Navigating the Deep Eutectic Solvent Landscape: Experimental and Machine Learning Solubility Explorations of Syringic, p-Coumaric, and Caffeic Acids
Abstract
1. Introduction
2. Results and Discussion
2.1. Experimental Solubility
2.2. Model Performance and Evaluation
3. Materials and Methods
3.1. Materials
3.2. Experimental Solubility Measurements
3.3. Molecular Descriptors
3.4. Machine Learning Protocol
3.4.1. Model Development Framework
3.4.2. Dual-Objective Optimization (DOO): Accuracy vs. Complexity
3.4.3. Iterative Feature Pruning and Candidate Selection
3.4.4. Information-Based Model Selection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Robbins, R.J. Phenolic acids in foods: An overview of analytical methodology. J. Agric. Food Chem. 2003, 51, 2866–2887. [Google Scholar] [CrossRef]
- Al Jitan, S.; Alkhoori, S.A.; Yousef, L.F. Phenolic Acids From Plants: Extraction and Application to Human Health. Stud. Nat. Prod. Chem. 2018, 58, 389–417. [Google Scholar]
- Kumar, N.; Goel, N. Phenolic acids: Natural versatile molecules with promising therapeutic applications. Biotechnol. Rep. 2019, 24, e00370. [Google Scholar] [CrossRef] [PubMed]
- Oracz, J.; Nebesny, E.; Zyzelewicz, D.; Budryn, G.; Luzak, B. Bioavailability and metabolism of selected cocoa bioactive compounds: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2020, 60, 1947–1985. [Google Scholar] [CrossRef] [PubMed]
- Majdan, M.; Bobrowska-Korczak, B. Active Compounds in Fruits and Inflammation in the Body. Nutrients 2022, 14, 2496. [Google Scholar] [CrossRef]
- Balasundram, N.; Sundram, K.; Samman, S. Phenolic compounds in plants and agri-industrial by-products: Antioxidant activity, occurrence, and potential uses. Food Chem. 2006, 99, 191–203. [Google Scholar] [CrossRef]
- Magnani, C.; Isaac, V.L.B.; Correa, M.A.; Salgado, H.R.N. Caffeic acid: A review of its potential use in medications and cosmetics. Anal. Methods 2014, 6, 3203–3210. [Google Scholar] [CrossRef]
- Sroka, Z.; Cisowski, W. Hydrogen peroxide scavenging, antioxidant and anti-radical activity of some phenolic acids. Food Chem. Toxicol. 2003, 41, 753–758. [Google Scholar] [CrossRef]
- Khan, F.A.; Maalik, A.; Murtaza, G. Inhibitory mechanism against oxidative stress of caffeic acid. J. Food Drug Anal. 2016, 24, 695–702. [Google Scholar] [CrossRef]
- Cizmarova, B.; Hubkova, B.; Bolerazska, B.; Marekova, M.; Birkova, A. Caffeic acid: A brief overview of its presence, metabolism, and bioactivity. Bioact. Compd. Health Dis. 2020, 3, 74–81. [Google Scholar]
- Srinivasulu, C.; Ramgopal, M.; Ramanjaneyulu, G.; Anuradha, C.M.; Suresh Kumar, C. Syringic acid (SA)—A Review of Its Occurrence, Biosynthesis, Pharmacological and Industrial Importance. Biomed. Pharmacother. 2018, 108, 547–557. [Google Scholar] [CrossRef]
- Ogut, E.; Armagan, K.; Gül, Z. The role of syringic acid as a neuroprotective agent for neurodegenerative disorders and future expectations. Metab. Brain Dis. 2022, 37, 859–880. [Google Scholar] [CrossRef]
- Shimsa, S.; Mondal, S.; Mini, S. Syringic acid: A promising phenolic phytochemical with extensive therapeutic applications. RD Funct. Food Prod. 2024, 1, 1–14. [Google Scholar]
- Szwajgier, D.; Borowiec, K.; Pustelniak, K. The Neuroprotective Effects of Phenolic Acids: Molecular Mechanism of Action. Nutrients 2017, 9, 477. [Google Scholar] [CrossRef]
- Güzelad, Ö.; Özkan, A.; Parlak, H.; Sinen, O.; Afşar, E.; Öğüt, E.; Yıldırım, F.B.; Bülbül, M.; Ağar, A.; Aslan, M. Protective mechanism of Syringic acid in an experimental model of Parkinson’s disease. Metab. Brain Dis. 2021, 36, 1003–1014. [Google Scholar] [CrossRef]
- Boz, H. p-Coumaric acid in cereals: Presence, antioxidant and antimicrobial effects. Int. J. Food Sci. Technol. 2015, 50, 2323–2328. [Google Scholar] [CrossRef]
- Pei, K.; Ou, J.; Huang, J.; Ou, S. p-Coumaric acid and its conjugates: Dietary sources, pharmacokinetic properties and biological activities. J. Sci. Food Agric. 2016, 96, 2952–2962. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Kaur, R. p-Coumaric Acid: A Naturally Occurring Chemical with Potential Therapeutic Applications. Curr. Org. Chem. 2022, 26, 1333–1349. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, X.; Wang, J.; Wang, F.; Mao, J. P-coumaric Acid: Advances in Pharmacological Research Based on Oxidative Stress. Curr. Top. Med. Chem. 2024, 24, 416–436. [Google Scholar] [CrossRef]
- Bouarab Chibane, L.; Degraeve, P.; Ferhout, H.; Bouajila, J.; Oulahal, N. Plant antimicrobial polyphenols as potential natural food preservatives. J. Sci. Food Agric. 2019, 99, 1457–1474. [Google Scholar] [CrossRef]
- Boudet, A.M. Evolution and current status of research in phenolic compounds. Phytochemistry 2007, 68, 2722–2735. [Google Scholar] [CrossRef]
- Kumar, K.; Debnath, P.; Singh, S.; Kumar, N. An Overview of Plant Phenolics and Their Involvement in Abiotic Stress Tolerance. Stresses 2023, 3, 570–585. [Google Scholar] [CrossRef]
- Hansen, B.B.; Spittle, S.; Chen, B.; Poe, D.; Zhang, Y.; Klein, J.M.; Horton, A.; Adhikari, L.; Zelovich, T.; Doherty, B.W.; et al. Deep Eutectic Solvents: A Review of Fundamentals and Applications. Chem. Rev. 2021, 121, 1232–1285. [Google Scholar] [CrossRef]
- Paiva, A.; Craveiro, R.; Aroso, I.; Martins, M.; Reis, R.L.; Duarte, A.R.C. Natural Deep Eutectic Solvents—Solvents for the 21st Century. ACS Sustain. Chem. Eng. 2014, 2, 1063–1071. [Google Scholar] [CrossRef]
- Cao, J.; Cao, J.; Wang, H.; Chen, L.; Cao, F.; Su, E. Solubility improvement of phytochemicals using (natural) deep eutectic solvents and their bioactivity evaluation. J. Mol. Liq. 2020, 318, 113997. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Chen, S.-N.; Friesen, J.B.; Nikolić, D.; Choules, M.P.; McAlpine, J.B.; Lankin, D.C.; Gemeinhart, R.A.; Pauli, G.F. The influence of natural deep eutectic solvents on bioactive natural products: Studying interactions between a hydrogel model and Schisandra chinensis metabolites. Fitoterapia 2018, 127, 212–219. [Google Scholar] [CrossRef]
- da Silva, D.T.; Smaniotto, F.A.; Costa, I.F.; Baranzelli, J.; Muller, A.; Somacal, S.; Monteiro, C.S.; Vizzotto, M.; Rodrigues, E.; Barcia, M.T.; et al. Natural deep eutectic solvent (NADES): A strategy to improve the bioavailability of blueberry phenolic compounds in a ready-to-use extract. Food Chem. 2021, 364, 130370. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Urios, C.; Viñas-Ospino, A.; Puchades-Colera, P.; Blesa, J.; López-Malo, D.; Frígola, A.; Esteve, M.J. Choline chloride-based natural deep eutectic solvents for the extraction and stability of phenolic compounds, ascorbic acid, and antioxidant capacity from Citrus sinensis peel. LWT 2023, 177, 114595. [Google Scholar] [CrossRef]
- Barbieri, J.B.; Goltz, C.; Batistão Cavalheiro, F.; Theodoro Toci, A.; Igarashi-Mafra, L.; Mafra, M.R. Deep eutectic solvents applied in the extraction and stabilization of rosemary (Rosmarinus officinalis L.) phenolic compounds. Ind. Crops Prod. 2020, 144, 112049, Erratum in Ind. Crops Prod. 2020, 154, 112722. [Google Scholar] [CrossRef]
- Usmani, Z.; Sharma, M.; Tripathi, M.; Lukk, T.; Karpichev, Y.; Gathergood, N.; Singh, B.N.; Thakur, V.K.; Tabatabaei, M.; Gupta, V.K. Biobased natural deep eutectic system as versatile solvents: Structure, interaction and advanced applications. Sci. Total Environ. 2023, 881, 163002. [Google Scholar] [CrossRef]
- Li, D. Natural deep eutectic solvents in phytonutrient extraction and other applications. Front. Plant Sci. 2022, 13, 1004332. [Google Scholar] [CrossRef]
- Ruesgas-Ramón, M.; Figueroa-Espinoza, M.C.; Durand, E. Application of Deep Eutectic Solvents (DES) for Phenolic Compounds Extraction: Overview, Challenges, and Opportunities. J. Agric. Food Chem. 2017, 65, 3591–3601. [Google Scholar] [CrossRef]
- Hikmawanti, N.P.E.; Ramadon, D.; Jantan, I.; Mun’im, A. Natural Deep Eutectic Solvents (NADES): Phytochemical Extraction Performance Enhancer for Pharmaceutical and Nutraceutical Product Development. Plants 2021, 10, 2091. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; van Spronsen, J.; Witkamp, G.-J.; Verpoorte, R.; Choi, Y.H. Natural deep eutectic solvents as new potential media for green technology. Anal. Chim. Acta 2013, 766, 61–68. [Google Scholar] [CrossRef]
- Prabhune, A.; Dey, R. Green and sustainable solvents of the future: Deep eutectic solvents. J. Mol. Liq. 2023, 379, 121676. [Google Scholar] [CrossRef]
- Lomba, L.; Ribate, M.P.; Zaragoza, E.; Concha, J.; Garralaga, M.P.; Errazquin, D.; García, C.B.; Giner, B. Deep Eutectic Solvents: Are They Safe? Appl. Sci. 2021, 11, 10061. [Google Scholar] [CrossRef]
- Rodrigues, L.A.; Cardeira, M.; Leonardo, I.C.; Gaspar, F.B.; Radojčić Redovniković, I.; Duarte, A.R.C.; Paiva, A.; Matias, A.A. Deep eutectic systems from betaine and polyols—Physicochemical and toxicological properties. J. Mol. Liq. 2021, 335, 116201. [Google Scholar] [CrossRef]
- Binnemans, K.; Jones, P.T. Ionic Liquids and Deep-Eutectic Solvents in Extractive Metallurgy: Mismatch Between Academic Research and Industrial Applicability. J. Sustain. Metall. 2023, 9, 423–438. [Google Scholar] [CrossRef]
- Usman, M.; Cheng, S.; Boonyubol, S.; Cross, J.S. Evaluating Green Solvents for Bio-Oil Extraction: Advancements, Challenges, and Future Perspectives. Energies 2023, 16, 5852. [Google Scholar] [CrossRef]
- Svärd, M.; Ma, C.; Forsberg, K.; Schiavi, P.G. Addressing the Reuse of Deep Eutectic Solvents in Li-Ion Battery Recycling: Insights into Dissolution Mechanism, Metal Recovery, Regeneration and Decomposition. ChemSusChem 2024, 17, e202400410. [Google Scholar] [CrossRef] [PubMed]
- Elizondo Sada, O.M.; Hiemstra, I.S.A.; Chorhirankul, N.; Eppink, M.; Wijffels, R.H.; Janssen, A.E.M.; Kazbar, A. Pressure-driven membrane processes for the recovery and recycling of deep eutectic solvents: A seaweed biorefinery case study. Biotechnol. Rep. 2024, 43, e00849. [Google Scholar] [CrossRef]
- Abdollahzadeh, M.; Khosravi, M.; Hajipour Khire Masjidi, B.; Samimi Behbahan, A.; Bagherzadeh, A.; Shahkar, A.; Tat Shahdost, F. Estimating the density of deep eutectic solvents applying supervised machine learning techniques. Sci. Rep. 2022, 12, 4954. [Google Scholar] [CrossRef]
- Halder, A.K.; Haghbakhsh, R.; Ferreira, E.S.C.; Duarte, A.R.C.; Cordeiro, M.N.D.S. Machine learning-driven prediction of deep eutectic solvents’ heat capacity for sustainable process design. J. Mol. Liq. 2025, 418, 126707. [Google Scholar] [CrossRef]
- Soleimani, R.; Saeedi Dehaghani, A.H. Unveiling CO2 capture in tailorable green neoteric solvents: An ensemble learning approach informed by quantum chemistry. J. Environ. Manag. 2024, 354, 120298. [Google Scholar] [CrossRef]
- Roosta, A.; Haghbakhsh, R.; Rita, C.; Duarte, A.; Raeissi, S. Deep eutectic solvent viscosity prediction by hybrid machine learning and group contribution. J. Mol. Liq. 2023, 388, 122747. [Google Scholar] [CrossRef]
- Darwish, A.S.; Abu Alwan, R.; Boublia, A.; Lemaoui, T.; Benguerba, Y.; AlNashef, I.M.; Banat, F. Machine learning approach for mapping the heat capacity of deep eutectic solvents for sustainable energy applications. Fuel 2025, 381, 133278. [Google Scholar] [CrossRef]
- Mohan, M.; Jetti, K.D.; Smith, M.D.; Demerdash, O.N.; Kidder, M.K.; Smith, J.C. Accurate Machine Learning for Predicting the Viscosities of Deep Eutectic Solvents. J. Chem. Theory Comput. 2024, 20, 3911–3926. [Google Scholar] [CrossRef]
- Jeliński, T.; Przybyłek, M.; Różalski, R.; Romanek, K.; Wielewski, D.; Cysewski, P. Tuning Ferulic Acid Solubility in Choline-Chloride- and Betaine-Based Deep Eutectic Solvents: Experimental Determination and Machine Learning Modeling. Molecules 2024, 29, 3841. [Google Scholar] [CrossRef]
- Cysewski, P.; Jeliński, T.; Przybyłek, M.; Mai, A.; Kułak, J. Experimental and Machine-Learning-Assisted Design of Pharmaceutically Acceptable Deep Eutectic Solvents for the Solubility Improvement of Non-Selective COX Inhibitors Ibuprofen and Ketoprofen. Molecules 2024, 29, 2296. [Google Scholar] [CrossRef]
- Mohan, M.; Demerdash, O.N.; Simmons, B.A.; Singh, S.; Kidder, M.K.; Smith, J.C. Physics-Based Machine Learning Models Predict Carbon Dioxide Solubility in Chemically Reactive Deep Eutectic Solvents. ACS Omega 2024, 9, 19548–19559. [Google Scholar] [CrossRef]
- Cysewski, P.; Jeliński, T.; Przybyłek, M. Exploration of the Solubility Hyperspace of Selected Active Pharmaceutical Ingredients in Choline- and Betaine-Based Deep Eutectic Solvents: Machine Learning Modeling and Experimental Validation. Molecules 2024, 29, 4894. [Google Scholar] [CrossRef]
- Cysewski, P.; Jeliński, T.; Przybyłek, M.; Nowak, W.; Olczak, M. Solubility Characteristics of Acetaminophen and Phenacetin in Binary Mixtures of Aqueous Organic Solvents: Experimental and Deep Machine Learning Screening of Green Dissolution Media. Pharmaceutics 2022, 14, 2828. [Google Scholar] [CrossRef]
- Wang, J.; Song, Z.; Chen, L.; Xu, T.; Deng, L.; Qi, Z. Prediction of CO2 solubility in deep eutectic solvents using random forest model based on COSMO-RS-derived descriptors. Green Chem. Eng. 2021, 2, 431–440. [Google Scholar] [CrossRef]
- Lemaoui, T.; Boublia, A.; Lemaoui, S.; Darwish, A.S.; Ernst, B.; Alam, M.; Benguerba, Y.; Banat, F.; AlNashef, I.M. Predicting the CO2 Capture Capability of Deep Eutectic Solvents and Screening over 1000 of their Combinations Using Machine Learning. ACS Sustain. Chem. Eng. 2023, 11, 9564–9580. [Google Scholar] [CrossRef]
- Cysewski, P.; Jeliński, T.; Przybyłek, M. Experimental and Theoretical Insights into the Intermolecular Interactions in Saturated Systems of Dapsone in Conventional and Deep Eutectic Solvents. Molecules 2024, 29, 1743. [Google Scholar] [CrossRef]
- Jeliński, T.; Przybyłek, M.; Mianowana, M.; Misiak, K.; Cysewski, P. Deep Eutectic Solvents as Agents for Improving the Solubility of Edaravone: Experimental and Theoretical Considerations. Molecules 2024, 29, 1261. [Google Scholar] [CrossRef] [PubMed]
- Triolo, A.; Lo Celso, F.; Brehm, M.; Di Lisio, V.; Russina, O. Liquid structure of a choline chloride-water natural deep eutectic solvent: A molecular dynamics characterization. J. Mol. Liq. 2021, 331, 115750. [Google Scholar] [CrossRef]
- Sarkar, S.; Maity, A.; Chakrabarti, R. In Silico Elucidation of Molecular Picture of Water-Choline Chloride Mixture. J. Phys. Chem. B 2021, 125, 13212–13228. [Google Scholar] [CrossRef] [PubMed]
- Noubigh, A.; Akermi, A. Solubility and Thermodynamic Behavior of Syringic Acid in Eight Pure and Water + Methanol Mixed Solvents. J. Chem. Eng. Data 2017, 62, 3274–3283. [Google Scholar] [CrossRef]
- Vilas-Boas, S.M.; Vieira, V.; Brandão, P.; Alves, R.S.; Coutinho, J.A.P.; Pinho, S.P.; Ferreira, O. Solvent and temperature effects on the solubility of syringic, vanillic or veratric acids: Experimental, modeling and solid phase studies. J. Mol. Liq. 2019, 289, 111089. [Google Scholar] [CrossRef]
- Noubigh, A.; Cherif, M.; Provost, E.; Abderrabba, M. Solubility of Gallic Acid, Vanillin, Syringic Acid, and Protocatechuic Acid in Aqueous Sulfate Solutions from (293.15 to 318.15) K. J. Chem. Eng. Data 2008, 53, 1675–1678. [Google Scholar] [CrossRef]
- Mota, F.L.; Queimada, A.J.; Pinho, S.P.; Macedo, E.A. Aqueous solubility of some natural phenolic compounds. Ind. Eng. Chem. Res. 2008, 47, 5182–5189. [Google Scholar] [CrossRef]
- Alevizou, E.I.; Voutsas, E.C. Solubilities of p-coumaric and caffeic acid in ionic liquids and organic solvents. J. Chem. Thermodyn. 2013, 62, 69–78. [Google Scholar] [CrossRef]
- Vilas-Boas, S.M.; Alves, R.S.; Brandão, P.; Campos, L.M.A.; Coutinho, J.A.P.; Pinho, S.P.; Ferreira, O. Solid-liquid phase equilibrium of trans-cinnamic acid, p-coumaric acid and ferulic acid in water and organic solvents: Experimental and modelling studies. Fluid Phase Equilibria 2020, 521, 112747. [Google Scholar] [CrossRef]
- Ji, W.; Meng, Q.; Li, P.; Yang, B.; Wang, F.; Ding, L.; Wang, B. Measurement and Correlation of the Solubility of p-Coumaric Acid in Nine Pure and Water + Ethanol Mixed Solvents at Temperatures from 293.15 to 333.15 K. J. Chem. Eng. Data 2016, 61, 3457–3465. [Google Scholar] [CrossRef]
- Prinos, N.; Dardavila, M.M.; Voutsas, E. Measurement and thermodynamic modelling of the solubilities of caffeic acid, p-coumaric acid and ferulic acid in three choline chloride-based deep eutectic solvents. J. Chem. Thermodyn. 2024, 197, 107335. [Google Scholar] [CrossRef]
- Ji, W.; Meng, Q.; Ding, L.; Wang, F.; Dong, J.; Zhou, G.; Wang, B. Measurement and correlation of the solubility of caffeic acid in eight mono and water+ethanol mixed solvents at temperatures from (293.15 to 333.15) K. J. Mol. Liq. 2016, 224, 1275–1281. [Google Scholar] [CrossRef]
- Manic, M.S.; Villanueva, D.; Fornari, T.; Queimada, A.J.; MacEdo, E.A.; Najdanovic-Visak, V. Solubility of high-value compounds in ethyl lactate: Measurements and modeling. J. Chem. Thermodyn. 2012, 48, 93–100. [Google Scholar] [CrossRef]
- Prinos, N.; Boli, E.; Louli, V.; Pappa, G.; Magoulas, K.; Voutsas, E. Solubilities of caffeic acid and tyrosol in two protic ionic liquids and one deep eutectic solvent. Fluid Phase Equilibria 2022, 559, 113462. [Google Scholar] [CrossRef]
- Tsarpali, V.; Dailianis, S. Toxicity of two imidazolium ionic liquids, [bmim][BF4] and [omim][BF4], to standard aquatic test organisms: Role of acetone in the induced toxicity. Ecotoxicol. Environ. Saf. 2015, 117, 62–71. [Google Scholar] [CrossRef]
- Bitencourt, R.G.; Cabral, F.A.; Meirelles, A.J.A. Ferulic acid solubility in supercritical carbon dioxide, ethanol and water mixtures. J. Chem. Thermodyn. 2016, 103, 285–291. [Google Scholar] [CrossRef]
- Shakeel, F.; Salem-Bekhit, M.M.; Haq, N.; Siddiqui, N.A. Solubility and thermodynamics of ferulic acid in different neat solvents: Measurement, correlation and molecular interactions. J. Mol. Liq. 2017, 236, 144–150. [Google Scholar] [CrossRef]
- Haq, N.; Siddiqui, N.A.; Shakeel, F. Solubility and molecular interactions of ferulic acid in various (isopropanol + water) mixtures. J. Pharm. Pharmacol. 2017, 69, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- António, J.Q.; Mota, F.L.; Pinho, S.P.; Macedo, E.A. Solubilities of biologically active phenolic compounds: Measurements and modeling. J. Phys. Chem. B 2009, 113, 3469–3476. [Google Scholar] [PubMed]
- COSMOtherm, Version 24.0.0; Dassault Systèmes; Biovia: San Diego, CA, USA, 2022.
- Klamt, A.; Eckert, F.; Arlt, W. COSMO-RS: An alternative to simulation for calculating thermodynamic properties of liquid mixtures. Annu. Rev. Chem. Biomol. Eng. 2010, 1, 101–122. [Google Scholar] [CrossRef]
- Klamt, A. COSMO-RS: From Quantum Chemistry to Fluid Phase Thermodynamics and Drug Design, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Klamt, A. Conductor-like Screening Model for Real Solvents: A New Approach to the Quantitative Calculation of Solvation Phenomena. J. Phys. Chem. 1995, 99, 2224–2235. [Google Scholar] [CrossRef]
- Klamt, A.; Eckert, F.; Hornig, M.; Beck, M.E.; Bürger, T. Prediction of aqueous solubility of drugs and pesticides with COSMO-RS. J. Comput. Chem. 2002, 23, 275–281. [Google Scholar] [CrossRef]
- Cordova, I.W.; Teixeira, G.; Ribeiro-Claro, P.J.A.; Abranches, D.O.; Pinho, S.P.; Ferreira, O.; Coutinho, J.A.P. Using Molecular Conformers in COSMO-RS to Predict Drug Solubility in Mixed Solvents. Ind. Eng. Chem. Res. 2024, 63, 9565–9575. [Google Scholar] [CrossRef]
- Vilas-Boas, S.M.; Abranches, D.O.; Crespo, E.A.; Ferreira, O.; Coutinho, J.A.P.; Pinho, S.P. Experimental solubility and density studies on aqueous solutions of quaternary ammonium halides, and thermodynamic modelling for melting enthalpy estimations. J. Mol. Liq. 2020, 300, 112281. [Google Scholar] [CrossRef]
- Freire, M.G.; Carvalho, P.J.; Santos, L.M.N.B.F.; Gomes, L.R.; Marrucho, I.M.; Coutinho, J.A.P. Solubility of water in fluorocarbons: Experimental and COSMO-RS prediction results. J. Chem. Thermodyn. 2010, 42, 213–219. [Google Scholar] [CrossRef]
- Miller, M.B.; Chen, D.-L.; Luebke, D.R.; Johnson, J.K.; Enick, R.M. Critical Assessment of CO2 Solubility in Volatile Solvents at 298.15 K. J. Chem. Eng. Data 2011, 56, 1565–1572. [Google Scholar] [CrossRef]
- Acree, W.; Chickos, J.S. Phase Transition Enthalpy Measurements of Organic and Organometallic Compounds. Sublimation, Vaporization and Fusion Enthalpies From 1880 to 2010. J. Phys. Chem. Ref. Data 2010, 39, 043101. [Google Scholar] [CrossRef]
- Vapnik, V.N. The Nature of Statistical Learning Theory, 2nd ed.; Springer Science & Business Media: New York, NY, USA, 2000. [Google Scholar]
- Schölkopf, B.; Smola, A.J.; Williamson, R.C.; Bartlett, P.L. New support vector algorithms. Neural Comput. 2000, 12, 1207–1245. [Google Scholar] [CrossRef]
- Gretton, A.; Borgwardt, K.M.; Rasch, M.J.; Schölkopf, B.; Smola, A. A kernel two-sample test. J. Mach. Learn. Res. 2012, 13, 723–773. [Google Scholar]
- Garreau, D.; Jitkrittum, W.; Kanagawa, M. Large sample analysis of the median heuristic. arXiv 2017, arXiv:1707.07269. [Google Scholar]
- Muandet, K.; Fukumizu, K.; Sriperumbudur, B.; Schölkopf, B. Kernel mean embedding of distributions: A review and beyond. Found. Trends Mach. Learn. 2017, 10, 1–141. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Akiba, T.; Sano, S.; Yanase, T.; Ohta, T.; Koyama, M. Optuna: A Next-generation Hyperparameter Optimization Framework. In Proceedings of the KDD ’19: The 25th ACM SIGKDD Conference on Knowledge Discovery and Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 2623–2631. [Google Scholar]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees, 1st ed.; Routledge: Boca Raton, FL, USA, 2017. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Akaike, H. A New Look at the Statistical Model Identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. Multimodel inference: Understanding AIC and BIC in model selection. Sociol. Methods Res. 2004, 33, 261–304. [Google Scholar] [CrossRef]
- Python, Version 3.13; Python Software Foundation: Wilmington, DE, USA, 2024.
- pandas Development Team. pandas, Version 2.3.0; Zenodo: Geneva, Switzerland, 2025.
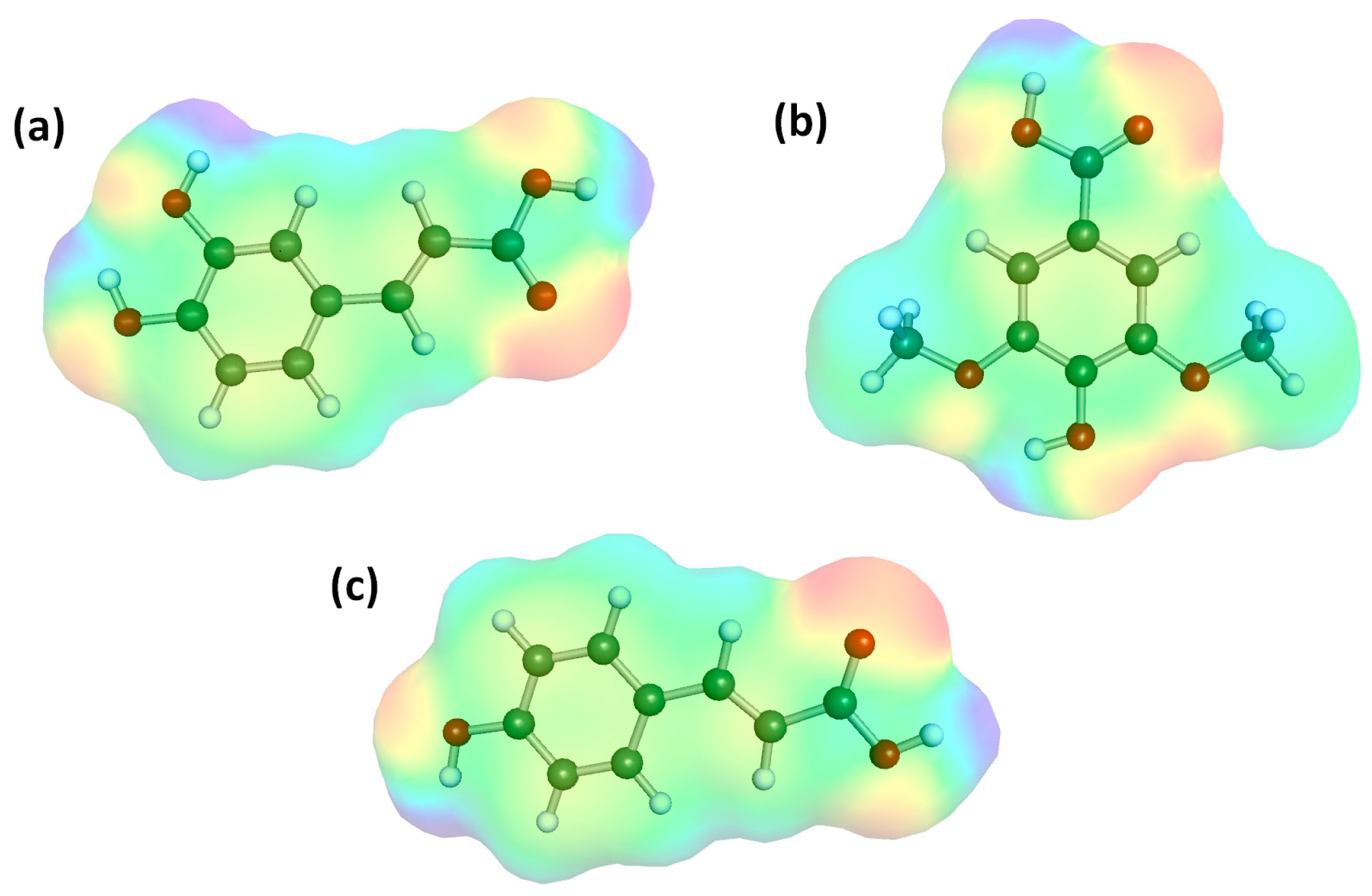

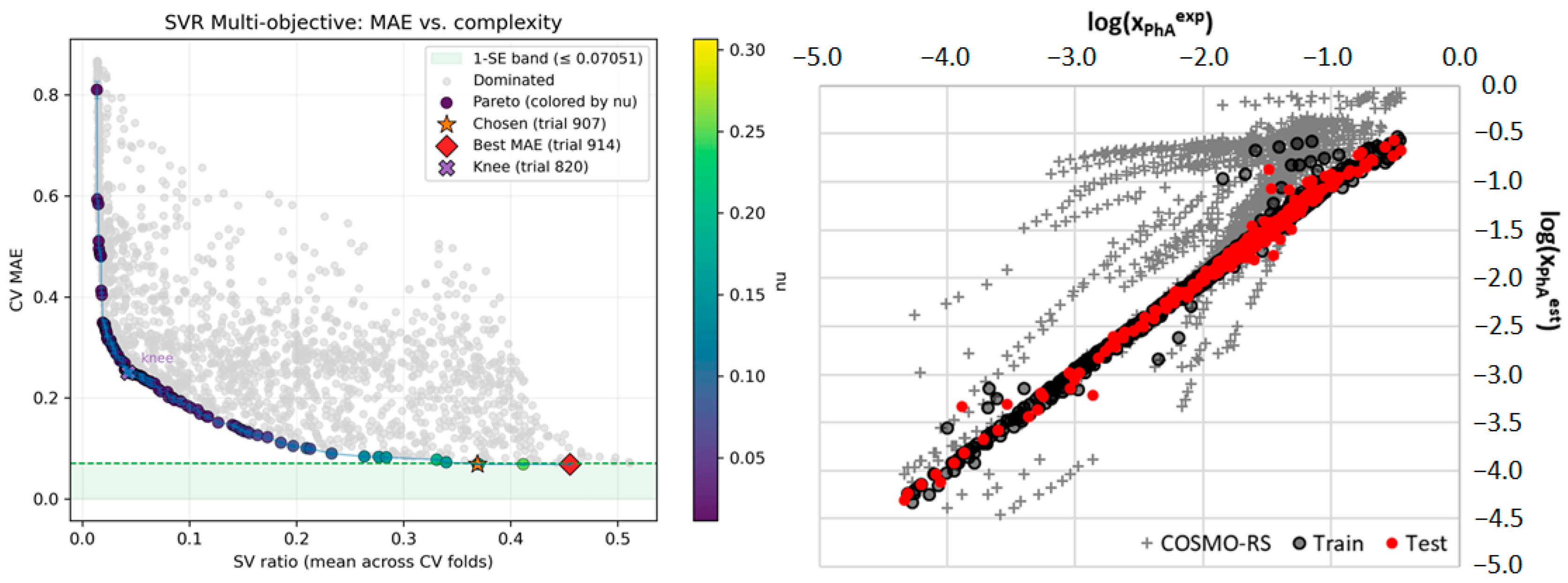
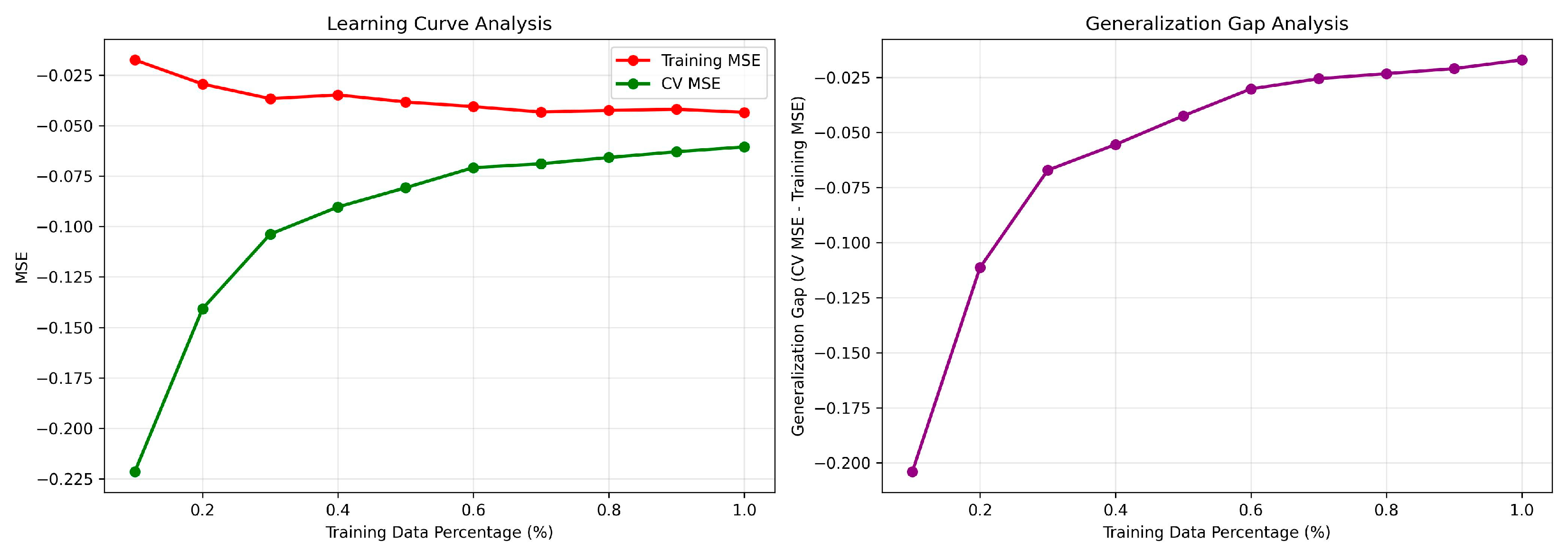
| Metric | 5-Fold Cross-Validation (on Training Set) | Held-Out Test Set (Final Evaluation) |
|---|---|---|
| R2 | 0.976 ± 0.004 | 0.984 |
| MAE | 0.072 ± 0.004 | 0.061 |
| RMSE | 0.116 ± 0.007 | 0.125 |
| Phenolic Acid | Regression Equation (C in [mg/mL]) | R2 | LOD [mg/mL] | LOQ [mg/mL] |
|---|---|---|---|---|
| caffeic | A = 62.51 × C − 0.0126 | 0.9993 | 0.00072 | 0.00215 |
| syringic | A = 111.64 × C − 0.0094 | 0.9987 | 0.00046 | 0.00138 |
| p-coumaric | A = 54.27 × C − 0.0106 | 0.9983 | 0.00139 | 0.00417 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cysewski, P.; Jeliński, T.; Przybyłek, M.; Gliniewicz, N.; Majkowski, M.; Wąs, M. Navigating the Deep Eutectic Solvent Landscape: Experimental and Machine Learning Solubility Explorations of Syringic, p-Coumaric, and Caffeic Acids. Int. J. Mol. Sci. 2025, 26, 10099. https://doi.org/10.3390/ijms262010099
Cysewski P, Jeliński T, Przybyłek M, Gliniewicz N, Majkowski M, Wąs M. Navigating the Deep Eutectic Solvent Landscape: Experimental and Machine Learning Solubility Explorations of Syringic, p-Coumaric, and Caffeic Acids. International Journal of Molecular Sciences. 2025; 26(20):10099. https://doi.org/10.3390/ijms262010099
Chicago/Turabian StyleCysewski, Piotr, Tomasz Jeliński, Maciej Przybyłek, Natalia Gliniewicz, Marcel Majkowski, and Michał Wąs. 2025. "Navigating the Deep Eutectic Solvent Landscape: Experimental and Machine Learning Solubility Explorations of Syringic, p-Coumaric, and Caffeic Acids" International Journal of Molecular Sciences 26, no. 20: 10099. https://doi.org/10.3390/ijms262010099
APA StyleCysewski, P., Jeliński, T., Przybyłek, M., Gliniewicz, N., Majkowski, M., & Wąs, M. (2025). Navigating the Deep Eutectic Solvent Landscape: Experimental and Machine Learning Solubility Explorations of Syringic, p-Coumaric, and Caffeic Acids. International Journal of Molecular Sciences, 26(20), 10099. https://doi.org/10.3390/ijms262010099









