes, A.S.; Howden, B.P.; Lapitan, C.K.; Lew, C.; Rose, W.E. Cell membrane adaptations mediate β-lactam-induced resensitization of daptomycin-resistant (DAP-R) Staphylococcus aureus in vitro. Microorganisms. 2021, 9, 1028. [Google Scholar] [CrossRef]
- CLSI. Performnace Standards for Antimicrobial Susceptibility Testing, 34th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2024. [Google Scholar]
- Thitiananpakorn, K.; Aiba, Y.; Tan, X.E.; Watanabe, S.; Kiga, K.; Sato'o, Y.; Boonsiri, T.; Li, F.Y.; Sasahara, T.; Taki, Y.; et al. Association of mprF mutations with cross-resistance to daptomycin and vancomycin in methicillin-resistant Staphylococcus aureus (MRSA). Sci. Rep. 2020, 10, 16107. [Google Scholar] [CrossRef]
- Jiang, H.; Lei, R.; Ding, S.W.; Zhu, S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014, 15, 1–12. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. Available online: https://raivokolde.r-universe.dev/pheatmap (accessed on 5 June 2025).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Taglialegna, A.; Varela, M.C.; Rosato, R.R.; Rosato, A.E. VraSR and virulence trait modulation during daptomycin resistance in methicillin-resistant Staphylococcus aureus infection. mSphere 2019, 4, e00557-18. [Google Scholar] [CrossRef] [PubMed]
- Ersoy, S.C.; Abdelhady, W.; Li, L.; Chambers, H.F.; Xiong, Y.Q.; Bayer, A.S. Bicarbonate resensitization of methicillin-resistant Staphylococcus aureus to β-Lactam antibiotics. Antimicrob. Agents Chemother. 2019, 63, e00496-19. [Google Scholar] [CrossRef]
- Vlaeminck, J.; Lin, Q.; Xavier, B.B.; De Backer, S.; Berkell, M.; De Greve, H.; Hernalsteens, J.P.; Kumar-Singh, S.; Goossens, H.; Malhotra-Kumar, S. The dynamic transcriptome during maturation of biofilms formed by methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2022, 13, 882346. [Google Scholar] [CrossRef]
- Elgharably, H.; Claesen, J.; Sangwan, N.; Etiwy, M.; Houghtaling, P.; Procop, G.; Shrestha, N.K.; Griffin, B.; Navia, J.L.; Svensson, L.G.; et al. In vivo virulence of Staphylococcus aureus in native versus prosthetic left-sided valve endocarditis. JTCVS Open 2024, 24, 156–169. [Google Scholar] [CrossRef]
- Fu, Z.; Liu, Y.; Chen, C.; Guo, Y.; Ma, Y.; Yang, Y.; Hu, F.; Xu, X.; Wang, M. Characterization of fosfomycin resistance gene, fosB, in methicillin-resistant Staphylococcus aureus isolates. PLoS ONE 2016, 11, e0154829. [Google Scholar] [CrossRef] [PubMed]
- Beltrame, C.O.; Côrtes, M.F.; Bonelli, R.R.; Côrrea, A.B.; Botelho, A.M.; Américo, M.A.; Fracalanzza, S.E.; Figueiredo, A.M. Inactivation of the autolysis-related genes lrgB and yycI in Staphylococcus aureus Increases cell lysis-dependent eDNA release and enhances biofilm development in vitro and in vivo. PLoS ONE 2015, 10, e0138924. [Google Scholar] [CrossRef]
- Ranjit, D.K.; Endres, J.L.; Bayles, K.W. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J Bacteriol. 2011, 193, 2468–2476. [Google Scholar] [CrossRef]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E.; Smeltzer, M.S.; Bayles, K.W. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, M.; Hang, T.; Wang, C.; Yang, Y.; Pan, W.; Zang, J.; Zhang, M.; Zhang, X. Staphylococcus aureus SdrE captures complement factor H’s C-terminus via a novel ‘close, dock, lock and latch’ mechanism for complement evasion. Biochem. J. 2017, 474, 1619–1631. [Google Scholar] [CrossRef]

| Polymorphisms in mprF, Amino Acid Changes (# of DAP-R Strains) | Polymorphisms Among Other Genes (# of DAP-R Strains) |
|---|---|
| Leu826Phe (11); Thr345Ala/Lys/Ile (5); Ser337Leu (2); Ser295Leu (2); Leu341Ser (4); Met347Arg (2); Val351Glu (1); Thr472Lys (1); Ile420Asn (1); Phe349_Asn352del (1), Pro314Leu (1); Leu42del (1); No mutation (1) | cls2 (5); cls1 (1); dltD (1); yycG (3); crtN (1); lytN (2); vraRS (4); fabFH (2) |
| Strain | Antibiotic Exposure for 10 Days | DAP MIC (µg/mL) | FOF MIC (µg/mL) | mprF or cls SNP | % of PL Composition (Mean ± SD) | ||
|---|---|---|---|---|---|---|---|
| L-PG | PG | CL | |||||
| DAP-S 1483 | Control ** | 0.25 (S) | 8 (S) | None | 14 ± 3 | 76 ± 3 | 9 ± 1 |
| DAP | 4 (R) | 8 (S) | mprF (L826F); cls2: (Leu52Phe) | 25 ± 6 * | 58 ± 9 * | 17 ± 6 | |
| FOF | 0.5 (S) | >256 (R) | None | 12 ± 3 | 76 ± 9 | 13 ± 6 | |
| DAP + FOF | 1 (S) | 8 (S) | None | 12 ± 1 | 74 ± 11 | 14 ± 12 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rose, W.E.; Ersoy, S.C.; Abdelhady, W.; Dominguez, A.R.; Muyah Manna, J.N.; Artaza, J.N.; Mishra, R.; Elsayed, A.M.; Proctor, R.A.; Baines, S.L.; et al. Genetic Correlates of Synergy Mechanisms of Daptomycin Plus Fosfomycin in Daptomycin-Susceptible and -Resistant Methicillin-Resistant Staphylococcus aureus (MRSA). Microorganisms 2025, 13, 1532. https://doi.org/10.3390/microorganisms13071532
Rose WE, Ersoy SC, Abdelhady W, Dominguez AR, Muyah Manna JN, Artaza JN, Mishra R, Elsayed AM, Proctor RA, Baines SL, et al. Genetic Correlates of Synergy Mechanisms of Daptomycin Plus Fosfomycin in Daptomycin-Susceptible and -Resistant Methicillin-Resistant Staphylococcus aureus (MRSA). Microorganisms. 2025; 13(7):1532. https://doi.org/10.3390/microorganisms13071532
Chicago/Turabian StyleRose, Warren E., Selvi C. Ersoy, Wessam Abdelhady, Alan R. Dominguez, Jedidiah Ndam Muyah Manna, Jorge N. Artaza, Reetakshi Mishra, Ahmed M. Elsayed, Richard A. Proctor, Sarah L. Baines, and et al. 2025. "Genetic Correlates of Synergy Mechanisms of Daptomycin Plus Fosfomycin in Daptomycin-Susceptible and -Resistant Methicillin-Resistant Staphylococcus aureus (MRSA)" Microorganisms 13, no. 7: 1532. https://doi.org/10.3390/microorganisms13071532
APA StyleRose, W. E., Ersoy, S. C., Abdelhady, W., Dominguez, A. R., Muyah Manna, J. N., Artaza, J. N., Mishra, R., Elsayed, A. M., Proctor, R. A., Baines, S. L., Howden, B. P., & Mishra, N. N. (2025). Genetic Correlates of Synergy Mechanisms of Daptomycin Plus Fosfomycin in Daptomycin-Susceptible and -Resistant Methicillin-Resistant Staphylococcus aureus (MRSA). Microorganisms, 13(7), 1532. https://doi.org/10.3390/microorganisms13071532





