The Characteristics of the Root-Zone Soil’s Biological Properties and Microbial Community Structure in Grafted Star Anise Plantations
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Soil Sampling
2.2. Analysis of Soil Biological Properties
2.3. Analysis of Soil Microbial Diversity
2.4. Statistical Analysis
3. Results
3.1. Soil Enzyme Activities
3.2. Soil Microbial Biomass
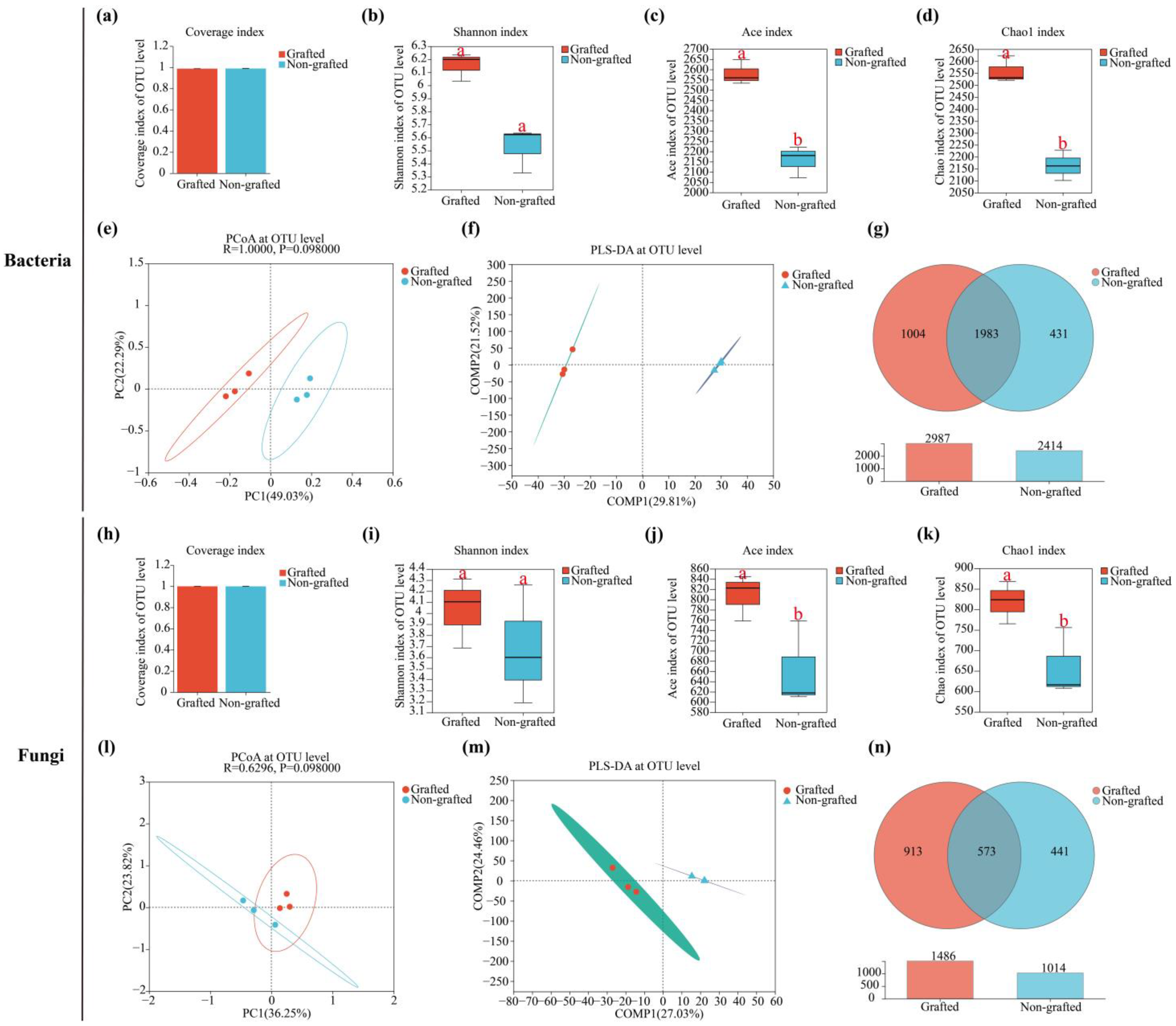
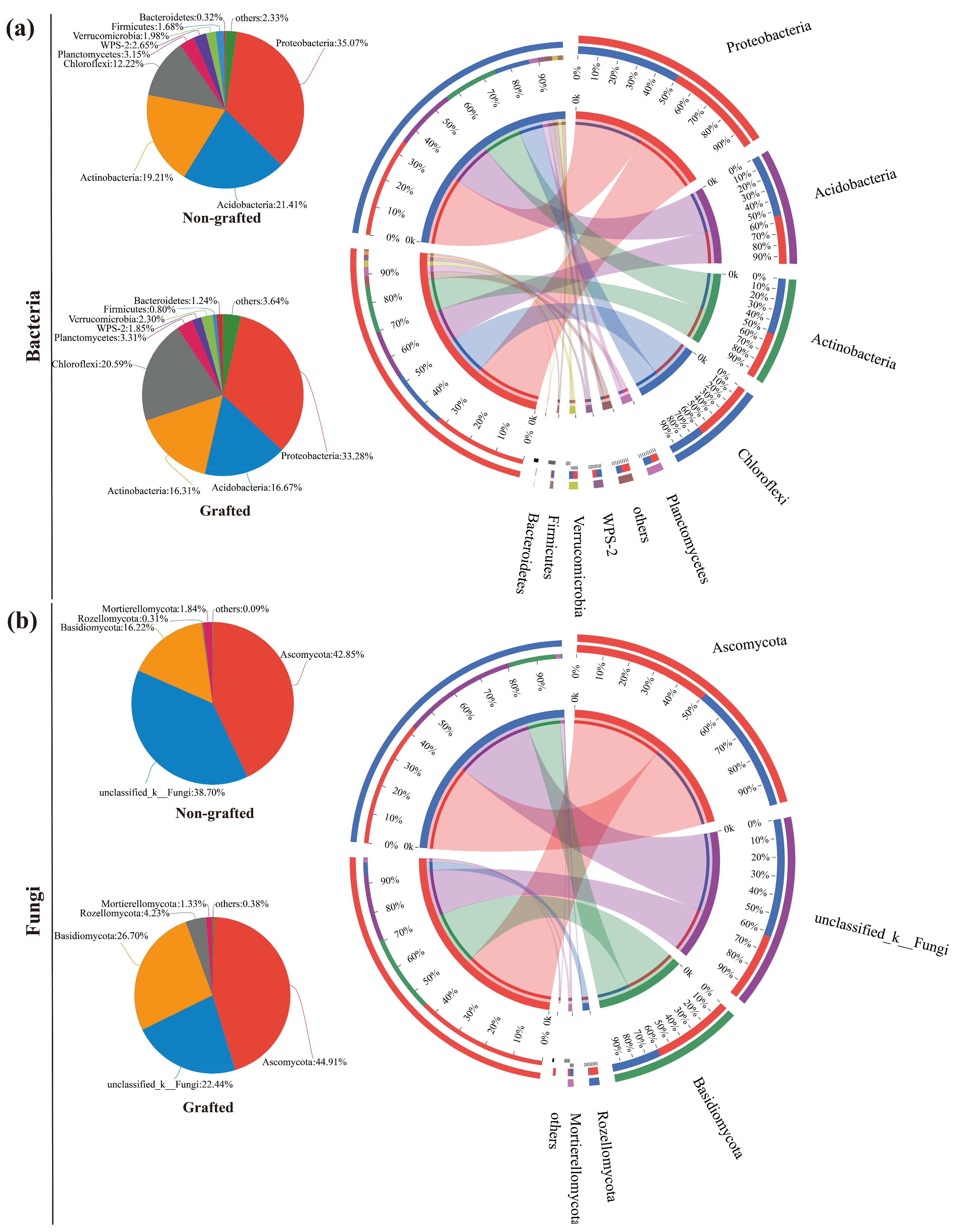
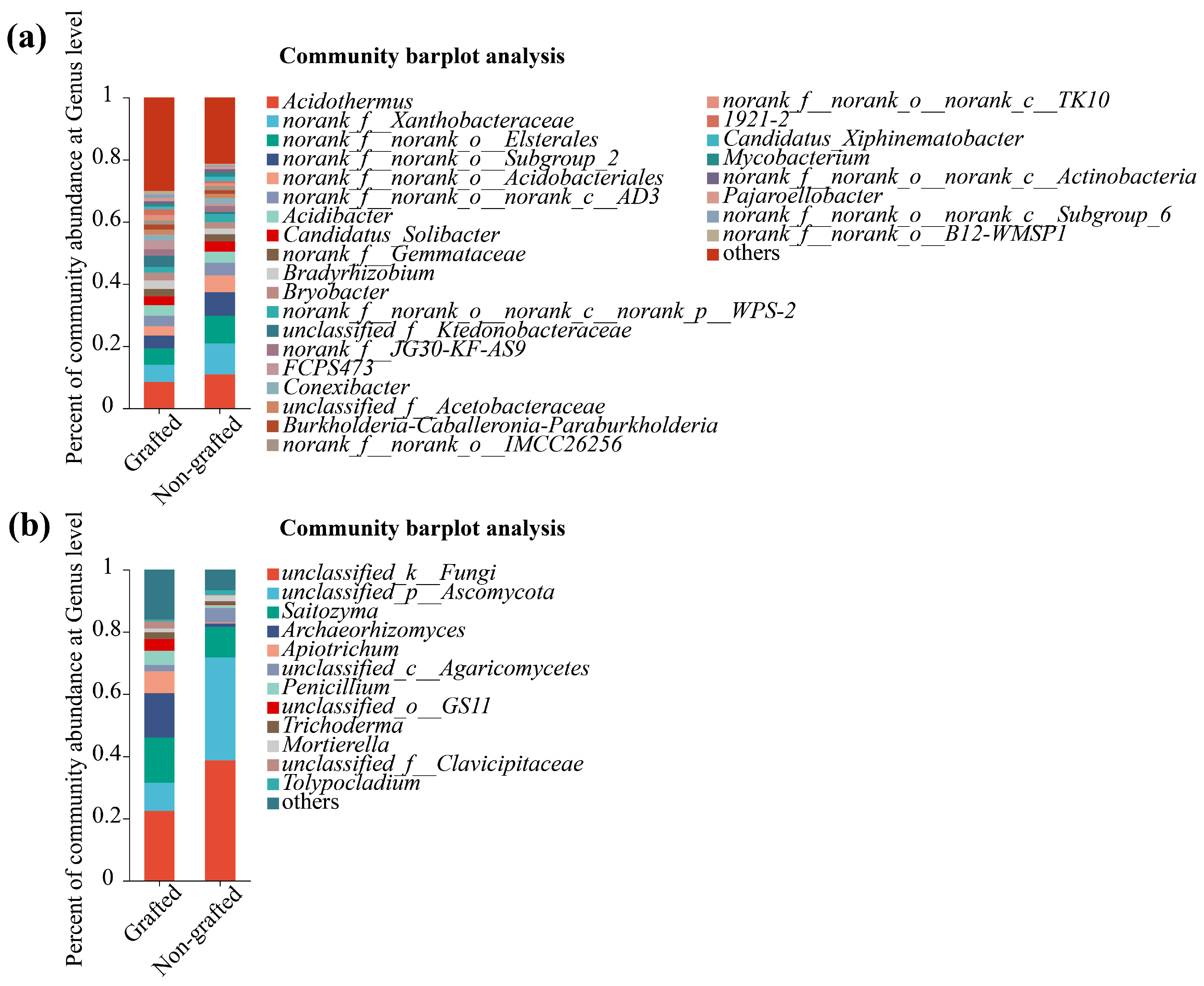
3.3. Soil Microbiological Diversity and Community Analysis
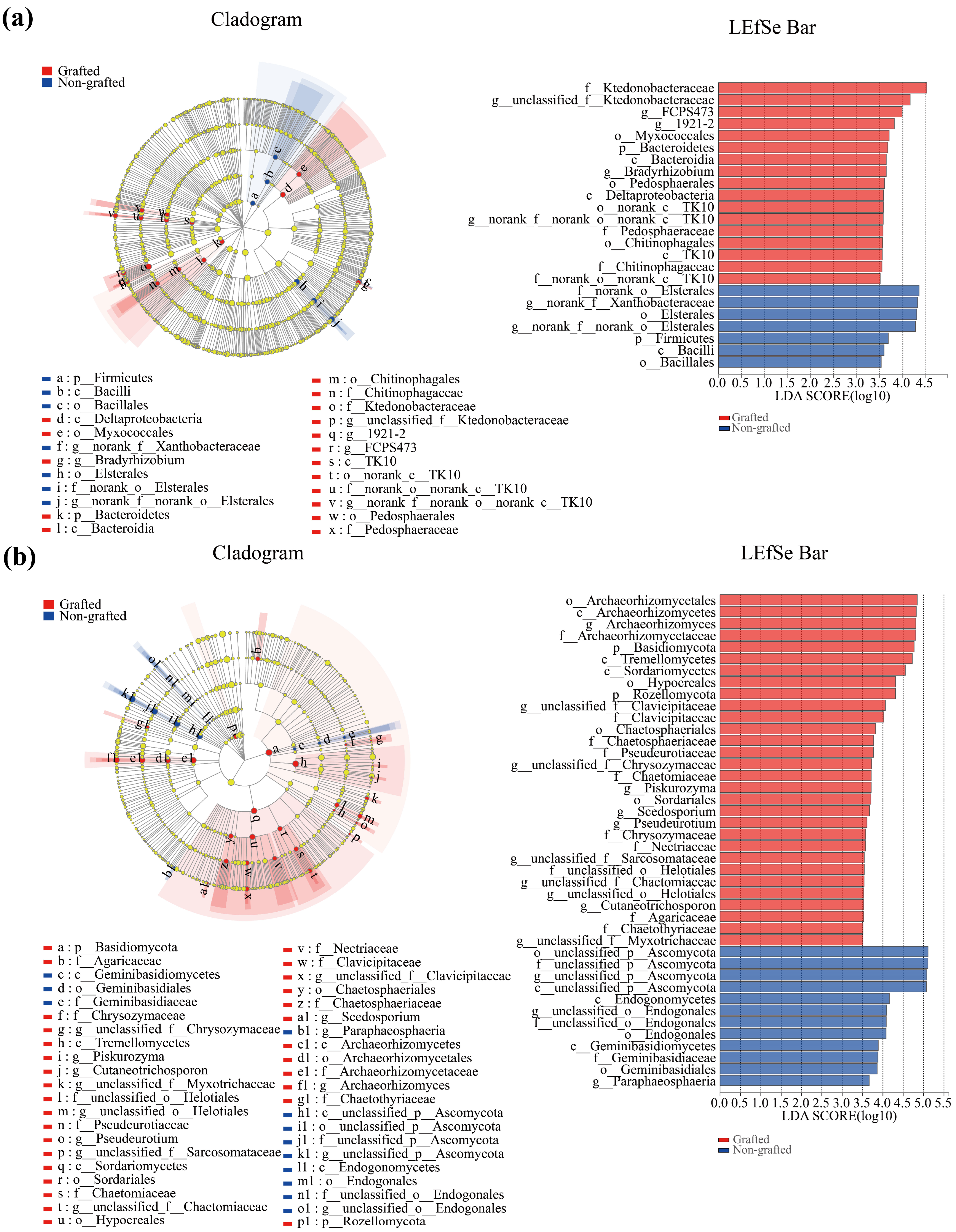
3.4. Soil Microbiological LEfSe Analysis and Function Prediction
3.5. Correlation Network Analysis
4. Discussion
4.1. Response of Soil Enzyme Activity and Microbial Biomass to Grafting in Star Anise Plantations
4.2. Response of Soil Microbial Diversity to Grafting in Star Anise Plantations
4.3. Response of Soil Microbial Community to Grafting in Star Anise Plantations
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, J.; Chen, S.; Sun, Y.; Wu, S.; Liang, W.; Yang, S. Effects of mechanical weeding on soil fertility and microbial community structure in star anise (Illicium verum Hook.f.) plantations. PLoS ONE 2022, 17, e0266949. [Google Scholar] [CrossRef] [PubMed]
- Assiry, A.A.; Karobari, M.I.; Bhavikatti, S.K.; Marya, A. Crossover analysis of the astringent, antimicrobial, and anti-inflammatory effects of Illicium verum/star anise in the oral cavity. BioMed Res. Int. 2021, 2021, 5510174. [Google Scholar] [CrossRef] [PubMed]
- Shahrajabian, M.H.; Sun, W.L.; Cheng, Q. Chinese star anise and anise, magic herbs in traditional Chinese medicine and modern pharmaceutical science. Asian J. Med. Biol. Res. 2019, 5, 162–179. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, G.; Bose, S.; Banerjee, S.; Vishnuprasad, C.N.; del Pilar Rodriguez-Torres, M.; Shin, H.S. Star anise (Illicium verum): Chemical compounds, antiviral properties, and clinical relevance. Phytother. Res. 2020, 34, 1248–1267. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, L.; Wu, R. Plant grafting: How genetic exchange promotes vascular reconnection. New Phytol. 2017, 214, 56–65. [Google Scholar] [CrossRef]
- Goldschmidt, E.E. Plant grafting: New mechanisms, evolutionary implications. Front. Plant Sci. 2014, 5, 727. [Google Scholar] [CrossRef]
- Lee, J.M.; Kubota, C.; Tsao, S.J.; Bie, Z.; Echevarria, P.H.; Morra, L.; Oda, M. Current status of vegetable grafting: Diffusion, grafting techniques, automation. Sci. Hortic. 2010, 127, 93–105. [Google Scholar] [CrossRef]
- Tsaballa, A.; Athanasiadis, C.; Pasentsis, K.; Ganopoulos, I.; Nianiou-Obeidat, I.; Tsaftaris, A. Molecular studies of inheritable grafting induced changes in pepper (Capsicum annuum) fruit shape. Sci. Hortic. 2013, 149, 2–8. [Google Scholar] [CrossRef]
- Zhao, X.; Guo, Y.; Huber, D.J.; Lee, J. Grafting effects on postharvest ripening and quality of 1-methylcyclopropene-treated muskmelon fruit. Sci. Hortic. 2011, 130, 581–587. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Wang, Z.; Guo, X.; Wang, F.; Xia, X.J.; Zhou, J.; Shi, K.; Yu, J.Q.; Zhou, Y.H. Microarray and genetic analysis reveals that csa-miR159b plays a critical role in abscisic acid-mediated heat tolerance in grafted cucumber plants. Plant Cell Environ. 2016, 39, 1790–1804. [Google Scholar] [CrossRef]
- Penella, C.; Landi, M.; Guidi, L.; Nebauer, S.G.; Pellegrini, E.; Bautista, A.S.; Remorini, D.; Nali, C.; López-Galarza, S.; Calatayud, A. Salt-tolerant rootstock increases yield of pepper under salinity through maintenance of photosynthetic performance and sinks strength. J. Plant Physiol. 2016, 193, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, R.M.; Upreti, K.K.; Divya, M.H.; Bhat, S.; Pavithra, C.B.; Sadashiva, A.T. Interspecific grafting to enhance physiological resilience to flooding stress in tomato (Solanum lycopersicum L.). Sci. Hortic. 2015, 182, 8–17. [Google Scholar] [CrossRef]
- Louws, F.J.; Rivard, C.L.; Kubota, C. Grafting fruiting vegetables to manage soilborne pathogens, foliar pathogens, arthropods and weeds. Sci. Hortic. 2010, 127, 127–146. [Google Scholar] [CrossRef]
- Schwarz, D.; Rouphael, Y.; Colla, G.; Venema, J.H. Grafting as a tool to improve tolerance of vegetables to abiotic stresses: Thermal stress, water stress and organic pollutants. Sci. Hortic. 2010, 127, 162–171. [Google Scholar] [CrossRef]
- Albacete, A.; Martínez-Andújar, C.; Martínez-Pérez, A.; Thompson, A.J.; Dodd, I.C.; Pérez-Alfocea, F. Unravelling rootstock×scion interactions to improve food security. J. Exp. Bot. 2015, 66, 2211–2226. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, J.; Hua, B.; Liu, Z.; Fan, M.; Bie, Z. Grafting onto different rootstocks as a means to improve watermelon tolerance to low potassium stress. Sci. Hortic. 2013, 149, 80–85. [Google Scholar] [CrossRef]
- Gregory, P.J.; Atkinson, C.J.; Bengough, A.G.; Else, M.A.; Fernández-Fernández, F.; Harrison, R.J.; Schmidt, S. Contributions of roots and rootstocks to sustainable, intensified crop production. J. Exp. Bot. 2013, 64, 1209–1222. [Google Scholar] [CrossRef]
- Zelles, L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: A review. Biol. Fert. Soils 1999, 29, 111–129. [Google Scholar] [CrossRef]
- Powlson, D.S.; Prookes, P.C.; Christensen, B.T. Measurement of soil microbial biomass provides an early indication of changes in total soil organic matter due to straw incorporation. Soil Biol. Biochem. 1987, 19, 159–164. [Google Scholar] [CrossRef]
- Dwivedi, V.; Soni, P. A review on the role of soil microbial biomass in eco-restoration of degraded ecosystem with special reference to mining areas. J. Appl. Nat. Sci. 2011, 3, 151–158. [Google Scholar] [CrossRef]
- Rice, C.W.; Moorman, T.B.; Beare, M. Role of Microbial Biomass Carbon and Nitrogen in Soil Quality. Methods Assess. Soil Qual. 1996, 49, 203–215. [Google Scholar] [CrossRef]
- Kwabiah, A.B.; Palm, C.A.; Stoskopf, N.C.; Voroney, R.P. Response of soil microbial biomass dynamics to quality of plant materials with emphasis on P availability. Soil Biol. Biochem. 2003, 35, 207–216. [Google Scholar] [CrossRef]
- Khan, K.S.; Joergensen, R.G. Changes in microbial biomass and P fractions in biogenic household waste compost amended with inorganic P fertilizers. Bioresour. Technol. 2009, 100, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.D.; Xiao, J.; Liang, T.; Tan, H.W. Response of bacterial compositions to the use of slow-release fertilizers with long-acting agents and synergists. Appl. Soil Ecol. 2023, 182, 104699. [Google Scholar] [CrossRef]
- Xiao, J.; Liang, T.; Yang, S.D.; Tan, H.W. Do full mechanized management strategies destroy soil health and fertility in sugarcane fields? Catena 2023, 224, 107000. [Google Scholar] [CrossRef]
- Gans, J.; Wolinsky, M.; Dunbar, J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 2005, 309, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Vance, G.F.; Pendall, E.; Stahl, P.D. Timber harvesting alters soil carbon mineralization and microbial community structure in coniferous forests. Soil Biol. Biochem. 2008, 40, 1901–1907. [Google Scholar] [CrossRef]
- Madsen, E.L. Microorganisms and their roles in fundamental biogeochemical cycles. Curr. Opin. Biotechnol. 2011, 22, 456–464. [Google Scholar] [CrossRef]
- Nacke, H.; Thürmer, A.; Wollherr, A.; Will, C.; Hodac, L.; Herold, N.; Schöning, I.; Schrumpf, M.; Daniel, R. Pyrosequencing based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE 2011, 6, e17000. [Google Scholar] [CrossRef]
- Koranda, M.; Kaiser, C.; Fuchslueger, L.; Kitzler, B.; Sessitsch, A.; Zechmeister-Boltenstern, S.; Richter, A. Seasonal variation in functional properties of microbial communities in beech forest soil. Soil Biol. Biochem. 2013, 60, 95–104. [Google Scholar] [CrossRef]
- Breulmann, M.; Schulz, E.; Weißhuhn, K.; Buscot, F. Impact of the plant community composition on labile soil organic carbon, soil microbial activity and community structure in semi-natural grassland ecosystems of different productivity. Plant Soil 2011, 352, 253–265. [Google Scholar] [CrossRef]
- Fang, X.; Yu, D.; Zhou, W.; Zhou, L.; Dai, L. The effects of forest type on soil microbial activity in Changbai Mountain, Northeast China. Ann. Forest Sci. 2016, 73, 473–482. [Google Scholar] [CrossRef]
- Gimenez, C.; Cabrera, R.; Reina, M.; Gonzalez-Coloma, A. Fungal endophytes and their role in plant protection. Curr. Org. Chem. 2007, 11, 707–720. [Google Scholar] [CrossRef]
- Ownley, B.H.; Gwinn, K.D.; Vega, F.E. Endophytic fungal entomopathogens with activity against plant pathogens: Ecology and evolution. BioControl 2010, 55, 113–128. [Google Scholar] [CrossRef]
- Boddington, C.L.; Dodd, J.C. The effect of agricultural practices on the development of indigenous arbuscular mycorrhizal fungi. I. Field studies in an Indonesian ultisol. Plant Soil 2000, 218, 137–144. [Google Scholar] [CrossRef]
- Chen, S.; Sun, Y.; Yang, D.; Yang, S.D.; Liang, T.; Tan, H.W. Using moss as a bio-indicator to evaluate soil quaility in litchi orchard. PLoS ONE 2022, 17, e0278303. [Google Scholar] [CrossRef] [PubMed]
- Hayano, K. A method for the determination of β-glucosidase activity in soil. Soil Sci. Plant Nutr. 1973, 19, 103–108. [Google Scholar] [CrossRef]
- Ladd, J.N. Properties of proteolytic enzymes extracted from soil. Soil Biol. Biochem. 1972, 4, 227–237. [Google Scholar] [CrossRef]
- Tabatabai, M.A.; Bremner, J.M. Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1969, 1, 301–307. [Google Scholar] [CrossRef]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Joergensen, R.G.; Brookes, P.C. Ninhydrin-reactive nitrogen measurements of microbial biomass in 0.5 M K2SO4 soil extracts. Soil Biol. Biochem. 1990, 22, 1023–1027. [Google Scholar] [CrossRef]
- Ling, N.; Song, Y.; Raza, W.; Huang, Q.; Guo, S.; Shen, Q. The response of root-associated bacterial community to the grafting of watermelon. Plant Soil 2015, 391, 253–264. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Zhang, Y.; Huang, Z.J.; Liu, S.H.; Liu, S.Q. Effects of grafting on cucumber rhizospheric soil microbial characteristics and enzyme activities under copper stress. Chin. J. Appl. Ecol. 2010, 21, 2317–2322. (In Chinese) [Google Scholar] [CrossRef]
- Wang, L.; Zhao, G.L.; Huang, J. Microbial biomass and enzyme activity of the rhizosphere soil under different grafted Xanthoceras sorbifolia cultivars. J. Beijing For. Univ. 2015, 37, 69–75. (In Chinese) [Google Scholar] [CrossRef]
- Ogundeji, A.O.; Meng, L.; Cheng, Z.; Hou, J.; Yin, T.; Zhang, S.; Liu, X.; Liu, X.; Li, S. Integrated crop practices management stimulates soil microbiome for Verticillium wilt suppression. Eur. J. Agron. 2022, 140, 126594. [Google Scholar] [CrossRef]
- Gaion, L.A.; Braz, L.T.; Carvalho, R.F. Grafting in vegetable crops: A great technique for agriculture. Int. J. Veg. Sci. 2018, 24, 85–102. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Wang, T.; Guo, S.; Ling, N.; Shen, Q. Plant grafting shapes complexity and co-occurrence of rhizobacterial assemblages. Microb. Ecol. 2020, 80, 643–655. [Google Scholar] [CrossRef]
- Bainard, L.D.; Koch, A.M.; Gordon, A.M.; Klironomos, J.N. Growth response of crops to soil microbial communities from conventional monocropping and tree-based intercropping systems. Plant Soil 2013, 363, 345–356. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef]
- Kang, S.; Mills, A.L. Soil bacterial community structure changes following disturbance of the overlying plant community. Soil Sci. 2004, 169, 55–65. [Google Scholar] [CrossRef]
- Yao, H.; Wu, F. Soil microbial community structure in cucumber rhizosphere of different resistance cultivars to fusarium wilt. FEMS Microbiol. Ecol. 2010, 72, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; Andreote, F.D.; Reinhold-Hurek, B.; Sessitsch, A.; van Overbeek, L.S.; van Elsas, J.D. Rice root-associated bacteria: Insights into community structures across 10 cultivars. FEMS Microbiol. Ecol. 2011, 77, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Marasco, R.; Alturkey, H.; Fusi, M.; Brandi, M.; Ghiglieno, I.; Valenti, L.; Daffonchio, D. Rootstock–scion combination contributes to shape diversity and composition of microbial communities associated with grapevine root system. Appl. Microbiol. Int. 2022, 24, 3791–3808. [Google Scholar] [CrossRef] [PubMed]
- Marschner, P.; Solaiman, Z.; Rengel, Z. Brassica genotypes differ in growth, phosphorus uptake and rhizosphere properties under P-limiting conditions. Soil Biol. Biochem. 2007, 39, 87–98. [Google Scholar] [CrossRef]
- Sanders-Smith, R.; Segovia, B.T.; Forbes, C.; Hessing-Lewis, M.; Morien, E.; Lemay, M.A.; O’Connor, M.I.; Parfrey, L.W. Host-Specificity and core taxa of seagrass leaf microbiome identified across tissue age and geographical regions. Front. Ecol. Evol. 2020, 8, 605304. [Google Scholar] [CrossRef]
- Pan, S.; Zhang, W.; Li, Y.; Zhou, P.; Zhang, H.; Niu, L.; Wang, L. Understanding the ecological processes governing hydrophyte-associated bacterial communities involved in hydrophyte growth and development. J. Environ. Manag. 2022, 312, 114952. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Castelle, C.J.; Wrighton, K.C.; Thomas, B.C.; Sharon, I.; Frischkorn, K.R.; Williams, K.H.; Tringe, S.G.; Banfield, J.F. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 2013, 1, 22. [Google Scholar] [CrossRef]
- Li, R.; Pang, Z.; Zhou, Y.; Fallah, N.; Hu, C.; Lin, W.; Yuan, Z. Metagenomic analysis exploring taxonomic and functional diversity of soil microbial communities in sugarcane fields applied with organic fertilizer. BioMed Res. Int. 2020, 2020, 9381506. [Google Scholar] [CrossRef]
- Larsbrink, J.; McKee, L.S. Bacteroidetes bacteria in the soil: Glycan acquisition, enzyme secretion, and gliding motility. Adv. Appl. Microbiol. 2020, 110, 63–98. [Google Scholar] [CrossRef] [PubMed]
- Kuramae, E.E.; Hillekens, R.H.E.; de Hollander, M.; van der Heijden, M.G.A.; van den Berg, M.; van Straalen, N.M.; Kowalchuk, G.A. Structural and functional variation in soil fungal communities associated with litter bags containing maize leaf. FEMS Microbiol. Ecol. 2013, 84, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Zhu, Y.G.; Nybroe, O.; Nicolaisen, M.H. The composition and phosphorus cycling potential of bacterial communities associated with hyphae of penicillium in soil are strongly affected by soil origin. Front. Microbiol. 2020, 10, 2951. [Google Scholar] [CrossRef] [PubMed]
- Inglis, P.W.; Mello, S.C.M.; Martins, I.; Silva, J.B.T.; Macêdo, K.; Sifuentes, D.N.; Valadares-Inglis, M.C. Trichoderma from Brazilian garlic and onion crop soils and description of two new species: Trichoderma azevedoi and Trichoderma peberdyi. PLoS ONE 2020, 15, e0228485. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Dong, M.; Cao, Y.; Wang, J.; Tang, M.; Ban, Y. Comparisons of soil properties, enzyme activities and microbial communities in heavy metal contaminated bulk and rhizosphere soils of Robinia pseudoacacia L. in the Northern foot of Qinling Mountain. Forests 2017, 8, 430. [Google Scholar] [CrossRef]
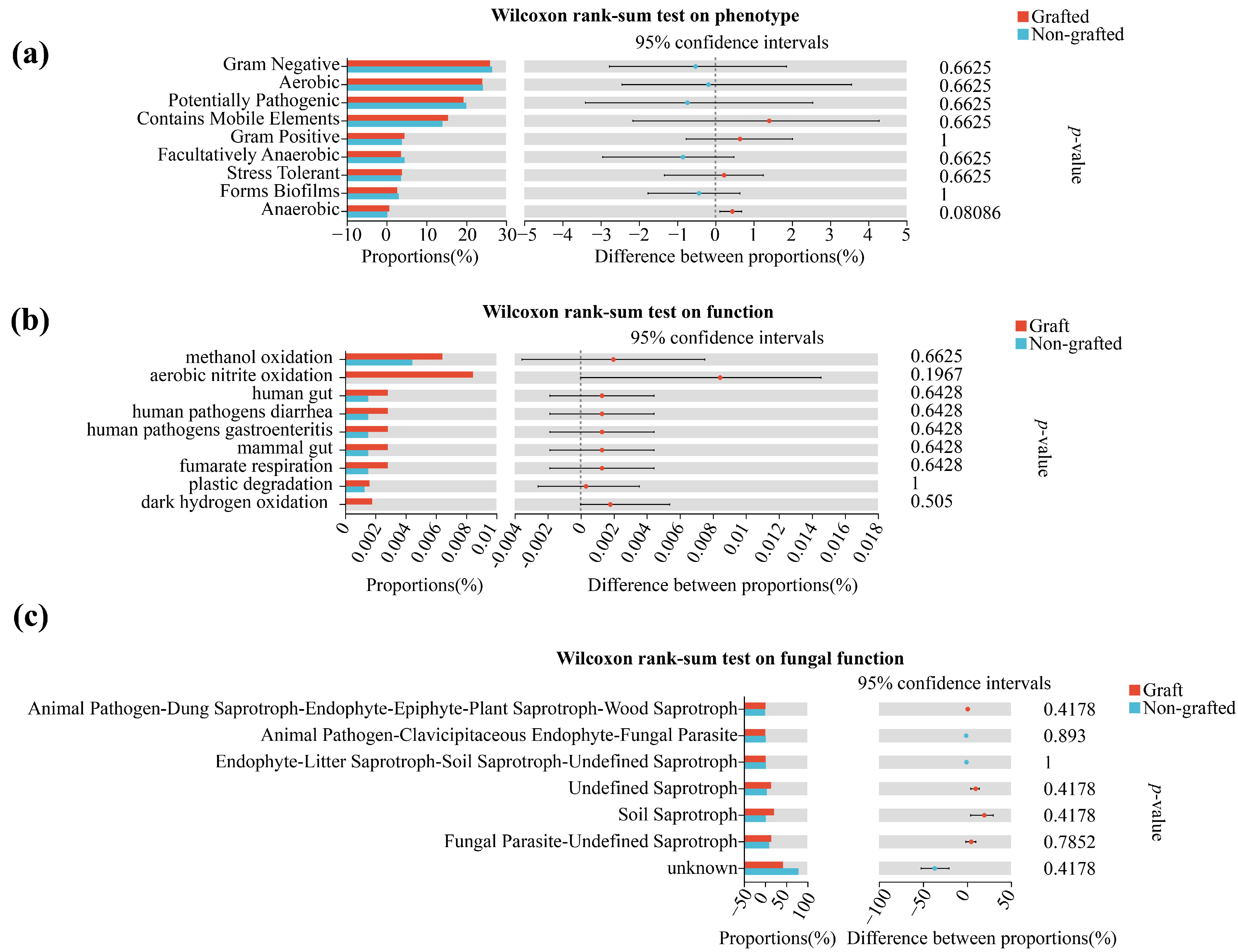
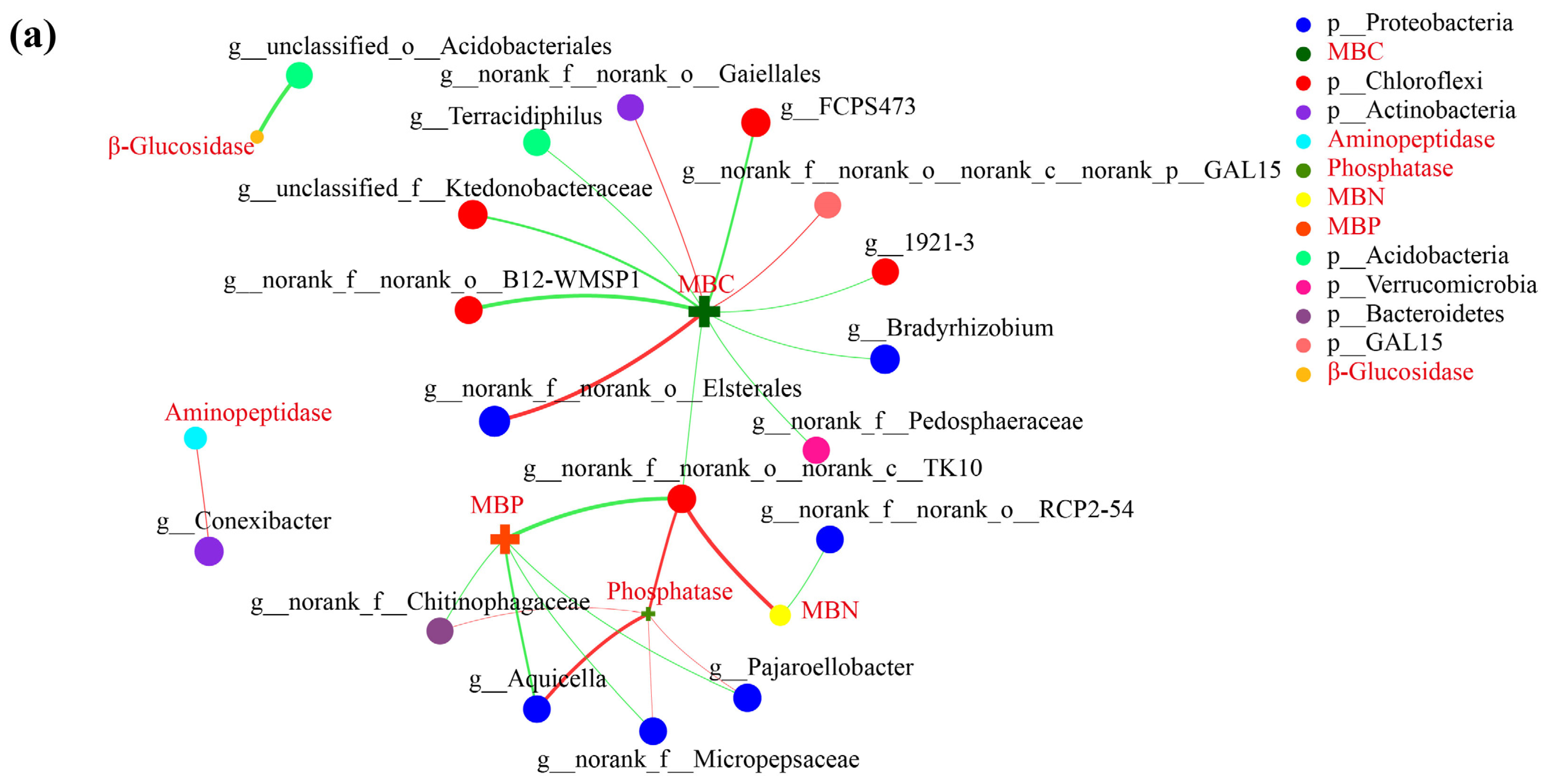

| Samples | β-Glucosidase | Aminopeptidase | Phosphatase |
|---|---|---|---|
| Grafted | 0.27 ± 0.03 a | 15.40 ± 0.87 b | 0.23 ± 0.03 a |
| Non-grafted | 0.26 ± 0.03 a | 16.67 ± 0.53 a | 0.08 ± 0.01 b |
| Samples | Microbial Biomass C | Microbial Biomass N | Microbial Biomass P |
|---|---|---|---|
| Grafted | 121.86 ± 7.61 b | 7.17 ± 0.77 a | 25.90 ± 1.44 b |
| Non-grafted | 144.64 ± 7.34 a | 6.55 ± 0.39 a | 52.40 ± 4.37 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, J.; Liu, J.; Wu, S.; Liang, W.; Yang, S. The Characteristics of the Root-Zone Soil’s Biological Properties and Microbial Community Structure in Grafted Star Anise Plantations. Microorganisms 2024, 12, 431. https://doi.org/10.3390/microorganisms12030431
Xiao J, Liu J, Wu S, Liang W, Yang S. The Characteristics of the Root-Zone Soil’s Biological Properties and Microbial Community Structure in Grafted Star Anise Plantations. Microorganisms. 2024; 12(3):431. https://doi.org/10.3390/microorganisms12030431
Chicago/Turabian StyleXiao, Jian, Junxian Liu, Siyu Wu, Wenhui Liang, and Shangdong Yang. 2024. "The Characteristics of the Root-Zone Soil’s Biological Properties and Microbial Community Structure in Grafted Star Anise Plantations" Microorganisms 12, no. 3: 431. https://doi.org/10.3390/microorganisms12030431
APA StyleXiao, J., Liu, J., Wu, S., Liang, W., & Yang, S. (2024). The Characteristics of the Root-Zone Soil’s Biological Properties and Microbial Community Structure in Grafted Star Anise Plantations. Microorganisms, 12(3), 431. https://doi.org/10.3390/microorganisms12030431






