Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, General Culture Conditions, and DNA Isolation
2.2. Genome Sequencing and Identification of azp BGC
2.3. Construction of Plasmid pJLH-RNAi-azpA for RNA-Mediated Silencing of azpA Gene
2.4. Construction of Plasmid pFC332-azpA for azpA Disruption by CRISPR-Cas9
2.5. Transformation of P. verrucosus FAE27 and Transformants Selection
2.6. RNA Extraction and qRT-PCR Experiments
2.7. Extraction of Red-Pigmented Metabolites and HPLC Analysis
3. Results
3.1. Identification and Characterization of azp BGC
3.2. RNAi-Mediated Silencing of azpA in P. verrucosus FAE27
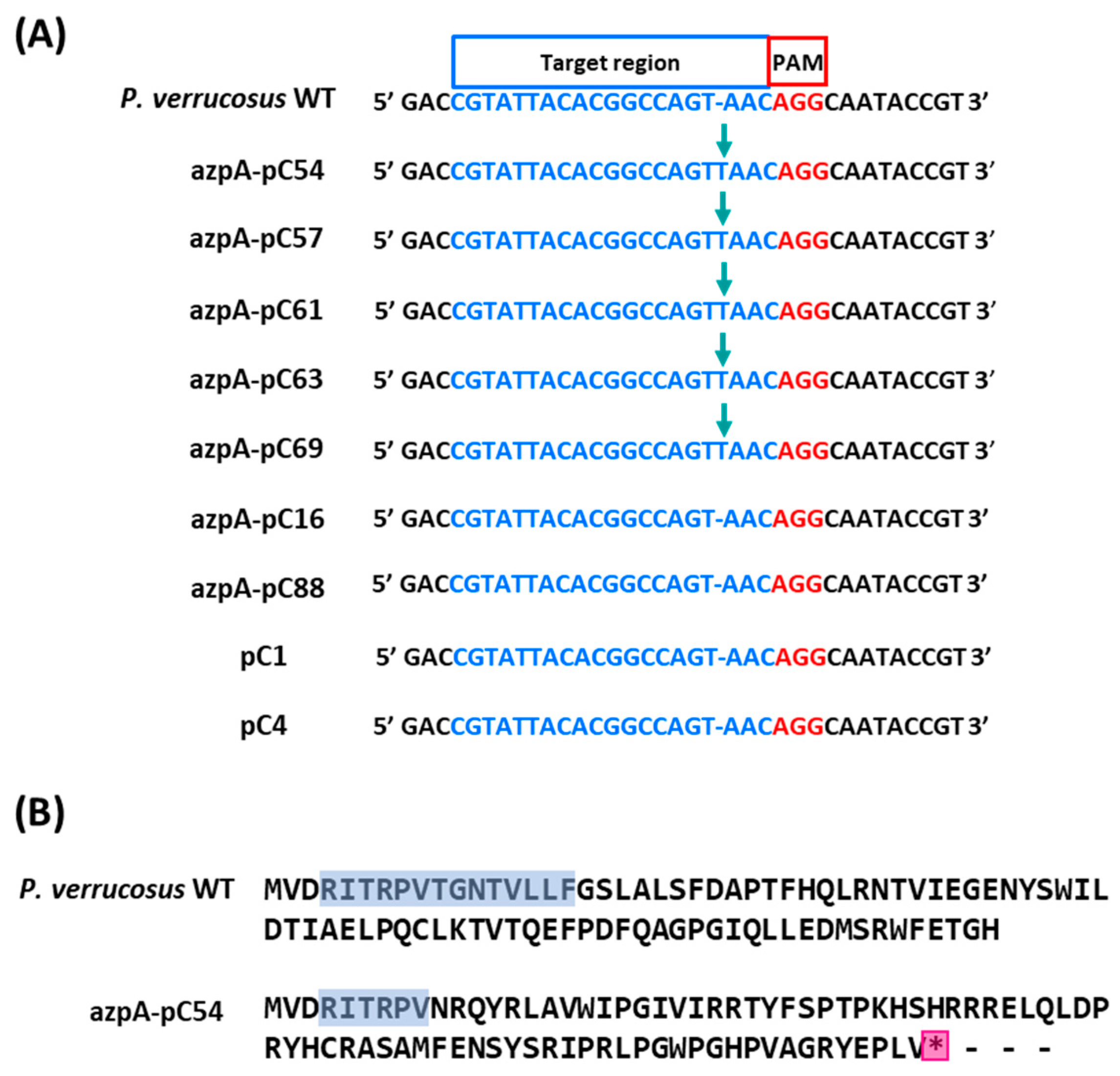
3.3. Disruption of azpA Gene in P. verrucosus FAE27 by CRISPR-Cas9
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, T.L.; Reichard, J.D.; Coleman, J.T.H.; Weller, T.J.; Thogmartin, W.E.; Reichert, B.E.; Bennett, A.B.; Broders, H.G.; Campbell, J.; Etchison, K.; et al. The scope and severity of white-nose syndrome on hibernating bats in North America. Conserv. Biol. 2021, 35, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Minnis, A.M.; Lindner, D.L. Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biol. 2013, 117, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, P.; Vásquez, G.; Gil-Durán, C.; Oliva, V.; Díaz, A.; Henríquez, M.; Álvarez, E.; Laich, F.; Chávez, R.; Vaca, I. Description of the first four species of the genus Pseudogymnoascus from Antarctica. Front. Microbiol. 2021, 12, 713189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Dong, C.; Chen, W.; Mou, Q.; Lu, X.; Han, Y.; Huang, J.; Liang, Z. The enigmatic Thelebolaceae (Thelebolales, Leotiomycetes): One new genus Solomyces and five new species. Front. Microbiol. 2020, 11, 572596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Shao, Q.Y.; Li, X.; Chen, W.H.; Liang, J.D.; Han, Y.F.; Huang, J.Z.; Liang, Z.Q. Culturable fungi from urban soils in China I: Description of 10 new taxa. Microbiol. Spectr. 2021, 9, e0086721. [Google Scholar] [CrossRef] [PubMed]
- Becker, P.; van den Eynde, C.; Baert, F.; D’hooge, E.; De Pauw, R.; Normand, A.C.; Piarroux, R.; Stubbe, D. Remarkable fungal biodiversity on northern Belgium bats and hibernacula. Mycologia 2023, 115, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Han, Y.F.; Chen, W.H.; Tao, G. Additions to Thelebolales (Leotiomycetes, Ascomycota): Pseudogeomyces lindneri gen. et sp. nov. and Pseudogymnoascus campensis sp. nov. MycoKeys 2023, 95, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Li, X.; Chen, W.H.; Liang, J.D.; Han, Y.F. Culturable fungi from urban soils in China II, with the description of 18 novel species in Ascomycota (Dothideomycetes, Eurotiomycetes, Leotiomycetes and Sordariomycetes). MycoKeys 2023, 98, 167–220. [Google Scholar] [CrossRef]
- Furbino, L.E.; Godinho, V.M.; Santiago, I.F.; Pellizari, F.M.; Alves, T.M.; Zani, C.L.; Junior, P.A.; Romanha, A.J.; Carvalho, A.G.; Gil, L.H.; et al. Diversity patterns, ecology and biological activities of fungal communities associated with the endemic macroalgae across the Antarctic peninsula. Microb. Ecol. 2014, 67, 775–787. [Google Scholar] [CrossRef]
- Henríquez, M.; Vergara, K.; Norambuena, J.; Beiza, A.; Maza, F.; Ubilla, P.; Araya, I.; Chávez, R.; San-Martín, A.; Darias, J.; et al. Diversity of cultivable fungi associated with Antarctic marine sponges and screening for their antimicrobial, antitumoral and antioxidant potential. World J. Microbiol. Biotechnol. 2014, 30, 65–76. [Google Scholar] [CrossRef]
- Gonçalves, V.N.; Carvalho, C.R.; Johann, S.; Mendes, G.; Alves, T.M.; Zani, C.L.; Junior, P.A.S.; Murta, S.M.F.; Romanha, A.J.; Cantrell, C.L.; et al. Antibacterial, antifungal and antiprotozoal activities of fungal communities present in different substrates from Antarctica. Polar Biol. 2015, 38, 1143–1152. [Google Scholar] [CrossRef]
- Gomes, E.C.Q.; Godinho, V.M.; Silva, D.A.S.; de Paula, M.T.R.; Vitoreli, G.A.; Zani, C.L.; Alves, T.M.A.; Junior, P.A.S.; Murta, S.M.F.; Barbosa, E.C.; et al. Cultivable fungi present in Antarctic soils: Taxonomy, phylogeny, diversity, and bioprospecting of antiparasitic and herbicidal metabolites. Extremophiles 2018, 22, 381–393. [Google Scholar] [CrossRef]
- Purić, J.; Vieira, G.; Cavalca, L.B.; Sette, L.D.; Ferreira, H.; Vieira, M.L.C.; Sass, D.C. Activity of Antarctic fungi extracts against phytopathogenic bacteria. Lett. Appl. Microbiol. 2018, 66, 530–536. [Google Scholar] [CrossRef]
- Vieira, G.; Purić, J.; Morão, L.G.; Dos Santos, J.A.; Inforsato, F.J.; Sette, L.D.; Ferreira, H.; Sass, D.C. Terrestrial and marine Antarctic fungi extracts active against Xanthomonas citri subsp. citri. Lett. Appl. Microbiol. 2018, 67, 64–71. [Google Scholar] [CrossRef]
- Shi, T.; Li, X.Q.; Zheng, L.; Zhang, Y.H.; Dai, J.J.; Shang, E.L.; Yu, Y.Y.; Zhang, Y.T.; Hu, W.P.; Shi, D.Y. Sesquiterpenoids from the Antarctic fungus Pseudogymnoascus sp. HSX2#-11. Front. Microbiol. 2021, 12, 688202. [Google Scholar] [CrossRef]
- Antipova, T.V.; Zaitsev, K.V.; Zhelifonova, V.P.; Tarlachkov, S.V.; Grishin, Y.K.; Kochkina, G.A.; Vainshtein, M.B. The potential of arctic Pseudogymnoascus fungi in the biosynthesis of natural products. Fermentation 2023, 9, 702. [Google Scholar] [CrossRef]
- Shi, T.; Yu, Y.-Y.; Dai, J.-J.; Zhang, Y.-T.; Hu, W.-P.; Zheng, L.; Shi, D.-Y. New polyketides from the Antarctic fungus Pseudogymnoascus sp. HSX2#-11. Mar. Drugs 2021, 19, 168. [Google Scholar]
- Han, X.; Gao, H.; Lai, H.; Zhu, W.; Wang, Y. Anti-Aβ42 Aggregative polyketides from the Antarctic psychrophilic fungus Pseudogymnoascus sp. OUCMDZ-3578. J. Nat. Prod. 2023, 86, 882–890. [Google Scholar] [CrossRef]
- Hou, X.; Li, C.; Zhang, R.; Li, Y.; Li, H.; Zhang, Y.; Tae, H.S.; Yu, R.; Che, Q.; Zhu, T.; et al. Unusual tetrahydropyridoindole-containing tetrapeptides with human nicotinic acetylcholine receptors targeting activity discovered from Antarctica-derived psychrophilic Pseudogymnoascus sp. HDN17-933. Mar. Drugs 2022, 20, 593. [Google Scholar] [CrossRef]
- Shi, T.; Zheng, L.; Li, X.-Q.; Dai, J.-J.; Zhang, Y.-T.; Yu, Y.-Y.; Hu, W.-P.; Shi, D.-Y. Nitrogenous compounds from the Antarctic fungus Pseudogymnoascus sp. HSX2#-11. Molecules 2021, 26, 2636. [Google Scholar]
- Figueroa, L.; Jiménez, C.; Rodríguez, J.; Areche, C.; Chávez, R.; Henríquez, M.; de la Cruz, M.; Díaz, C.; Segade, Y.; Vaca, I. 3-Nitroasterric acid derivatives from an Antarctic sponge-derived Pseudogymnoascus sp. fungus. J. Nat. Prod. 2015, 78, 919–923. [Google Scholar] [CrossRef]
- Zhang, T.; Ren, P.; Chaturvedi, V.; Chaturvedi, S. Development of an agrobacterium-mediated transformation system for the cold-adapted fungi Pseudogymnoascus destructans and P. pannorum. Fungal Genet. Biol. 2015, 81, 73–81. [Google Scholar] [CrossRef]
- Díaz, A.; Villanueva, P.; Oliva, V.; Gil-Durán, C.; Fierro, F.; Chávez, R.; Vaca, I. Genetic transformation of the filamentous fungus Pseudogymnoascus verrucosus of Antarctic origin. Front. Microbiol. 2019, 10, 2675. [Google Scholar] [CrossRef]
- Lax, C.; Tahiri, G.; Patiño-Medina, J.A.; Cánovas-Márquez, J.T.; Pérez-Ruiz, J.A.; Osorio-Concepción, M.; Navarro, E.; Calo, S. The evolutionary significance of RNAi in the Fungal Kingdom. Int. J. Mol. Sci. 2020, 21, 9348. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.S.; Zhang, Z.; Liu, Y. RNA interference pathways in fungi: Mechanisms and functions. Annu. Rev. Microbiol. 2012, 66, 305–323. [Google Scholar] [CrossRef]
- Nakayashiki, H.; Nguyen, Q.B. RNA interference: Roles in fungal biology. Curr. Opin. Microbiol. 2008, 11, 494–502. [Google Scholar] [CrossRef]
- Van Leeuwe, T.M.; Arentshorst, M.; Ernst, T.; Alazi, E.; Punt, P.J.; Ram, A.F. Efficient marker free CRISPR/Cas9 genome editing for functional analysis of gene families in filamentous fungi. Fungal Biol. Biotechnol. 2009, 6, 13. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N. Multiplex genome engineering using CRISPR-Cas systems. Science 2013, 339, 197–217. [Google Scholar] [CrossRef]
- O’Connell, M.R.; Oakes, B.L.; Sternberg, S.H.; Eastseletsky, A.; Kaplan, M.; Doudna, J.A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.V.; Currah, R.S. Two new species of Pseudogymnoascus with Geomyces anamorphs and their phylogenetic relationship with Gymnostellatospora. Mycologia 2006, 98, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.; Menezes, G.; Silva, T.; Bicas, J.; Oliveira, V.; Rosa, L. Antarctic fungi as producers of pigments. In Fungi of Antarctica; Rosa, L., Ed.; Springer: Cham, Switzerland, 2019; pp. 305–318. [Google Scholar]
- Mahuku, G. A simple extraction method suitable for PCR-based analysis of plant, fungal, and bacterial DNA. Plant Mol. Biol. Rep. 2004, 22, 71–81. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Ullán, R.V.; Godio, R.P.; Teijeira, F.; Vaca, I.; García-Estrada, C.; Feltrer, R.; Kosalková, K.; Martín, J.F. RNA-silencing in Penicillium chrysogenum and Acremonium chrysogenum: Validation studies using β-lactam genes expression. J. Microbiol. Methods 2008, 75, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Punt, P.J.; Oliver, R.P.; Dingemanse, M.A.; Pouwels, P.H.; van den Hondel, C.A. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 1987, 56, 117–124. [Google Scholar] [CrossRef]
- Gietz, R.D.; Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. In Methods in Enzymology; Guthrie, C., Fink, G.R., Eds.; Academic Press: Cambridge, MA, USA, 2002; Volume 350, pp. 87–96. [Google Scholar]
- Xie, S.; Shen, B.; Zhang, C.; Huang, X.; Zhang, Y. sgRNAcas9: A software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE 2014, 9, e100448. [Google Scholar] [CrossRef]
- Marcano, Y.; Montanares, M.; Gil-Durán, C.; González, K.; Levicán, G.; Vaca, I.; Chávez, R. PrlaeA affects the production of roquefortine C, mycophenolic acid, and andrastin A in Penicillium roqueforti, but it has little impact on asexual development. J. Fungi 2023, 9, 954. [Google Scholar] [CrossRef]
- Gil-Durán, C.; Palma, D.; Marcano, Y.; Palacios, J.-L.; Martínez, C.; Rojas-Aedo, J.F.; Levicán, G.; Vaca, I.; Chávez, R. CRISPR/Cas9-mediated disruption of the pcz1 gene and its impact on growth, development, and penicillin production in Penicillium rubens. J. Fungi 2023, 9, 1010. [Google Scholar] [CrossRef]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- Schumann, U.; Smith, N.A.; Wang, M.B. A fast and efficient method for preparation of high-quality RNA from fungal mycelia. BMC Res. Notes 2013, 6, 71. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, B.; Karki, S.; Chiu, S.H.; Kim, H.-J.; Suh, J.-W.; Nam, B.; Yoon, Y.-M.; Chen, C.-C.; Kwon, H.-J. Genetic localization and in vivo characterization of a Monascus azaphilone pigment biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 2013, 97, 6337–6345. [Google Scholar] [CrossRef] [PubMed]
- Zabala, A.O.; Xu, W.; Chooi, Y.H.; Tang, Y. Characterization of a silent azaphilone gene cluster from Aspergillus niger ATCC 1015 reveals a hydroxylation-mediated pyran-ring formation. Chem. Biol. 2012, 19, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Pavesi, C.; Flon, V.; Mann, S.; Leleu, S.; Prado, S.; Franck, X. Biosynthesis of azaphilones: A review. Nat. Prod. Rep. 2021, 38, 1058–1071. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Shao, Y.; Chen, F. Monascus pigments. Appl. Microbiol. Biotechnol. 2012, 96, 1421–1440. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chen, R.; Liu, Q.; He, Y.; He, K.; Ding, X.; Kang, L.; Guo, X.; Xie, N.; Zhou, Y.; et al. Orange, red, yellow: Biosynthesis of azaphilone pigments in Monascus fungi. Chem. Sci. 2017, 8, 4917–4925. [Google Scholar] [CrossRef]
- Chen, C.; Tao, H.; Chen, W.; Yang, B.; Zhou, X.; Luo, X.; Liu, Y. Recent advances in the chemistry and biology of azaphilones. RSC Adv. 2020, 10, 10197–10220. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, B.; Du, X.; Li, P.; Liang, B.; Cheng, X.; Du, L.; Huang, D.; Wang, L.; Wang, S. Complete genome sequence and transcriptomics analyses reveal pigment biosynthesis and regulatory mechanisms in an industrial strain, Monascus purpureus YY-1. Sci. Rep. 2015, 5, 8331. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Feng, Y.; Molnár, I.; Chen, F. Nature and nurture: Confluence of pathway determinism with metabolic and chemical serendipity diversifies Monascus azaphilone pigments. Nat. Prod. Rep. 2019, 36, 561–572. [Google Scholar] [CrossRef]
- Stadler, M.; Fournier, J. Pigment chemistry, taxonomy and phylogeny of the Hypoxyloideae (Xylariaceae). Rev. Iberoam. Micol. 2006, 23, 160–170. [Google Scholar] [CrossRef]
- Kuhnert, E.; Surup, F.; Sir, E.B.; Lambert, C.; Hyde, K.D.; Hladki, A.I.; Romero, A.I.; Stadler, M. Lenormandins A—G, new azaphilones from Hypoxylon lenormandii and Hypoxylon jaklitschii sp. nov., recognised by chemotaxonomic data. Fungal Div. 2015, 71, 165–184. [Google Scholar] [CrossRef]
- Yang, M.Y.; Wang, Y.X.; Chang, Q.H.; Li, L.F.; Liu, Y.F.; Cao, F. Cytochalasans and azaphilones: Suitable chemotaxonomic markers for the Chaetomium species. Appl. Microbiol. Biotechnol. 2021, 105, 8139–8155. [Google Scholar] [CrossRef]
- Osmanova, N.; Schultze, W.; Ayoub, N. Azaphilones: A class of fungal metabolites with diverse biological activities. Phytochem. Rev. 2010, 9, 315–342. [Google Scholar] [CrossRef]
- Sousa, T.F.; de Araújo Júnior, M.B.; Peres, E.G.; Souza, M.P.; da Silva, F.M.A.; de Medeiros, L.S.; de Souza, A.D.L.; de Souza, A.Q.L.; Yamagishi, M.E.B.; da Silva, G.F.; et al. Discovery of dual PKS involved in sclerotiorin biosynthesis in Penicillium meliponae using genome mining and gene knockout. Arch. Microbiol. 2023, 205, 75. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.R.; Han, S.; Shin, S.C.; Yeom, S.C.; Kim, H.J. Improved natural food colorant production in the filamentous fungus Monascus ruber using CRISPR-based engineering. Food Res. Int. 2023, 167, 112651. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Yang, Q.; Xue, Z.; Liu, Y. RNA interference in fungi: Pathways, functions, and applications. Eukaryot. Cell 2011, 10, 1148–1155. [Google Scholar] [CrossRef]
- Moriwaki, A.; Ueno, M.; Arase, S.; Kihara, J. RNA-mediated gene silencing in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 2007, 269, 85–89. [Google Scholar] [CrossRef]
- Hu, Y.; Hao, X.; Lou, J.; Zhang, P.; Pan, J.; Zhu, X. A PKS gene, pks-1, is involved in chaetoglobosin biosynthesis, pigmentation and sporulation in Chaetomium globosum. Sci. China Life Sci. 2012, 55, 1100–1108. [Google Scholar] [CrossRef]
- Hu, Y.; Hao, X.; Chen, L.; Akhberdi, O.; Yu, X.; Liu, Y.; Zhu, X. Gα-cAMP/PKA pathway positively regulates pigmentation, chaetoglobosin A biosynthesis and sexual development in Chaetomium globosum. PLoS ONE 2018, 13, e0195553. [Google Scholar] [CrossRef]
- Alhawatema, M.S.; Gebril, S.; Cook, D.; Creamer, R. RNAi-mediated down-regulation of a melanin polyketide synthase (pks1) gene in the fungus Slafractonia leguminicola. World J. Microbiol. Biotechnol. 2017, 33, 179. [Google Scholar] [CrossRef]
- Voigt, O.; Knabe, N.; Nitsche, S.; Erdmann, E.A.; Schumacher, J.; Gorbushina, A.A. An advanced genetic toolkit for exploring the biology of the rock-inhabiting black fungus Knufia petricola. Sci Rep. 2020, 10, 12021. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Yilmaz, N.; Thrane, U.; Rasmussen, K.B.; Houbraken, J.; Samson, R.A. Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS ONE 2013, 8, e84102. [Google Scholar] [CrossRef]
- Fernández, P.; Fañanás, F.J.; Rodríguez, F. Nitrogenated azaphilone derivatives through a silver-catalysed reaction of imines from ortho-alkynylbenzaldehydes. Chemistry 2017, 23, 3002–3006. [Google Scholar] [CrossRef] [PubMed]
- Sledzinski, P.; Nowaczyk, M.; Olejniczak, M. Computational tools and supporting CRISPR-Cas experiments. Cells 2020, 9, 1288. [Google Scholar] [CrossRef] [PubMed]
- Chibucos, M.C.; Crabtree, J.; Nagaraj, S.; Chaturvedi, S.; Chaturvedi, V. Draft genome sequences of human pathogenic fungus Geomyces pannorum sensu lato and bat white nose syndrome pathogen Geomyces (Pseudogymnoascus) destructans. Genome Announc. 2013, 1, e01045-13. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Drees, K.P.; Foster, J.T.; Lindner, D.L. Extreme sensitivity to ultraviolet light in the fungal pathogen causing white-nose syndrome of bats. Nat. Commun. 2018, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Davy, C.M.; Donaldson, M.E.; Bandouchova, H.; Breit, A.M.; Dorville, N.A.S.; Dzal, Y.A.; Kovacova, V.; Kunkel, E.L.; Martínková, N.; Norquay, K.J.O.; et al. Transcriptional host-pathogen responses of Pseudogymnoascus destructans and three species of bats with white-nose syndrome. Virulence 2020, 11, 781–794. [Google Scholar] [CrossRef]
- Kim, S.; Lee, R.; Jeon, H.; Lee, N.; Park, J.; Moon, H.; Shin, J.; Min, K.; Kim, J.E.; Yang, J.W.; et al. Identification of essential genes for the establishment of spray-induced gene silencing-based disease control in Fusarium graminearum. J. Agric. Food Chem. 2023, 71, 19302–19311. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, K.; Li, M.; Hu, H.; Zhang, X.; Liu, J.; Pan, H.; Zhang, Y. SsAGM1-mediated uridine diphosphate-N-acetylglucosamine synthesis is essential for development, stress response, and pathogenicity of Sclerotinia sclerotiorum. Front. Microbiol. 2022, 13, 938784. [Google Scholar] [CrossRef]
- Del-Cid, A.; Gil-Durán, C.; Vaca, I.; Rojas-Aedo, J.F.; García-Rico, R.O.; Levicán, G.; Chávez, R. Identification and functional analysis of the mycophenolic acid gene cluster of Penicillium roqueforti. PLoS ONE 2016, 11, e0147047. [Google Scholar] [CrossRef]



| Closest Characterized Homologues | |||||
|---|---|---|---|---|---|
| Protein Name | Size (Aminoacids) | Putative Function | Protein Name (Organism) | GenBank Accession Number | Identity (%) |
| AzpA | 2698 | Non-reducing polyketide synthase | Conidial yellow pigment biosynthesis polyketide synthase (Monascus pilosus) | AGN71604 | 61 |
| AzpB | 2315 | Highly reducing polyketide synthase | Polyketide synthase (Aspergillus niger) | EHA28244 | 45 |
| AzpC | 374 | Ketoreductase | Aldehyde reductase (Monascus pilosus) | AGN71608 | 50 |
| AzpD | 455 | O-acetyltransferase | Acetyltransferase (Monascus pilosus) | AGN71607 | 43 |
| AzpE | 445 | FAD monooxygenase | Monooxygenase (Phoma sp.) | QCO93109 | 53 |
| AzpF | 268 | Serine hydrolase | Amino oxidase/esterase (Monascus pilosus) | AGN71609 | 58 |
| AzpG | 364 | Enoyl reductase | Putative quinone-oxidoreductase-like protein (Monascus pilosus) | AGN71610 | 50 |
| AzpH | 644 | FAD oxidase | Isoamyl alcohol oxidase (Penicillium expansum) | AIG62142 | 36 |
| AzpI | 368 | Cytochrome P450 | BuaG cytochrome P450 (Aspergillus burnettii) | QBE85647 | 49 |
| AzpJ | 482 | FAD oxidase | FAD oxidase (Aspergillus niger) | EHA28243 | 47 |
| AzpK | 216 | Transporter | AflT transporter (Aspergillus flavus) | AAS90069 | 45 |
| AzpL | 119 | Transcription factor | Putative citrinin biosynthesis transcriptional activator CtnR (Monascus pilosus) | AGN71605 | 51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palma, D.; Oliva, V.; Montanares, M.; Gil-Durán, C.; Travisany, D.; Chávez, R.; Vaca, I. Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus. J. Fungi 2024, 10, 157. https://doi.org/10.3390/jof10020157
Palma D, Oliva V, Montanares M, Gil-Durán C, Travisany D, Chávez R, Vaca I. Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus. Journal of Fungi. 2024; 10(2):157. https://doi.org/10.3390/jof10020157
Chicago/Turabian StylePalma, Diego, Vicente Oliva, Mariana Montanares, Carlos Gil-Durán, Dante Travisany, Renato Chávez, and Inmaculada Vaca. 2024. "Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus" Journal of Fungi 10, no. 2: 157. https://doi.org/10.3390/jof10020157
APA StylePalma, D., Oliva, V., Montanares, M., Gil-Durán, C., Travisany, D., Chávez, R., & Vaca, I. (2024). Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus. Journal of Fungi, 10(2), 157. https://doi.org/10.3390/jof10020157







