Research Progresses on the Function and Detection Methods of Insect Gut Microbes
Abstract
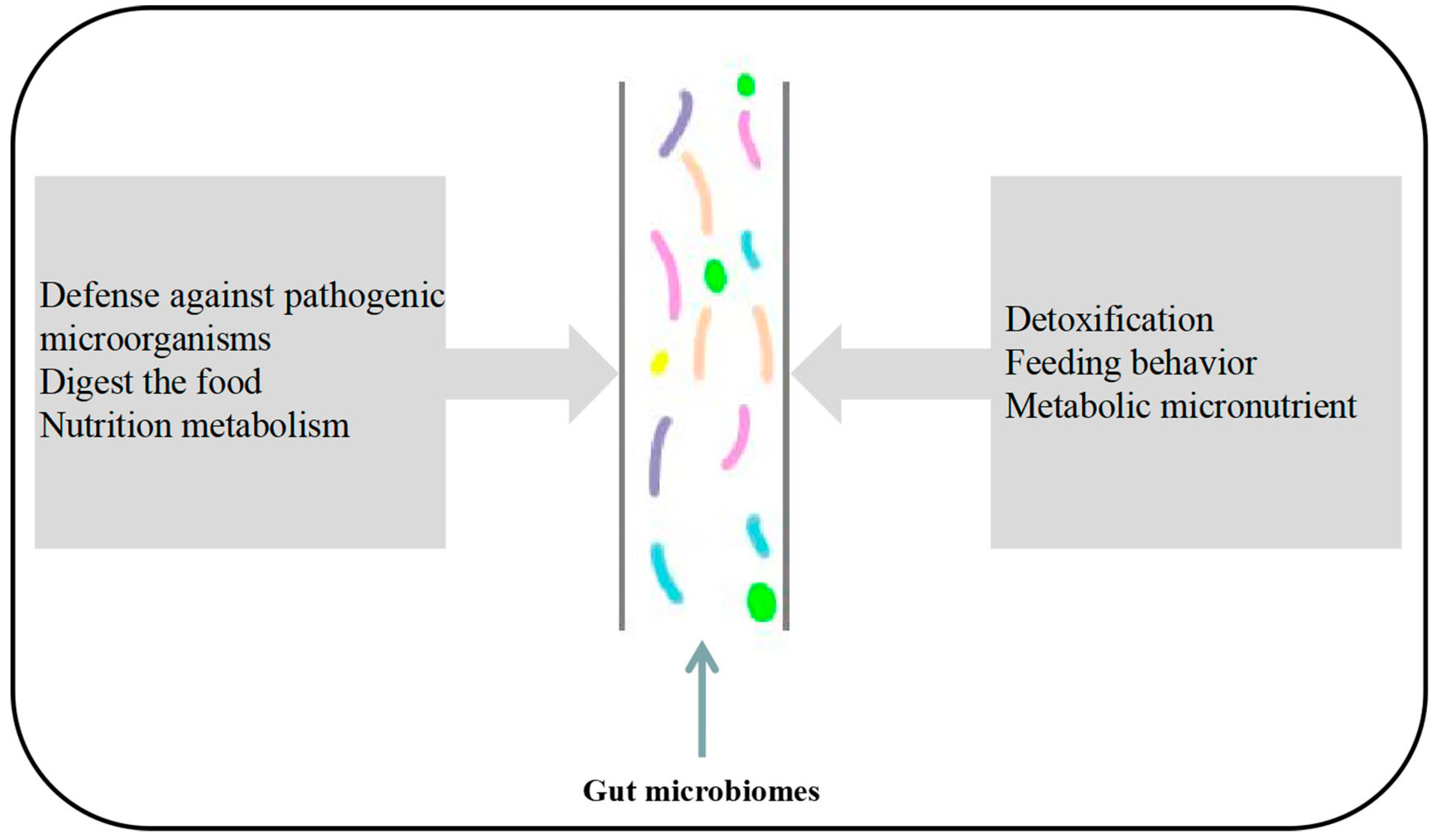
:1. Introduction
2. Functional Roles of the Insect Gut Microbiota
2.1. Nutrient Metabolism
2.2. Immune Defense
2.3. Antioxidation Function
2.4. Enhance Host Drug Resistance
3. Factors Affecting the Gut Microbiome of Insects
3.1. Effects of Feed, Antibiotics and Culture Temperature on Insect Gut Microbes
3.2. Effects of Sex and Developmental Stage on Insect Gut Microbes
3.3. Effects of Pesticides on Insect Gut Microbes
4. Negative Effects of Gut Microbes on the Host
5. Detection Methods of Insect Gut Microbes
5.1. Traditional Identification Methods
5.2. Electrophoresis
5.3. Molecular Detection Method Based on 16S rRNA Gene
5.4. qRT-PCR
| Techniques | Advantages | Disadvantages | Application | References |
|---|---|---|---|---|
| Traditional culture method | Low-cost. | Many bacteria cannot be cultured because of the limitations of culture medium and culture condition. | Staphylococcus, Bacillus, Enterococcus, Corynebacterium, Micrococcus Dothideomycetes, Eurotiomycetes, Mucormycotina, Saccharomycetes, Zygomycetes, Yeasts, Molds | [8,87,88,89] |
| Electrophoresis | Rapid analysis of a large number of specimens. | Suitable for small DNA fragments, and can only reflect the dominant genus. | Lactobacillus, Campylobacter jejuni | [93,95,97] |
| Molecular detection method based on 16S rRNA gene | Generally accurate to the level of genus, a few can be identified to species. For some bacteria, the similarity of sequences in hypervariable regions is very high. | Other methods and techniques are needed to identify species with small interspecific differences. | Bacillus, Enterococcus, Klebsiella, Hamiltonella | [98,100,101,102] |
| qRT-PCR | The method is simple, rapid, quantifiable, repeatable, sensitive, and specific and can be used to detect live and dead bacteria. | High cost, strict requirements for the operation of laboratory personnel, the need for specific instruments and reagents. | Serratia marcescens, Bacteroides fragilis, Enterococcus faecalis | [60,104,105] |
| Metagenomics | Metagenomics overcomes the technical limitations of traditional pure culture methods and can provide information about low abundance or even trace microorganisms in the environment, which can more accurately reflect the true state of microbial survival. | The sequencing data are large, the price is much more expensive than 16S sequencing, and the computing resources required for subsequent data processing are high. | Major Phylum: Firmicutes, Proteobacteria, Elusimicrobia, Mycobacterium, Bacteroidetes Major Genus: Enterococcus, Pantoea, Acinetobacter, Enterobacteriales, Lactobacillales | [106,107,108,109,110] |
5.5. Metagenomics
6. Application of Insect Intestinal Microflora in Pest Management
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krams, I.A.; Kecko, S.; Joers, P.; Trakimas, G.; Elferts, D.; Krams, R.; Luoto, S.; Rantala, M.J.; Inashkina, I.; Gudra, D.; et al. Microbiome symbionts and diet diversity incur costs on the immune system of insect larvae. J. Exp. Biol. 2017, 220, 4204–4212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, F.; Lu, X. Diversity and functional roles of the gut microbiota in lepidopteran insects. Microorganisms. 2022, 10, 1234. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Kikuchi, Y. Impact of the insect gut microbiota on ecology, evolution, and industry. Curr. Opin. Insect Sci. 2020, 41, 33–39. [Google Scholar] [CrossRef]
- Lee, W.J.; Hase, K. Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 2014, 10, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Whitten, M.M.; Facey, P.D.; Del, S.R.; Fernandez-Martinez, L.T.; Evans, M.C.; Mitchell, J.J.; Bodger, O.G.; Dyson, P.J. Symbiont-mediated RNA interference in insects. Proc. Biol. Sci. 2016, 283, 20160042. [Google Scholar] [CrossRef]
- Blow, F.; Bueno, E.; Clark, N.; Zhu, D.T.; Chung, S.H.; Gullert, S.; Schmitz, R.A.; Douglas, A.E. B-vitamin nutrition in the pea aphid-Buchnera symbiosis. J. Insect Physiol. 2020, 126, 104092. [Google Scholar] [CrossRef]
- Wier, A.; Dolan, M.; Grimaldi, D.; Guerrero, R.; Wagensberg, J.; Margulis, L. Spirochete and protist symbionts of a termite (Mastotermes electrodominicus) in Miocene amber. Proc. Natl. Acad. Sci. USA 2002, 99, 1410–1413. [Google Scholar] [CrossRef]
- Khan, K.A.; Al-Ghamdi, A.A.; Ghramh, H.A.; Ansari, M.J.; Ali, H.; Alamri, S.A.; Al-Kahtani, S.N.; Adgaba, N.; Qasim, M.; Hafeez, M. Structural diversity and functional variability of gut microbial communities associated with honey bees. Microb. Pathog. 2020, 138, 103793. [Google Scholar] [CrossRef]
- Broderick, N.A.; Raffa, K.F.; Goodman, R.M.; Handelsman, J. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 2004, 70, 293–300. [Google Scholar] [CrossRef]
- Settanni, L.; Guarcello, R.; Gaglio, R.; Francesca, N.; Aleo, A.; Felis, G.E.; Moschetti, G. Production, stability, gene sequencing and in situ anti-Listeria activity of mundticin KS expressed by three Enterococcus mundtii strains. Food Control. 2014, 35, 311–322. [Google Scholar] [CrossRef]
- Jing, T.Z.; Qi, F.H.; Wang, Z.Y. Most dominant roles of insect gut bacteria: Digestion, detoxification, or essential nutrient provision? Microbiome 2020, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Yang, L.; Pang, X.; Zhang, R.; Zhu, Y.; Wang, P.; Gao, G.; Cheng, G. A Mesh-Duox pathway regulates homeostasis in the insect gut. Nat. Microbiol. 2017, 2, 17020. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Sanders, J.G.; Lukasik, P.; D’Amelio, C.L.; Millar, J.S.; Vann, D.R.; Lan, Y.; Newton, J.A.; Schotanus, M.; Kronauer, D.; et al. Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome. Nat. Commun. 2018, 9, 964. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.A.; Vennison, S.J.; Sankar, S.G.; Prabhu, D.I.; Vasan, P.T.; Raghuraman, T.; Geoffrey, C.J.; Vendan, S.E. Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J. Insect Sci. 2010, 10, 107. [Google Scholar] [CrossRef]
- Su, L.; Liu, H.; Li, Y.; Zhang, H.; Chen, M.; Gao, X.; Wang, F.; Song, A. Cellulolytic activity and structure of symbiotic bacteria in locust guts. Genet. Mol. Res. 2014, 13, 7926–7936. [Google Scholar] [CrossRef]
- Guégan, M.; Tran Van, V.; Martin, E.; Minard, G.; Tran, F.H.; Fel, B.; Hay, A.E.; Simon, L.; Barakat, M.; Potier, P.; et al. Who is eating fructose within the Aedes albopictus gut microbiota? Environ. Microbiol. 2020, 22, 1193–1206. [Google Scholar] [CrossRef]
- Tokuda, G.; Lo, N.; Watanabe, H. Marked variations in patterns of cellulase activity against crystalline-vs. carboxymethyl-cellulose in the digestive systems of diverse, wood-feeding termites. Physiol. Entomol. 2005, 30, 372–380. [Google Scholar] [CrossRef]
- Dantur, K.I.; Enrique, R.; Welin, B.; Castagnaro, A.P. Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express 2015, 5, 15. [Google Scholar] [CrossRef]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects—diversity in structure and function. Fems Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Duplais, C.; Sarou-Kanian, V.; Massiot, D.; Hassan, A.; Perrone, B.; Estevez, Y.; Wertz, J.T.; Martineau, E.; Farjon, J.; Giraudeau, P.; et al. Gut bacteria are essential for normal cuticle development in herbivorous turtle ants. Nat. Commun. 2021, 12, 676. [Google Scholar] [CrossRef] [PubMed]
- Ludington, W.B.; Ja, W.W. Drosophila as a model for the gut microbiome. PLoS Pathog. 2020, 16, e1008398. [Google Scholar] [CrossRef]
- Wong, A.C.; Wang, Q.P.; Morimoto, J.; Senior, A.M.; Lihoreau, M.; Neely, G.G.; Simpson, S.J.; Ponton, F. Gut microbiota modifies olfactory-guided microbial preferences and foraging decisions in Drosophila. Curr. Biol. 2017, 27, 2397–2404. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Keesey, I.W.; Hansson, B.S.; Knaden, M. Gut microbiota affects development and olfactory behavior in Drosophila melanogaster. J. Exp. Biol. 2019, 222, jeb192500. [Google Scholar] [CrossRef] [PubMed]
- Habineza, P.; Muhammad, A.; Ji, T.; Xiao, R.; Yin, X.; Hou, Y.; Shi, Z. The promoting effect of gut microbiota on growth and development of red palm weevil, Rhynchophorus ferrugineus (Olivier) (Coleoptera: Dryophthoridae) by modulating its nutritional metabolism. Front. Microbiol. 2019, 10, 1212. [Google Scholar] [CrossRef] [PubMed]
- Ben-Yosef, M.; Pasternak, Z.; Jurkevitch, E.; Yuval, B. Symbiotic bacteria enable olive flies (Bactrocera oleae) to exploit intractable sources of nitrogen. J. Evol. Biol. 2014, 27, 2695–2705. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.W.; Lu, Z.Q. Midgut immune responses induced by bacterial infection in the silkworm, Bombyx mori. J. Zhejiang Univ. Sci. B 2015, 16, 875–882. [Google Scholar] [CrossRef]
- Zeng, T.; Jaffar, S.; Xu, Y.; Qi, Y. The intestinal immune defense system in insects. Int. J. Mol. Sci. 2022, 23, 15132. [Google Scholar] [CrossRef]
- Shao, Y.; Chen, B.; Sun, C.; Ishida, K.; Hertweck, C.; Boland, W. Symbiont-derived antimicrobials contribute to the control of the Lepidopteran gut microbiota. Cell Chem. Biol. 2017, 24, 66–75. [Google Scholar] [CrossRef]
- Akbar, N.; Siddiqui, R.; Iqbal, M.; Sagathevan, K.; Khan, N.A. Gut bacteria of cockroaches are a potential source of antibacterial compound(s). Lett. Appl. Microbiol. 2018, 66, 416–426. [Google Scholar] [CrossRef]
- Knight, C.A.; Bowman, M.J.; Frederick, L.; Day, A.; Lee, C.; Dunlap, C.A. The first report of antifungal lipopeptide production by a Bacillus subtilis subsp. inaquosorum strain. Microbiol. Res. 2018, 216, 40–46. [Google Scholar] [CrossRef]
- Douglas, A.E. The molecular basis of bacterial-insect symbiosis. J. Mol. Biol. 2014, 426, 3830–3837. [Google Scholar] [CrossRef]
- Pandey, N.; Rajagopal, R. Tissue damage induced midgut stem cell proliferation and microbial dysbiosis in Spodoptera litura. FEMS Microbiol. Ecol. 2017, 93, fix132. [Google Scholar] [CrossRef]
- Muhammad, A.; Habineza, P.; Ji, T.; Hou, Y.; Shi, Z. Intestinal microbiota confer protection by priming the immune system of red palm weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae). Front. Physiol. 2019, 10, 1303. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Song, X.; Wang, J. Gut microbiota is essential in PGRP-LA regulated immune protection against Plasmodium berghei infection. Parasites Vectors. 2020, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kim, K.M.; Lee, J.H.; Seo, S.J.; Lee, I.H. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect. Immun. 2007, 75, 1861–1869. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Y.; Wang, Y.; Xu, H.; Mei, X.; Yu, D.; Wang, Y.; Li, W. Antioxidant properties of probiotic bacteria. Nutrients. 2017, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, B.J.; Liu, K.H.; Owens, J.A.; Hunter-Chang, S.; Camacho, M.C.; Eboka, R.U.; Chandrasekharan, B.; Baker, N.F.; Darby, T.M.; Robinson, B.S.; et al. Gut-resident Lactobacilli activate hepatic Nrf2 and protect against oxidative liver injury. Cell. Metab. 2020, 31, 956–968. [Google Scholar] [CrossRef] [PubMed]
- Elzeini, H.M.; Ali, A.; Nasr, N.F.; Hassan, M.; Hassan, A.; Elenany, Y.E. Probiotic capability of novel lactic acid bacteria isolated from worker honey bees gut microbiota. FEMS Microbiol. Lett. 2021, 368, fnab030. [Google Scholar] [CrossRef]
- Barretto, D.A.; Vootla, S.K. In vitro anticancer activity of Staphyloxanthin pigment extracted from Staphylococcus gallinarum KX912244, a gut microbe of Bombyx mori. Indian J. Microbiol. 2018, 58, 146–158. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Hayatsu, M.; Hosokawa, T.; Nagayama, A.; Tago, K.; Fukatsu, T. Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. USA. 2012, 109, 8618–8622. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, N.; Xie, S.; Zhang, X.; He, J.; Muhammad, A.; Sun, C.; Lu, X.; Shao, Y. Gut bacteria of the silkworm Bombyx mori facilitate host resistance against the toxic effects of organophosphate insecticides. Environ. Int. 2020, 143, 105886. [Google Scholar] [CrossRef] [PubMed]
- Trinder, M.; Mcdowell, T.W.; Daisley, B.A.; Ali, S.N.; Leong, H.S.; Sumarah, M.W.; Reid, G. Probiotic Lactobacillus rhamnosus reduces organophosphate pesticide absorption and toxicity to Drosophila melanogaster. Appl. Environ. Microbiol. 2016, 82, 6204–6213. [Google Scholar] [CrossRef]
- Danilenko, V.N.; Devyatkin, A.V.; Marsova, M.V.; Shibilova, M.U.; Ilyasov, R.A.; Shmyrev, V.I. Common inflammatory mechanisms in COVID-19 and parkinson’s diseases: The Role of microbiome, pharmabiotics and postbiotics in their prevention. J. Iamm. Res. 2021, 14, 6349–6381. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Jang, S.; Takeshita, K.; Itoh, H.; Koike, H.; Tago, K.; Hayatsu, M.; Hori, T.; Kikuchi, Y. Insecticide resistance by a host-symbiont reciprocal detoxification. Nat. Commun. 2021, 12, 6432. [Google Scholar] [CrossRef]
- Itoh, H.; Hori, T.; Sato, Y.; Nagayama, A.; Tago, K.; Hayatsu, M.; Kikuchi, Y. Infection dynamics of insecticide-degrading symbionts from soil to insects in response to insecticide spraying. ISME J. 2018, 12, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.A.; Moreau, C.S.; Goldman-Huertas, B.; Fujiwara, M.; Lohman, D.J.; Pierce, N.E. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc. Natl. Acad. Sci. USA 2009, 106, 21236–21241. [Google Scholar] [CrossRef]
- Thakur, A.; Dhammi, P.; Saini, H.S.; Kaur, S. Effect of antibiotic on survival and development of Spodoptera litura (Lepidoptera: Noctuidae) and its gut microbial diversity. Bull. Entomol. Res. 2016, 106, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Raza, M.F.; Wang, Y.; Cai, Z.; Bai, S.; Yao, Z.; Awan, U.A.; Zhang, Z.; Zheng, W.; Zhang, H. Gut microbiota promotes host resistance to low-temperature stress by stimulating its arginine and proline metabolism pathway in adult Bactrocera dorsalis. PLoS Pathog. 2020, 16, e1008441. [Google Scholar] [CrossRef]
- Lv, D.; Liu, X.; Dong, Y.; Yan, Z.; Zhang, X.; Wang, P.; Yuan, X.; Li, Y. Comparison of Gut Bacterial Communities of Fall Armyworm (Spodoptera frugiperda) Reared on Different Host Plants. Int. J. Mol. Sci. 2021, 22, 11266. [Google Scholar] [CrossRef]
- Liang, X.; Fu, Y.; Tong, L.; Liu, H. Microbial shifts of the silkworm larval gut in response to lettuce leaf feeding. Appl. Microbiol. Biotechnol. 2014, 98, 3769–3776. [Google Scholar] [CrossRef]
- Priya, N.G.; Ojha, A.; Kajla, M.K.; Raj, A.; Rajagopal, R. Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS ONE 2012, 7, e30768. [Google Scholar]
- Wang, J.; Chen, H.; Tang, M. Community structure of gut bacteria of Dendroctonus armandi (Coleoptera: Curculionidae: Scolytinae) larvae during overwintering stage. Sci. Rep. 2017, 7, 14242. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Tada, A.; Musolin, D.L.; Hari, N.; Hosokawa, T.; Fujisaki, K.; Fukatsu, T. Collapse of insect gut symbiosis under simulated climate change. Mbio 2016, 7, e01578-16. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Nair, S. Dynamics of insect-microbiome interaction influence host and microbial symbiont. Front. Microbiol. 2020, 11, 1357. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Lu, Y.; Zhang, H.; Kumar, D.; Liu, B.; Gong, Y.; Zhu, M.; Zhu, L.; Liang, Z.; Kuang, S.; et al. Effects of BmCPV infection on silkworm Bombyx mori intestinal bacteria. PLoS ONE 2016, 11, e146313. [Google Scholar] [CrossRef]
- Chen, B.; Du, K.; Sun, C.; Vimalanathan, A.; Liang, X.; Li, Y.; Wang, B.; Lu, X.; Li, L.; Shao, Y. Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J. 2018, 12, 2252–2262. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Li, W.; Luo, J.; Zhang, L.; Ji, J.; Zhu, X.; Wang, L.; Zhang, S.; Cui, J. Biodiversity of the microbiota in Spodoptera exigua (Lepidoptera: Noctuidae). J. Appl. Microbiol. 2019, 126, 1199–1208. [Google Scholar] [CrossRef]
- Wang, Z.L.; Wang, T.Z.; Zhu, H.F.; Pan, H.B.; Yu, X.P. Diversity and dynamics of microbial communities in brown planthopper at different developmental stages revealed by high-throughput amplicon sequencing. Insect Sci. 2020, 27, 883–894. [Google Scholar] [CrossRef]
- Wei, G.; Lai, Y.; Wang, G.; Chen, H.; Li, F.; Wang, S. Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc. Natl. Acad. Sci. USA 2017, 114, 5994–5999. [Google Scholar] [CrossRef]
- Gu, Z.; Li, F.; Hu, J.; Ding, C.; Wang, C.; Tian, J.; Xue, B.; Xu, K.; Shen, W.; Li, B. Sublethal dose of phoxim and Bombyx mori nucleopolyhedrovirus interact to elevate silkworm mortality. Pest Manag. Sci. 2017, 73, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hu, J.; Tian, J.; Xu, K.; Ni, M.; Wang, B.; Shen, W.; Li, B. Effects of phoxim on nutrient metabolism and insulin signaling pathway in silkworm midgut. Chemosphere 2016, 146, 478–485. [Google Scholar] [CrossRef]
- Sun, S.; Cheng, Z.; Fan, J.; Cheng, X.; Pang, Y. The utility of camptothecin as a synergist of Bacillus thuringiensis var. kurstaki and nucleopolyhedroviruses against Trichoplusia ni and Spodoptera exigua. J. Econ. Entomol. 2012, 105, 1164–1170. [Google Scholar] [CrossRef]
- Kumar, D.; Sun, Z.; Cao, G.; Xue, R.; Hu, X.; Gong, C. Bombyx mori bidensovirus infection alters the intestinal microflora of fifth instar silkworm (Bombyx mori) larvae. J. Invertebr. Pathol. 2019, 163, 48–63. [Google Scholar] [CrossRef]
- Motta, E.; Mak, M.; De Jong, T.K.; Powell, J.E.; O’Donnell, A.; Suhr, K.J.; Riddington, I.M.; Moran, N.A. Oral or topical exposure to glyphosate in herbicide formulation impacts the gut microbiota and survival rates of Honey Bees. Appl. Environ. Microbiol. 2020, 86, e01150-20. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Yan, Z.; Ma, S.; Yang, Y.; Wang, Q.; Hou, C.; Wu, Y.; Liu, Y.; Diao, Q. The herbicide glyphosate negatively affects midgut bacterial communities and survival of honey bee during larvae reared in vitro. J. Agric. Food Chem. 2018, 66, 7786–7793. [Google Scholar] [CrossRef]
- Zhu, L.; Qi, S.; Xue, X.; Niu, X.; Wu, L. Nitenpyram disturbs gut microbiota and influences metabolic homeostasis and immunity in honey bee (Apis mellifera L.). Environ. Pollut. 2020, 258, 113671. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.R.; Liu, Q.Z. Evaluation and entomopathogenicity of gut bacteria associated with dauer juveniles of Oscheius chongmingensis (Nematoda: Rhabditidae). Microbiol. Open 2019, 8, e823. [Google Scholar] [CrossRef]
- Li, F.; Li, M.; Mao, T.; Wang, H.; Chen, J.; Lu, Z.; Qu, J.; Fang, Y.; Gu, Z.; Li, B. Effects of phoxim exposure on gut microbial composition in the silkworm, Bombyx mori. Ecotox. Environ. Saf. 2020, 189, 110011. [Google Scholar] [CrossRef]
- Raymann, K.; Bobay, L.M.; Moran, N.A. Antibiotics reduce genetic diversity of core species in the honeybee gut microbiome. Mol. Ecol. 2018, 27, 2057–2066. [Google Scholar] [CrossRef]
- Li, S.; Xu, X.; De Mandal, S.; Shakeel, M.; Hua, Y.; Shoukat, R.F.; Fu, D.; Jin, F. Gut microbiota mediate Plutella xylostella susceptibility to Bt Cry1Ac protoxin is associated with host immune response. Environ. Pollut. 2021, 271, 116271. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Q.; Yin, Y.; Cao, K.L.; Zhao, X.X.; Wang, X.Y.; Zhang, Y.X.; Shi, W.P. Effects of a combined infection with Paranosema locustae and Beauveria bassiana on Locusta migratoria and its gut microflora. Insect Sci. 2021, 28, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, M.; Zhu, Q.; Mao, T.; Dai, M.; Ye, W.; Bian, D.; Su, W.; Feng, P.; Ren, Y.; et al. Imbalance of intestinal microbial homeostasis caused by acetamiprid is detrimental to resistance to pathogenic bacteria in Bombyx mori. Environ. Pollut. 2021, 289, 117866. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, M.; Wang, H.; Mao, T.; Chen, J.; Lu, Z.; Qu, J.; Fang, Y.; Li, B. Effects of phoxim pesticide on the immune system of silkworm midgut. Pest. Biochem. Physiol. 2020, 164, 58–64. [Google Scholar] [CrossRef]
- Silva, F.; Fernandes, K.M.; Freitas, L.L.; Cascardo, R.S.; Bernardes, R.C.; Oliveira, L.L.; Martins, G.F.; Vanetti, M. Effects of sub-lethal doses of nisin on the virulence of Salmonella enterica in Galleria mellonella larvae. Res. Microbiol. 2021, 172, 103836. [Google Scholar] [CrossRef]
- Raymann, K.; Shaffer, Z.; Moran, N.A. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017, 15, e2001861. [Google Scholar] [CrossRef]
- Van Meyel, S.; Devers, S.; Dupont, S.; Dedeine, F.; Meunier, J. Alteration of gut microbiota with a broad-spectrum antibiotic does not impair maternal care in the European earwig. J. Evol. Biol. 2021, 34, 1034–1045. [Google Scholar] [CrossRef]
- Powell, J.E.; Carver, Z.; Leonard, S.P.; Moran, N.A. Field-realistic tylosin exposure impacts honey bee microbiota and pathogen susceptibility, which is ameliorated by native gut probiotics. Microbiol. Spectr. 2021, 9, e10321. [Google Scholar] [CrossRef]
- Schretter, C.E.; Vielmetter, J.; Bartos, I.; Marka, Z.; Marka, S.; Argade, S.; Mazmanian, S.K. A gut microbial factor modulates locomotor behaviour in Drosophila. Nature 2018, 563, 402–406. [Google Scholar] [CrossRef]
- Stathopoulos, S.; Neafsey, D.E.; Lawniczak, M.K.; Muskavitch, M.A.; Christophides, G.K. Genetic dissection of Anopheles gambiae gut epithelial responses to Serratia marcescens. PLoS Pathog. 2014, 10, e1003897. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, D.; Wu, H.; Ji, Y.; Liu, Z.; Guo, X.; Guo, W.; Bi, Y. Bt GS57 interaction with gut microbiota accelerates Spodoptera exigua mortality. Front. Microbiol. 2022, 13, 835227. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.M.; Weeks, E.; Lovullo, E.D.; Shirk, P.D.; Geden, C.J. Mortality effects of three bacterial pathogens and Beauveria bassiana when topically applied or injected into house flies (Diptera: Muscidae). J. Med. Entomol. 2019, 56, 774–783. [Google Scholar] [CrossRef]
- Wu, P.; Sun, P.; Nie, K.; Zhu, Y.; Shi, M.; Xiao, C.; Liu, H.; Liu, Q.; Zhao, T.; Chen, X.; et al. A gut commensal bacterium promotes mosquito permissiveness to arboviruses. Cell Host Microbe 2019, 25, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Eski, A.; Demir, İ.; Güllü, M.; Demirbağ, Z. Biodiversity and pathogenicity of bacteria associated with the gut microbiota of beet armyworm, Spodoptera exigua Hübner (Lepidoptera: Noctuidae). Microb. Pathog. 2018, 121, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Secil, E.S.; Sevim, A.; Demirbag, Z.; Demir, I. Isolation, characterization and virulence of bacteria from Ostrinia nubilalis (Lepidoptera: Pyralidae). Biologia 2012, 67, 767–776. [Google Scholar] [CrossRef]
- Galeano-Castaneda, Y.; Urrea-Aguirre, P.; Piedrahita, S.; Bascunan, P.; Correa, M.M. Composition and structure of the culturable gut bacterial communities in Anopheles albimanus from Colombia. PLoS ONE 2019, 14, e225833. [Google Scholar] [CrossRef] [PubMed]
- Pace, N.R. A molecular view of microbial diversity and the biosphere. Science 1997, 276, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef]
- Anjum, S.I.; Shah, A.H.; Aurongzeb, M.; Kori, J.; Azim, M.K.; Ansari, M.J.; Bin, L. Characterization of gut bacterial flora of Apis mellifera from north-west Pakistan. Saudi J. Biol. Sci. 2018, 25, 388–392. [Google Scholar] [CrossRef]
- Sklarz, M.Y.; Angel, R.; Gillor, O.; Soares, M.I. Evaluating amplified rDNA restriction analysis assay for identification of bacterial communities. Antonie Leeuwenhoek 2009, 96, 659–664. [Google Scholar] [CrossRef]
- Tabar, E.A.; Staji, H.; Mahdavi, A. Comparative restriction enzyme mapping of Campylobacter jejuni isolates from turkeys and broilers based on flaA flagellar gene using HpyF3I endonuclease. Folia Microbiol. 2019, 64, 189–195. [Google Scholar] [CrossRef]
- He, H.; Wei, C.; Wheeler, D.E. The gut bacterial communities associated with lab-raised and field-collected ants of Camponotus fragilis (Formicidae: Formicinae). Curr. Microbiol. 2014, 69, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Puchalski, A.; Urban-Chmiel, R.; Wernicki, A. 16S-ARDRA and MALDI-TOF mass spectrometry as tools for identification of Lactobacillus bacteria isolated from poultry. BMC Microbiol. 2016, 16, 105. [Google Scholar] [CrossRef] [PubMed]
- van Burgh, S.; Maghdid, D.M.; Ganjo, A.R.; Mansoor, I.Y.; Kok, D.J.; Fatah, M.H.; Alnakshabandi, A.A.; Asad, D.; Hammerum, A.M.; Ng, K.; et al. PME and other ESBL-positive multiresistant Pseudomonas aeruginosa isolated from hospitalized patients in the region of kurdistan, Iraq. Microb. Drug Resist. 2019, 25, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Lindstedt, B.A.; Heir, E.; Vardund, T.; Melby, K.K.; Kapperud, G. Comparative fingerprinting analysis of Campylobacter jejuni subsp. jejuni strains by amplified-fragment length polymorphism genotyping. J. Clin. Microbiol. 2000, 38, 3379–3387. [Google Scholar] [CrossRef] [PubMed]
- Al-Soud, W.A.; Bennedsen, M.; On, S.; Ouis, I.S.; Vandamme, P.; Nilsson, H.O.; Ljungh, A.; Wadstrom, T. Assessment of PCR-DGGE for the identification of diverse Helicobacter species, and application to faecal samples from zoo animals to determine Helicobacter prevalence. J. Med. Microbiol. 2003, 52, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.L.; Pan, Q.J.; Tian, H.G.; Douglas, A.E.; Liu, T.X. Bacteria abundance and diversity of different life stages of Plutella xylostella (Lepidoptera: Plutellidae), revealed by bacteria culture-dependent and PCR-DGGE methods. Insect Sci. 2015, 22, 375–385. [Google Scholar] [CrossRef]
- Snyman, M.; Gupta, A.K.; Bezuidenhout, C.C.; Claassens, S.; van den Berg, J. Gut microbiota of Busseola fusca (Lepidoptera: Noctuidae). World J. Microbiol. Biotechnol. 2016, 32, 115. [Google Scholar] [CrossRef]
- Yadav, K.K.; Bora, A.; Datta, S.; Chandel, K.; Gogoi, H.K.; Prasad, G.B.; Veer, V. Molecular characterization of midgut microbiota of Aedes albopictus and Aedes aegypti from Arunachal Pradesh, India. Parasites Vectors 2015, 8, 641. [Google Scholar] [CrossRef]
- Lv, Z.H.; Wei, X.Y.; Tao, Y.L.; Chu, D. Differential susceptibility of whitefly-associated bacteria to antibiotic as revealed by metagenomics analysis. Infect. Genet. Evol. 2018, 63, 24–29. [Google Scholar] [CrossRef]
- Chiel, E.; Gottlieb, Y.; Zchori-Fein, E.; Mozes-Daube, N.; Katzir, N.; Inbar, M.; Ghanim, M. Biotype-dependent secondary symbiont communities in sympatric populations of Bemisia tabaci. Bull. Entomol. Res. 2007, 97, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Gao, C.S.; De Barro, P.; Zhang, Y.J.; Wan, F.H.; Khan, I.A. Further insights into the strange role of bacterial endosymbionts in whitefly, Bemisia tabaci: Comparison of secondary symbionts from biotypes B and Q in China. Bull. Entomol. Res. 2011, 101, 477–486. [Google Scholar] [CrossRef]
- Su, M.M.; Guo, L.; Tao, Y.L.; Zhang, Y.J.; Wan, F.H.; Chu, D. Effects of host plant factors on the bacterial communities associated with two whitefly sibling species. PLoS ONE 2016, 11, e152183. [Google Scholar] [CrossRef]
- Tong, J.; Liu, C.; Summanen, P.; Xu, H.; Finegold, S.M. Application of quantitative real-time PCR for rapid identification of Bacteroides fragilis group and related organisms in human wound samples. Anaerobe 2011, 17, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Sedgley, C.M.; Nagel, A.C.; Shelburne, C.E.; Clewell, D.B.; Appelbe, O.; Molander, A. Quantitative real-time PCR detection of oral Enterococcus faecalis in humans. Arch. Oral Biol. 2005, 50, 575–583. [Google Scholar] [CrossRef]
- Telmadarrehei, T.; Tang, J.D.; Raji, O.; Rezazadeh, A.; Narayanan, L.; Shmulsky, R.; Jeremic, D. A Study of the gut bacterial community of Reticulitermes virginicus exposed to chitosan treatment. Insects 2020, 11, 681. [Google Scholar] [CrossRef] [PubMed]
- Bapatla, K.G.; Singh, A.; Yeddula, S.; Patil, R.H. Annotation of gut bacterial taxonomic and functional diversity in Spodoptera litura and Spilosoma obliqua. J. Basic Microbiol. 2018, 58, 217–226. [Google Scholar] [CrossRef]
- Xia, X.; Lan, B.; Tao, X.; Lin, J.; You, M. Characterization of Spodoptera litura Gut Bacteria and Their Role in Feeding and Growth of the Host. Front. Microbiol. 2020, 11, 1492. [Google Scholar] [CrossRef]
- Gao, X.; Li, W.; Luo, J.; Zhang, L.; Ji, J.; Zhu, X.; Wang, L.; Zhang, K.; Zhang, S.; Cui, J. DNA sequencing reveals bacterial communities in midgut and other parts of the larvae of Spodoptera exigua Hubner (Lepidoptera: Noctuidae). FEMS Microbiol. Lett. 2020, 367, fnaa002. [Google Scholar] [CrossRef]
- Chen, B.; Teh, B.S.; Sun, C.; Hu, S.; Lu, X.; Boland, W.; Shao, Y. Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 2016, 6, 29505. [Google Scholar] [CrossRef]
- Shan, Y.; Shu, C.; Crickmore, N.; Liu, C.; Xiang, W.; Song, F.; Zhang, J. Cultivable gut bacteria of scarabs (Coleoptera: Scarabaeidae) inhibit Bacillus thuringiensis multiplication. Environ. Entomol. 2014, 43, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Broderick, N.A.; Raffa, K.F.; Handelsman, J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc. Natl. Acad. Sci. USA 2006, 103, 15196–15199. [Google Scholar] [CrossRef] [PubMed]
- Paramasiva, I.; Sharma, H.C.; Krishnayya, P.V. Antibiotics influence the toxicity of the delta endotoxins of Bacillus thuringiensis towards the cotton bollworm, Helicoverpa armigera. BMC Microbiol. 2014, 14, 200. [Google Scholar] [CrossRef] [PubMed]
- Raymond, B.; Johnston, P.R.; Wright, D.J.; Ellis, R.J.; Crickmore, N.; Bonsall, M.B. A mid-gut microbiota is not required for the pathogenicity of Bacillus thuringiensis to diamondback moth larvae. Environ. Microbiol. 2009, 11, 2556–2563. [Google Scholar] [CrossRef]
- Ren, X.L.; Chen, R.R.; Zhang, Y.; Ma, Y.; Cui, J.J.; Han, Z.J.; Mu, L.L.; Li, G.Q. A Spodoptera exigua cadherin serves as a putative receptor for Bacillus thuringiensis Cry1Ca toxin and shows differential enhancement of Cry1Ca and Cry1Ac toxicity. Appl. Environ. Microbiol. 2013, 79, 5576–5583. [Google Scholar] [CrossRef]
- Lucena, W.A.; Pelegrini, P.B.; Martins-De-Sa, D.; Fonseca, F.C.; Gomes, J.J.; de Macedo, L.L.; Da, S.M.; Oliveira, R.S.; Grossi-De-Sa, M.F. Molecular approaches to improve the insecticidal activity of Bacillus thuringiensis Cry toxins. Toxins 2014, 6, 2393–2423. [Google Scholar] [CrossRef]
- Pigott, C.R.; Ellar, D.J. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 2007, 71, 255–281. [Google Scholar] [CrossRef]
- Broderick, N.A.; Raffa, K.F.; Handelsman, J. Chemical modulators of the innate immune response alter gypsy moth larval susceptibility to Bacillus thuringiensis. BMC Microbiol. 2010, 10, 129. [Google Scholar] [CrossRef]
- Caccia, S.; Di Lelio, I.; La Storia, A.; Marinelli, A.; Varricchio, P.; Franzetti, E.; Banyuls, N.; Tettamanti, G.; Casartelli, M.; Giordana, B.; et al. Midgut microbiota and host immunocompetence underlie Bacillus thuringiensis killing mechanism. Proc. Natl. Acad. Sci. USA 2016, 113, 9486–9491. [Google Scholar] [CrossRef]
- Mason, K.L.; Stepien, T.A.; Blum, J.E.; Holt, J.F.; Labbe, N.H.; Rush, J.S.; Raffa, K.F.; Handelsman, J. From commensal to pathogen: Translocation of Enterococcus faecalis from the midgut to the hemocoel of Manduca sexta. mBio 2011, 2, e11–e65. [Google Scholar] [CrossRef]
| Insect | Bacteria Genera | Function | References |
|---|---|---|---|
| Bombyx mori | Pseudomonas, Klebsiella pneumoniae, Clostridium flexneri | It secretes cellulase, which breaks down carbohydrates to provide energy for the host. | [14] |
| Locusta migratoria manilensis | Klebsiella pneumoniae | It breaks down grass into carbohydrates, amino acids and sugars, providing energy to the host. | [15] |
| Aedes albopictus | Lelliottia, Cladosporium, Aspergillus, Ampullimonas, Cyberlindnera | It can digest food and provide nutrients for insects. | [16] |
| Herbivorous turtle ants Cephalotes | Burkholderiales, Opitutales | It can participate in nitrogen recycling to provide amino acids for the host. | [13] |
| Acyrthosiphon pisum | Buchnera aphidicola | It can provide the host with vitamins B2 and B5. | [6] |
| Honeybee | Gilliamella apicola and Lactobacillus sp. | It produces SCFAs that promote host growth. | [19] |
| Drosophila | Acetobacter pomorum, Lactobacillus plantarum, Saccharomyces cerevisiae, Lactobacillus plantarum, Acetobacter malorum | It can influence the feeding behavior of the host. | [23] |
| Rhynchophorus ferrugineus Olivier | Lactococcus lactis, Enterobacter cloacae | It can affect protein, glucose, and triglyceride levels in the host’s hemolymph. | [25] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Chang, L.; Xu, K.; Zhang, S.; Gao, F.; Fan, Y. Research Progresses on the Function and Detection Methods of Insect Gut Microbes. Microorganisms 2023, 11, 1208. https://doi.org/10.3390/microorganisms11051208
Li Y, Chang L, Xu K, Zhang S, Gao F, Fan Y. Research Progresses on the Function and Detection Methods of Insect Gut Microbes. Microorganisms. 2023; 11(5):1208. https://doi.org/10.3390/microorganisms11051208
Chicago/Turabian StyleLi, Yazi, Liyun Chang, Ke Xu, Shuhong Zhang, Fengju Gao, and Yongshan Fan. 2023. "Research Progresses on the Function and Detection Methods of Insect Gut Microbes" Microorganisms 11, no. 5: 1208. https://doi.org/10.3390/microorganisms11051208





