Eliciting Clavulanic Acid Biosynthesis: The Impact of Bacillus velezensis FZB42 on the Metabolism of Streptoyces clavuligerus ATCC 27064
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.2. Co-Culture Preparation
2.3. Analytical Techniques
2.4. RNA Sequencing (RNA-Seq)
2.5. Data Processing
2.6. Statistical Analysis
3. Results
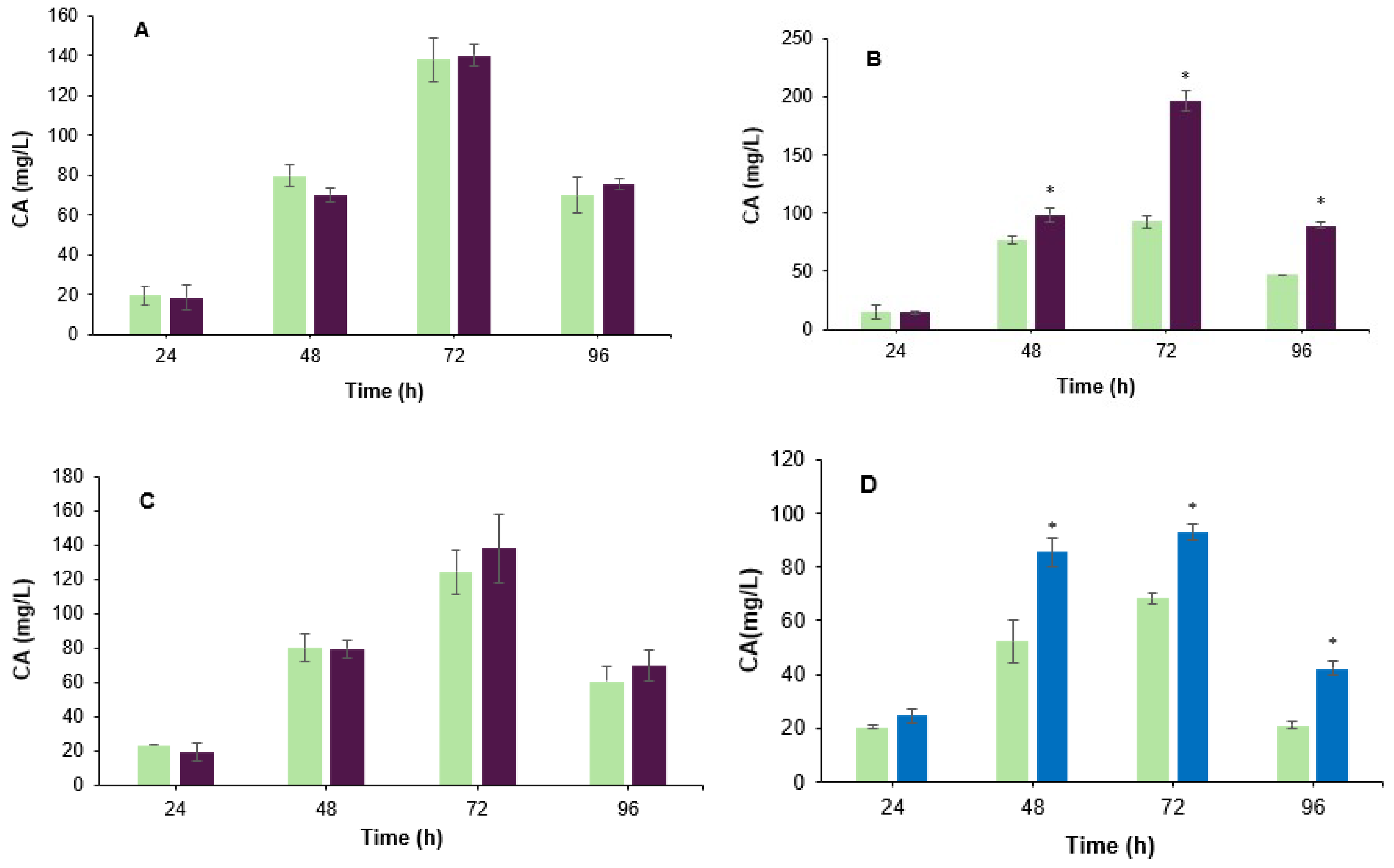
3.1. Effect of the Co-Culture on the Biosynthesis of Clavulanic Acid
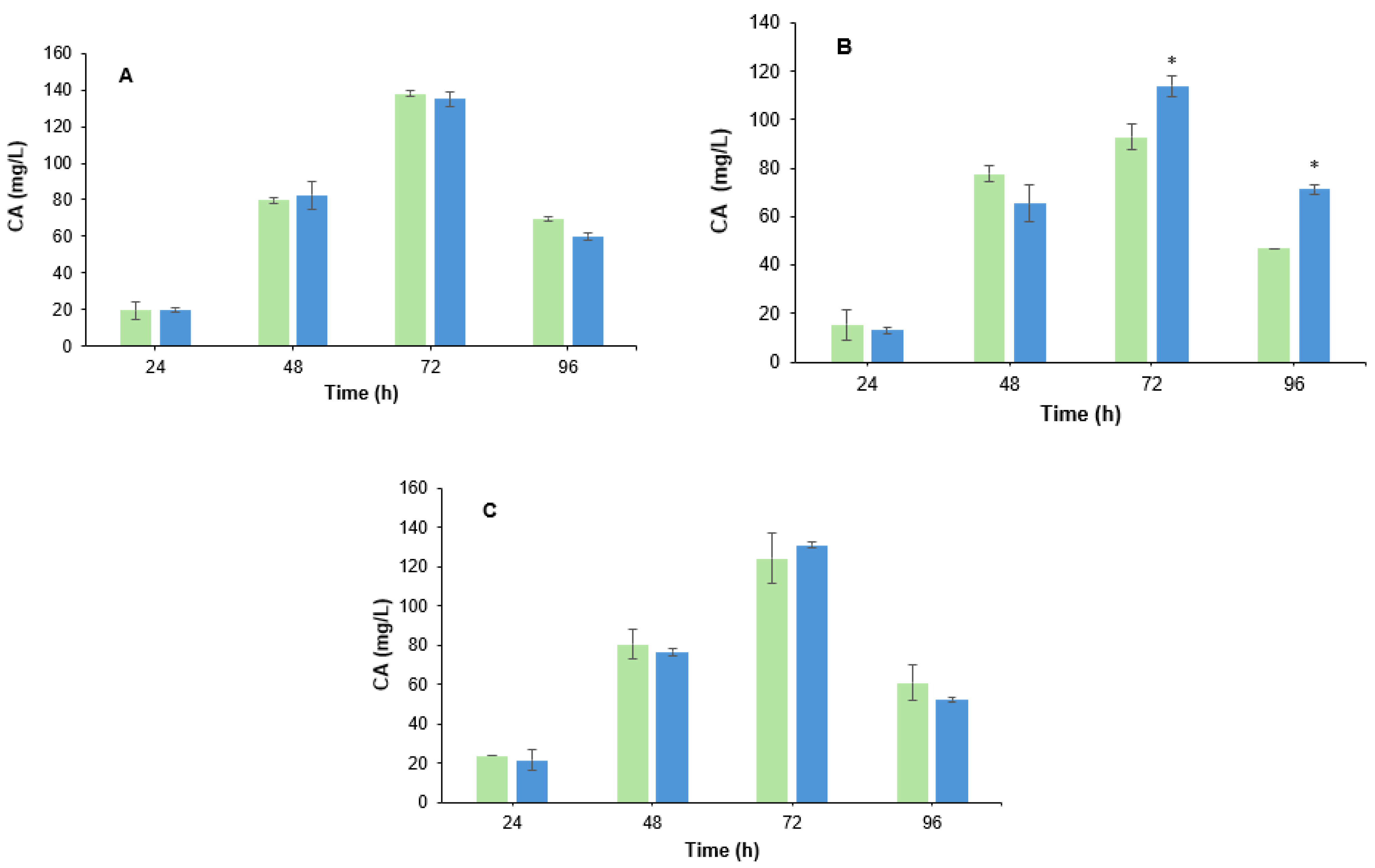
3.2. Effect of B. velezensis Cell Filtrate on Clavulanic Acid Biosynthesis in S. clavuligerus
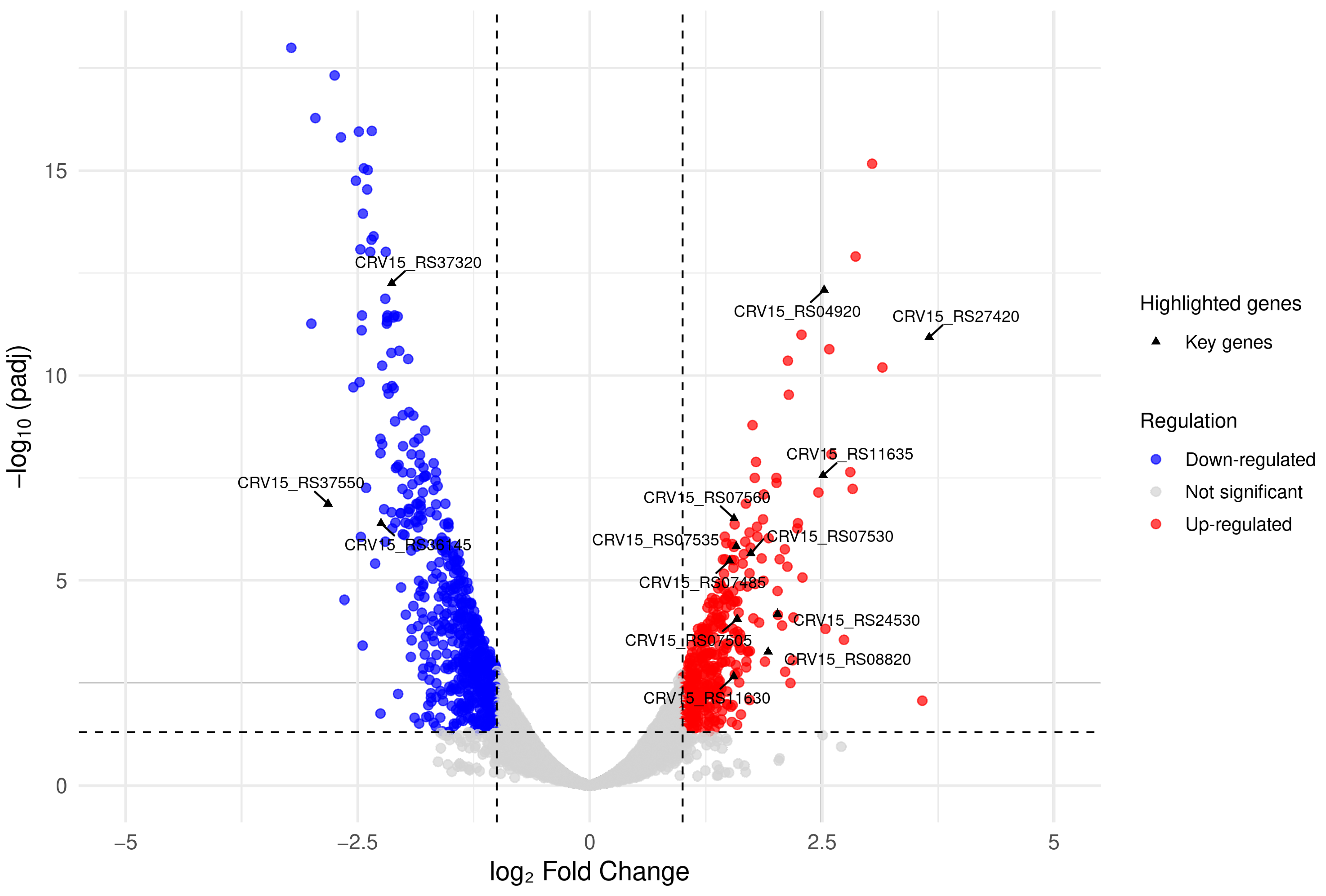
3.3. Differential Expression Analysis
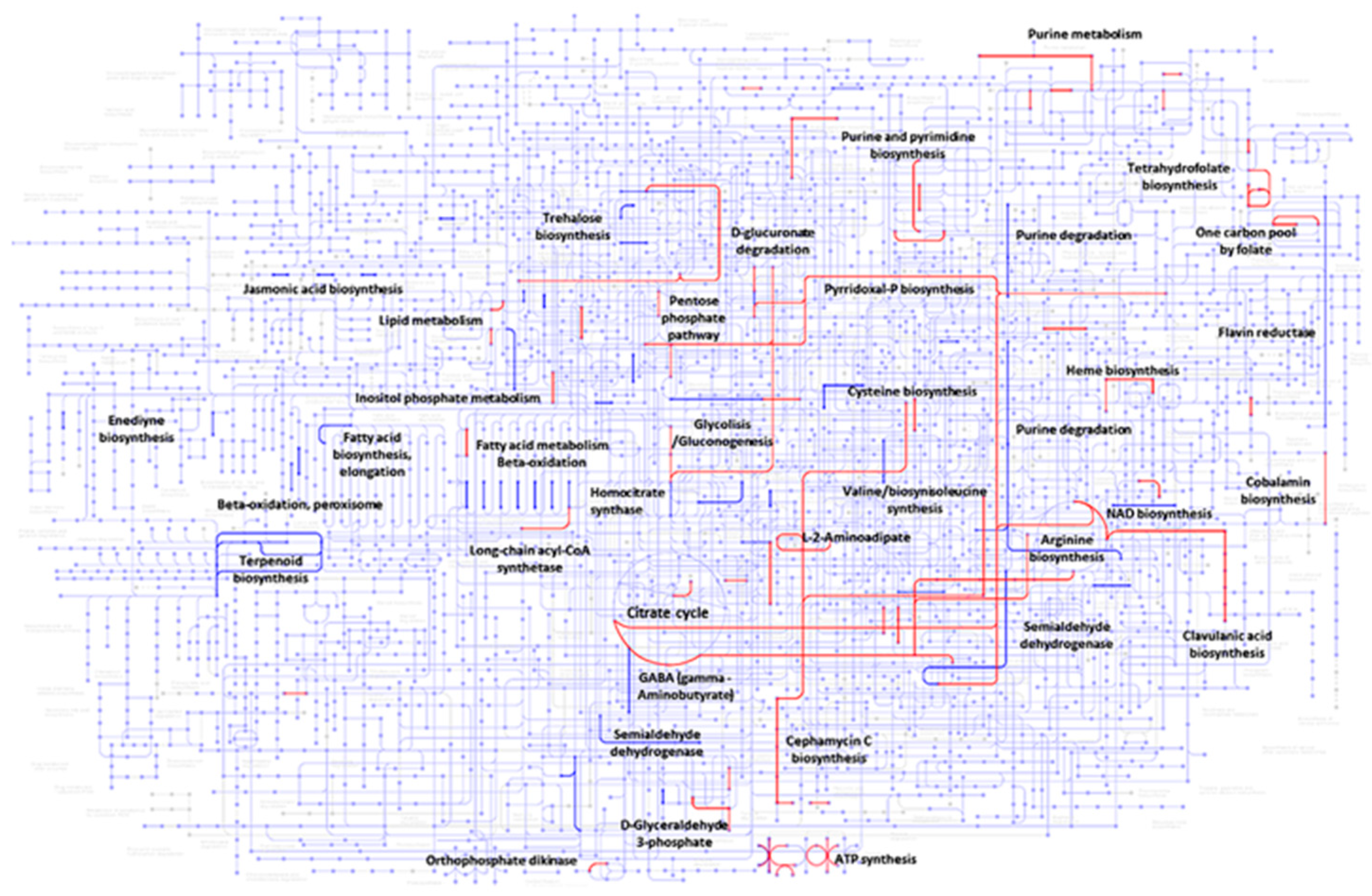
3.4. Gene Set Enrichment Analysis During Co-Culture
3.5. Up-Regulated Genes
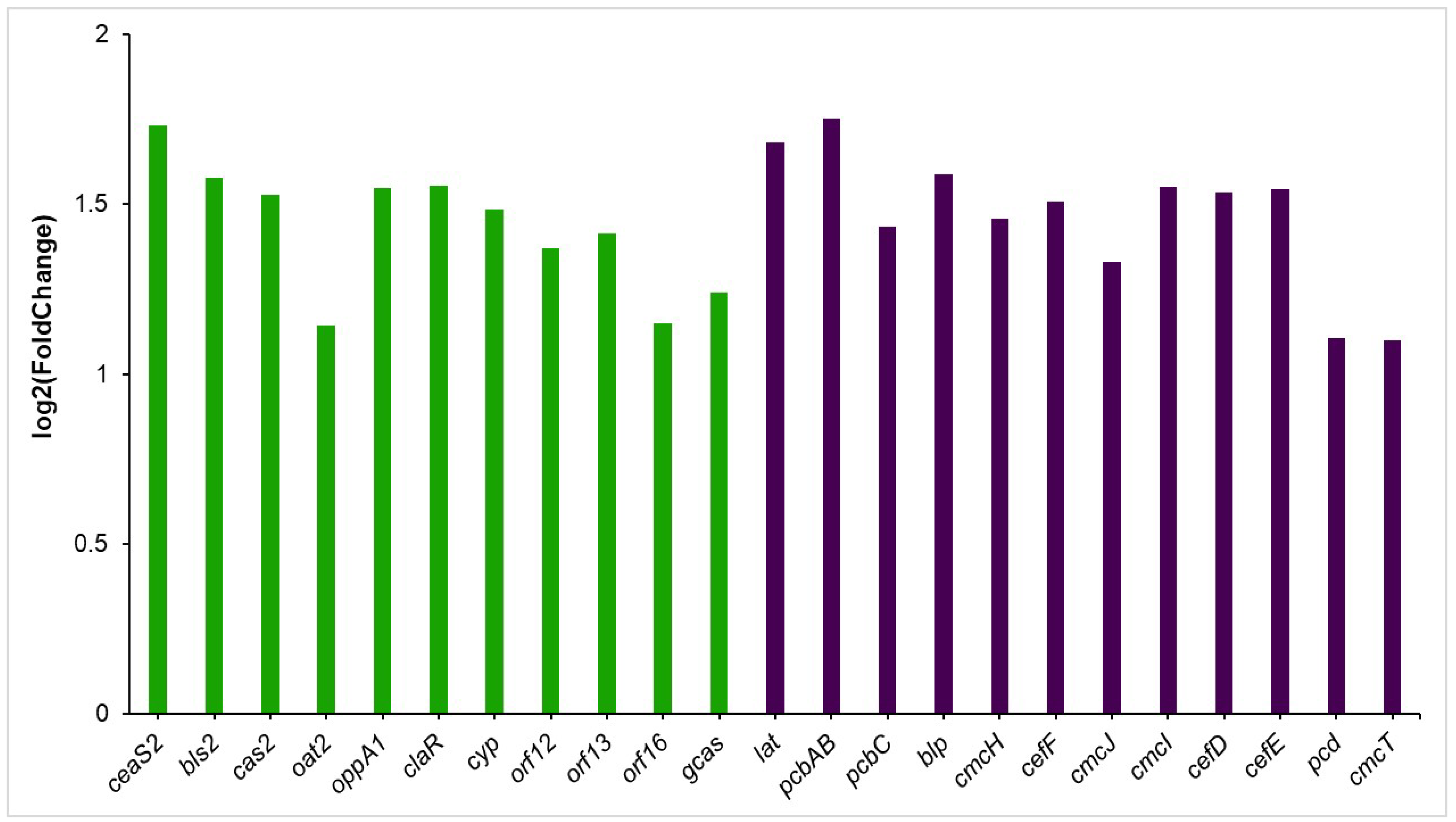
3.5.1. Clavulanic Acid and Cephamycin C Clusters
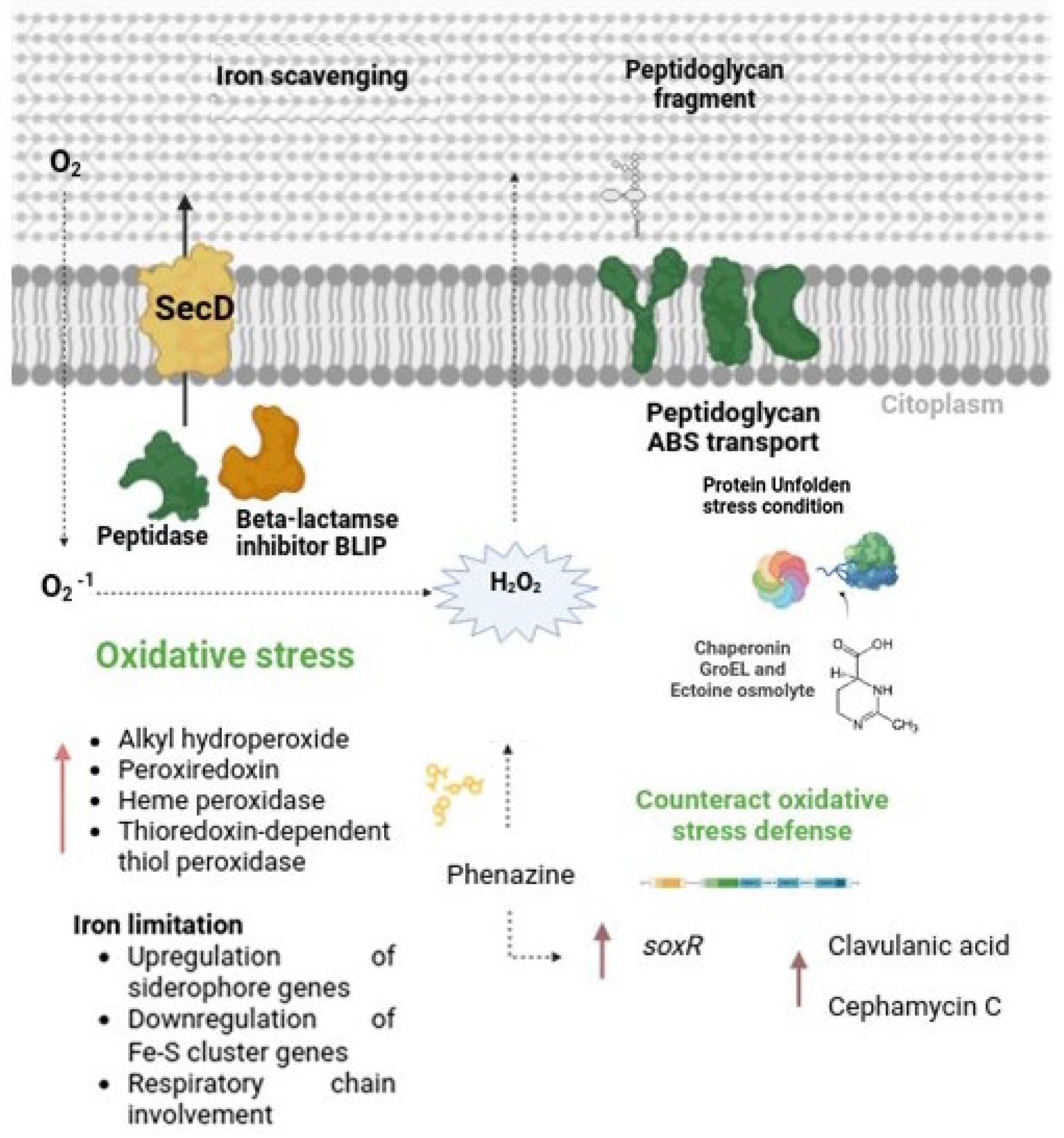
3.5.2. Differentially Expressed Genes Associated with Stress Response
3.5.3. Transcriptional Regulators and Transporters
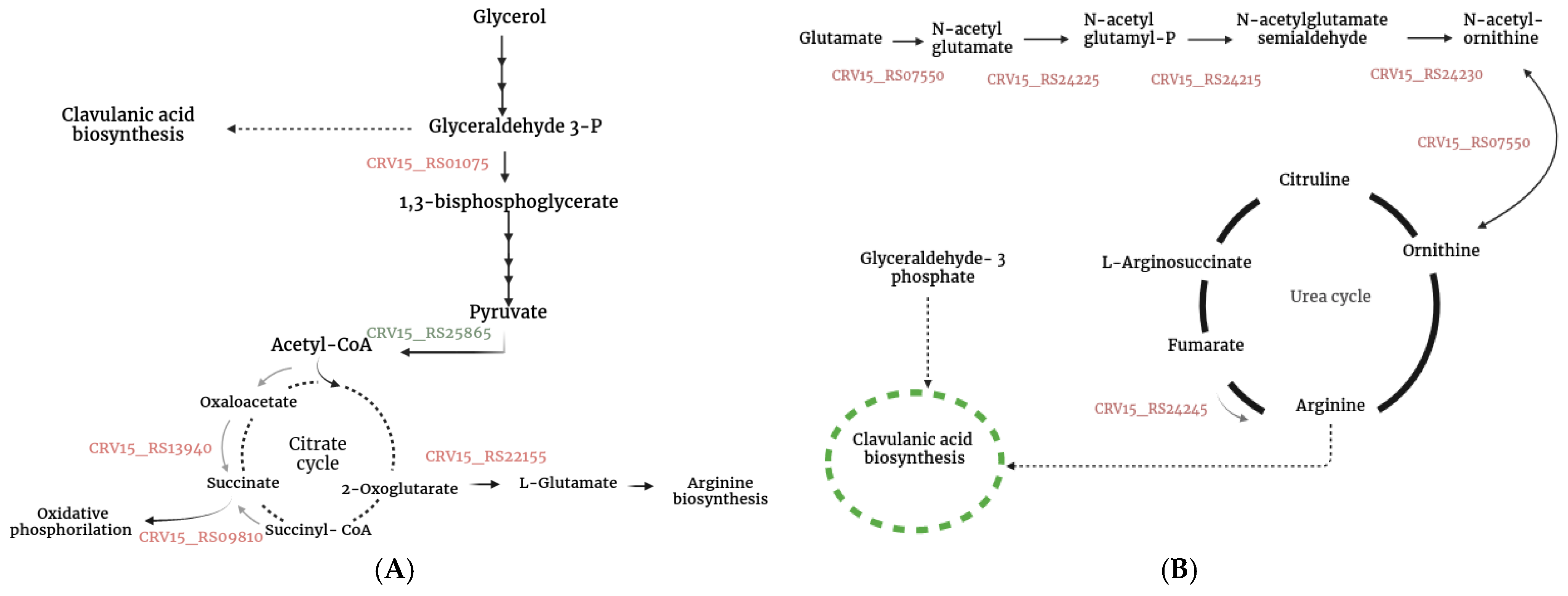
3.5.4. Additional Genes Associated with S. clavuligerus Metabolism That Were Differentially Expressed During Co-Culture
3.6. Down-Regulated Genes
4. Discussion
4.1. Co-Cultured-Mediated Alterations in Specialized Metabolite Synthesis in S. clavuligerus
4.2. Triggering of Clavulanic Acid Biosynthesis: Role of Oxidative Stress and Peptide Signaling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- López-Agudelo, V.A.; Gómez-Ríos, D.; Ramirez-Malule, H. Clavulanic Acid Production by Streptomyces clavuligerus: Insights from Systems Biology, Strain Engineering, and Downstream Processing. Antibiotics 2021, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Kizildoğan, A.K.; Jaccard, G.V.; Mutlu, A.; Sertdemïr, I.; Özcengiz, G. Genetic engineering of an industrial strain of Streptomyces clavuligerus for further enhancement of clavulanic acid production. Turk. J. Biol. 2017, 41, 342–353. [Google Scholar] [CrossRef]
- Law, K.-H.; Tsang, M.-W.; Wong, Y.-K.; Tsang, M.-S.; Lau, P.-Y.; Wong, K.-Y.; Ho, K.-P.; Leung, Y.-C. Efficient production of secretory Streptomyces clavuligerus β-lactamase inhibitory protein (BLIP) in Pichia pastoris. AMB Express 2018, 8, 64. [Google Scholar] [CrossRef]
- Santamarta, I.; Rodríguez-García, A.; Pérez-Redondo, R.; Martín, J.F.; Liras, P. CcaR Is an Autoregulatory Protein That Binds to the ccaR and cefD-cmcI Promoters of the Cephamycin C-Clavulanic Acid Cluster in Streptomyces clavuligerus. J. Bacteriol. 2002, 184, 3106–3113. [Google Scholar] [CrossRef]
- Santamarta, I.; López-García, M.T.; Kurt, A.; Nárdiz, N.; Álvarez-Álvarez, R.; Pérez-Redondo, R.; Martín, J.F.; Liras, P. Characterization of DNA-binding sequences for CcaR in the cephamycin–clavulanic acid supercluster of Streptomyces clavuligerus. Mol. Microbiol. 2011, 81, 968–981. [Google Scholar] [CrossRef]
- Fu, J.; Qin, R.; Zong, G.; Liu, C.; Kang, N.; Zhong, C.; Cao, G. The CagRS Two-Component System Regulates Clavulanic Acid Metabolism via Multiple Pathways in Streptomyces clavuligerus F613-1. Front. Microbiol. 2019, 10, 244. [Google Scholar] [CrossRef]
- Lee, N.; Kim, W.; Chung, J.; Lee, Y.; Cho, S.; Jang, K.-S.; Kim, S.C.; Palsson, B.; Cho, B.-K. Iron competition triggers antibiotic biosynthesis in Streptomyces coelicolor during coculture with Myxococcus xanthus. ISME J. 2020, 14, 1111–1124. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Ali, M.D.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K.-H. Bacillus velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Liu, X.; Li, Y.; Wang, Y.; Li, Y.; Wang, Y.; Li, Y.; Wang, Y.; Li, Y.; et al. Biocontrol potential of Bacillus velezensis EM-1 associated with suppressive rhizosphere soil microbes against tobacco bacterial wilt. Front. Microbiol. 2022, 13, 940156. [Google Scholar] [CrossRef]
- Yu, C.; Qiao, J.; Ali, Q.; Jiang, Q.; Song, Y.; Zhu, L.; Gu, Q.; Borriss, R.; Dong, S.; Gao, X.; et al. degQ associated with the degS/degU two-component system regulates biofilm formation, antimicrobial metabolite production, and biocontrol activity in Bacillus velezensis DMW1. Mol. Plant Pathol. 2023, 24, 1510–1521. [Google Scholar] [CrossRef]
- Shepherd, M.D.; Kharel, M.K.; Bosserman, M.A.; Rohr, J. Laboratory Maintenance of Streptomyces Species. Curr. Protoc. Microbiol. 2010, 18, 10E.1.1–10E.1.8. [Google Scholar] [CrossRef] [PubMed]
- Patiño, L.F.; Aguirre-Hoyos, V.; Pinilla, L.I.; Toro, L.F.; Ríos-Estepa, R. Environmental Factors Modulate the Role of orf21 Sigma Factor in Clavulanic Acid Production in Streptomyces Clavuligerus ATCC27064. Bioengineering 2022, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Malule, H.; Junne, S.; López, C.; Zapata, J.; Sáez, A.; Neubauer, P.; Rios-Estepa, R. An improved HPLC-DAD method for clavulanic acid quantification in fermentation broths of Streptomyces clavuligerus. J. Pharm. Biomed. Anal. 2016, 120, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 February 2025).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 15 February 2025).
- Nicault, M.; Zaiter, A.; Dumarcay, S.; Chaimbault, P.; Gelhaye, E.; Leblond, P.; Bontemps, C. Elicitation of Antimicrobial Active Compounds by Streptomyces-Fungus Co-Cultures. Microorganisms 2021, 9, 178. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Baños, S.; Pérez-Redondo, R.; Koekman, B.; Liras, P. Glycerol Utilization Gene Cluster in Streptomyces clavuligerus. Appl. Environ. Microbiol. 2009, 75, 2991–2995. [Google Scholar] [CrossRef]
- Ramirez-Malule, H.; Junne, S.; Cruz-Bournazou, M.N.; Neubauer, P.; Ríos-Estepa, R. Streptomyces clavuligerus shows a strong association between TCA cycle intermediate accumulation and clavulanic acid biosynthesis. Appl. Microbiol. Biotechnol. 2018, 102, 4009–4023. [Google Scholar] [CrossRef] [PubMed]
- Zhihan, Z.; Yanping, W. Application of lat gene disruption to increase the clavulanic acid production of Streptomyces clavuligerus. J. Mol. Catal. B Enzym. 2006, 43, 102–107. [Google Scholar] [CrossRef]
- Hahn, J.-S.; Oh, S.-Y.; Roe, J.-H. Role of OxyR as a Peroxide-Sensing Positive Regulator in Streptomyces coelicolor A3(2). J. Bacteriol. 2002, 184, 5214–5222. [Google Scholar] [CrossRef]
- Sadeghi, A.; Soltani, B.M.; Nekouei, M.K.; Jouzani, G.S.; Mirzaei, H.H.; Sadeghizadeh, M. Diversity of the ectoines biosynthesis genes in the salt tolerant Streptomyces and evidence for inductive effect of ectoines on their accumulation. Microbiol. Res. 2014, 169, 699–708. [Google Scholar] [CrossRef]
- Martínez-Hackert, E.; Stock, A.M. Structural Relationships in the OmpR Family of Winged-Helix Transcription Factors. J. Mol. Biol. 1997, 269, 301–312. [Google Scholar] [CrossRef]
- Seo, S.W.; Gao, Y.; Kim, D.; Szubin, R.; Yang, J.; Cho, B.-K.; Palsson, B.O. Revealing genome-scale transcriptional regulatory landscape of OmpR highlights its expanded regulatory roles under osmotic stress in Escherichia coli K-12 MG1655. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Hamed, M.B.; Anné, J.; Karamanou, S.; Economou, A. Streptomyces Protein Secretion and Its Application in Biotechnology. FEMS Microbiol. Lett. 2018, 365, fny250. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Cecchini, G. Function and Structure of Complex II of the Respiratory Chain. Annu. Rev. Biochem. 2003, 72, 77–109. [Google Scholar] [CrossRef]
- Wang, X.; Chen, N.; Cruz-Morales, P.; Zhong, B.; Zhang, Y.; Acharya, S.; Li, Z.; Deng, H.; Luo, X.; Keasling, J.D. The Natural Product Co-Evolved Pyrroloquinoline Quinone Gene Enhances Their Production When Heterologously Expressed in a Variety of Streptomycetes. Preprints 2023, 2023040045. [Google Scholar] [CrossRef]
- Noushahi, H.A.; Khan, A.H.; Noushahi, U.F.; Hussain, M.; Javed, T.; Zafar, M.; Batool, M.; Ahmed, U.; Liu, K.; Harrison, M.T.; et al. Biosynthetic pathways of triterpenoids and strategies to improve their Biosynthetic Efficiency. Plant Growth Regul. 2022, 97, 439–454. [Google Scholar] [CrossRef]
- Liras, P.; Martín, J.F. Streptomyces clavuligerus: The Omics Era. J. Ind. Microbiol. Biotechnol. 2021, 48. [Google Scholar] [CrossRef] [PubMed]
- Bothwell, I.R.; Caetano, T.; Sarksian, R.; Mendo, S.; van der Donk, W.A. Structural Analysis of Class I Lanthipeptides from Pedobacter lusitanus NL19 Reveals an Unusual Ring Pattern. ACS Chem. Biol. 2021, 16, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Bierbaum, G.; Sahl, H.-G. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Burgo, Y.; Álvarez-Álvarez, R.; Pérez-Redondo, R.; Liras, P. Heterologous expression of Streptomyces clavuligerus ATCC 27064 cephamycin C gene cluster. J. Biotechnol. 2014, 186, 21–29. [Google Scholar] [CrossRef]
- Deng, R.-X.; Zhang, Z.; Li, H.-L.; Wang, W.; Hu, H.-B.; Zhang, X.-H. Identification of a Novel Bioactive Phenazine Derivative and Regulation of phoP on Its Production in Streptomyces lomondensis S015. J. Agric. Food Chem. 2021, 69, 974–981. [Google Scholar] [CrossRef]
- Pratiwi, R.H.; Hidayat, I.; Hanafi, M.; Mangunwardoyo, W. Isolation and Structure Elucidation of Phenazine Derivative from Streptomyces sp. Strain UICC B-92 Isolated from Neesia altissima (Malvaceae). Iran. J. Microbiol. 2020, 12, 126–133. Available online: https://ijm.tums.ac.ir/index.php/ijm/article/view/2107 (accessed on 15 January 2025).
- Widderich, N.; Höppner, A.; Pittelkow, M.; Heider, J.; Smits, S.H.J.; Bremer, E. Biochemical Properties of Ectoine Hydroxylases from Extremophiles and Their Wider Taxonomic Distribution among Microorganisms. PLoS ONE 2014, 9, e93809. [Google Scholar] [CrossRef]
- Bursy, J.; Kuhlmann, A.U.; Pittelkow, M.; Hartmann, H.; Jebbar, M.; Pierik, A.J.; Bremer, E. Synthesis and Uptake of the Compatible Solutes Ectoine and 5-Hydroxyectoine by Streptomyces coelicolor A3(2) in Response to Salt and Heat Stresses. Appl. Environ. Microbiol. 2008, 74, 7286–7296. [Google Scholar] [CrossRef]
- Amoroso, A.; Boudet, J.; Berzigotti, S.; Duval, V.; Teller, N.; Mengin-Lecreulx, D.; Luxen, A.; Simorre, J.-P.; Joris, B. A Peptidoglycan Fragment Triggers β-lactam Resistance in Bacillus licheniformis. PLOS Pathog. 2012, 8, e1002571. [Google Scholar] [CrossRef]
- Gilmore, M.C.; Cava, F. Peptidoglycan recycling mediated by an ABC transporter in the plant pathogen Agrobacterium tumefaciens. Nat. Commun. 2022, 13, 7927. [Google Scholar] [CrossRef]
| Gene Annotation | Gene Product | Signal Peptide (Sec/SPI) Likelihood | Fold-Change |
|---|---|---|---|
| CRV15_RS06310 | beta_lactamase_inhibitory_protein_BLIP | 0.9089 | 2.2 |
| CRV15_RS07505 | Biotin transporter BioY | 0.9804 | 1.6 |
| CRV15_RS10680 | C40 family peptidase | 0.8146 | 2.5 |
| CRV15_RS11710 | Hypothetical protein | 0.9984 | 2.6 |
| CRV15_RS29805 | Peptidase inhibitor family | 0.9943 | 1.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patiño, L.F.; Caicedo-Montoya, C.; Pinilla-Mendoza, L.; Cuartas, J.H.; Ríos-Estepa, R. Eliciting Clavulanic Acid Biosynthesis: The Impact of Bacillus velezensis FZB42 on the Metabolism of Streptoyces clavuligerus ATCC 27064. Metabolites 2025, 15, 337. https://doi.org/10.3390/metabo15050337
Patiño LF, Caicedo-Montoya C, Pinilla-Mendoza L, Cuartas JH, Ríos-Estepa R. Eliciting Clavulanic Acid Biosynthesis: The Impact of Bacillus velezensis FZB42 on the Metabolism of Streptoyces clavuligerus ATCC 27064. Metabolites. 2025; 15(5):337. https://doi.org/10.3390/metabo15050337
Chicago/Turabian StylePatiño, Luisa F., Carlos Caicedo-Montoya, Laura Pinilla-Mendoza, Jaison H. Cuartas, and Rigoberto Ríos-Estepa. 2025. "Eliciting Clavulanic Acid Biosynthesis: The Impact of Bacillus velezensis FZB42 on the Metabolism of Streptoyces clavuligerus ATCC 27064" Metabolites 15, no. 5: 337. https://doi.org/10.3390/metabo15050337
APA StylePatiño, L. F., Caicedo-Montoya, C., Pinilla-Mendoza, L., Cuartas, J. H., & Ríos-Estepa, R. (2025). Eliciting Clavulanic Acid Biosynthesis: The Impact of Bacillus velezensis FZB42 on the Metabolism of Streptoyces clavuligerus ATCC 27064. Metabolites, 15(5), 337. https://doi.org/10.3390/metabo15050337






