Genome-Wide Identification of Ginkgo biloba SPL Gene Family and Expression Analysis in Flavonoid Biosynthesis and Water Stress
Abstract
1. Introduction
2. Results
2.1. Whole-Genome Identification of the GbSPL Genes in Ginkgo biloba
2.2. Phylogenetic Analysis of GbSPL Genes
2.3. Motif, Conserved Domain, and Gene Structure Analysis of GbSPLs
2.4. Multiple Sequence Alignment Analysis and MicroRNA Target Prediction
2.5. The Expansion and Collinearity Analysis of the GbSPL Gene Family
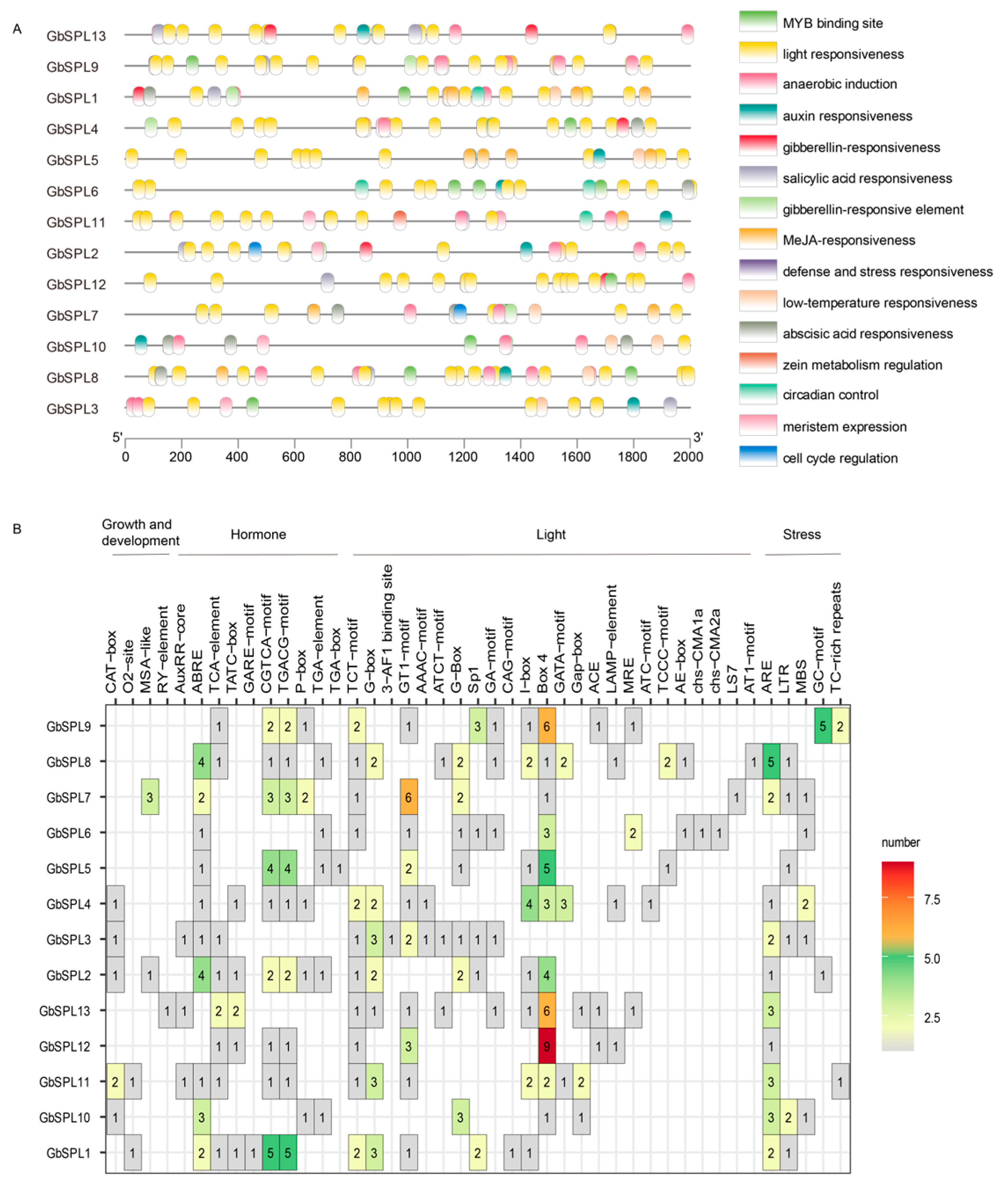
2.6. Cis-Acting Element Analysis in the Promoter of GbSPL Genes
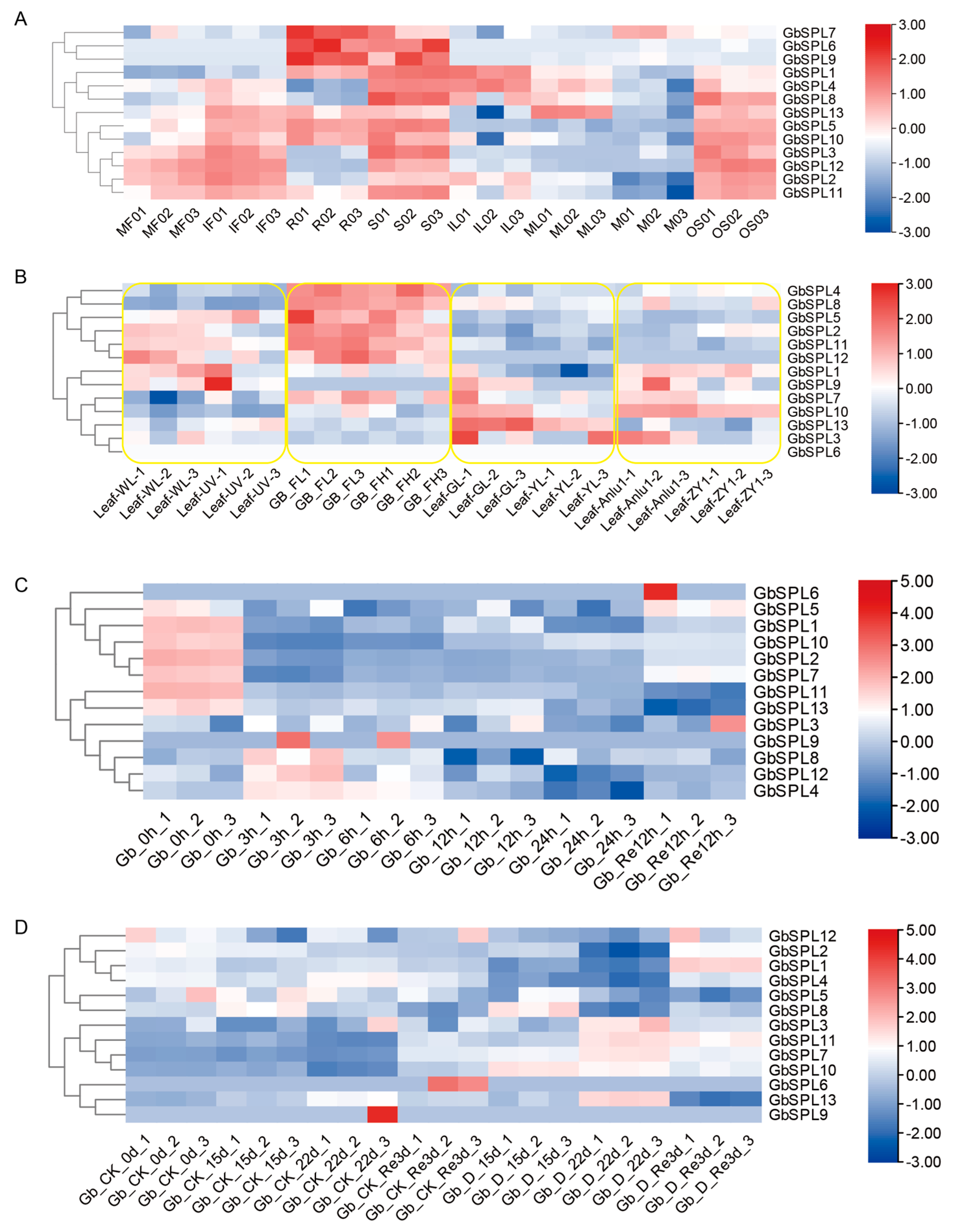
2.7. Expression Profiles of GbSPL Genes in Various Tissues, Low- and High-Flavonoid Leaves, and Water Stress in Ginkgo biloba
2.8. Quantitative Real-Time PCR (qRT-PCR) Analysis and Subcellular Localization Assay of the GbSPL Genes
3. Discussion
3.1. Characteristics and Evolution of SPL Genes in Ginkgo biloba
3.2. Expression Patterns and Potential Functions of the GbSPL Genes
4. Materials and Methods
4.1. Whole-Genome Identification of SPL Genes in Ginkgo biloba
4.2. Physicochemical Properties, MicroRNA Target Prediction, and Chromosomal Location Analysis of SPL Genes
4.3. Phylogenetic Analysis
4.4. Motif, Conserved Domain, Gene Structure, and Cis-Elements Analysis
4.5. Gene Duplication and Syntenic Analysis of the GbSPL Gene Family
4.6. RNA-Seq and Quantitative Real-Time PCR (qRT-PCR)-Based Expression Analysis of GbSPL Genes
4.7. Subcellular Localization Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plant 2021, 172, 847–868. [Google Scholar] [CrossRef] [PubMed]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. MGG 1996, 250, 7–16. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef]
- Salinas, M.; Xing, S.; Hoehmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hou, H.; Li, X.; Xiang, J.; Yin, X.; Gao, H.; Zheng, Y.; Bassett, C.L.; Wang, X. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus domestica Borkh.). Plant Physiol. Biochem. 2013, 70, 100–114. [Google Scholar] [CrossRef]
- Liu, M.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification of the SPL gene family in Tartary Buckwheat (Fagopyrum tataricum) and expression analysis during fruit development stages. BMC Plant Biol. 2019, 19, 299. [Google Scholar] [CrossRef]
- Hu, X.; Liu, C.; Tian, J.; Zhang, Y.; Xin, Q.; Chen, A.; Li, D.; Liu, X. Identification, molecular characterization, and expression analysis of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family in Betula platyphylla Suk. Trees 2019, 34, 229–241. [Google Scholar] [CrossRef]
- Qian, M.; Ni, J.; Niu, Q.; Bai, S.; Bao, L.; Li, J.; Sun, Y.; Zhang, D.; Teng, Y. Response of miR156-SPLModule during the Red Peel Coloration of Bagging-Treated Chinese Sand Pear (Pyrus pyrifolia Nakai). Front. Physiol. 2017, 8, 550. [Google Scholar] [CrossRef]
- Wang, H.; Lu, Z.; Xu, Y.; Kong, L.; Shi, J.; Liu, Y.; Fu, C.; Wang, X.; Wang, Z.Y.; Zhou, C.; et al. Genome-wide characterization of SPL family in Medicago truncatula reveals the novel roles of miR156/SPL module in spiky pod development. BMC Genom. 2019, 20, 552. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Wang, Y.; Zhang, N.; Zhang, X.; Wu, J.; Huang, Y.; Ruan, M.; Zhang, J.; Qi, Y. Systematic Identification, Evolution and Expression Analysis of the SPL Gene Family in Sugarcane (Saccharum spontaneum). Trop. Plant Biol. 2021, 14, 313–328. [Google Scholar] [CrossRef]
- Ren, Y.; Ma, R.; Fan, Y.; Zhao, B.; Cheng, P.; Fan, Y.; Wang, B. Genome-wide identification and expression analysis of the SPL transcription factor family and its response to abiotic stress in Quinoa (Chenopodium quinoa). BMC Genom. 2022, 23, 773. [Google Scholar] [CrossRef]
- De Paola, C.; Garcia-Carpintero, V.; Vazquez-Vilar, M.; Kaminski, K.; Fernandez-Del-Carmen, A.; Sierro, N.; Ivanov, N.V.; Giuliano, G.; Waterhouse, P.; Orzaez, D. Comparative analysis of the Squamosa Promoter Binding-Like (SPL) gene family in Nicotiana benthamiana and Nicotiana tabacum. Plant Sci. 2023, 335, 111797. [Google Scholar] [CrossRef] [PubMed]
- Jadhao, K.R.; Kale, S.S.; Chavan, N.S.; Janjal, P.H. Genome-wide analysis of the SPL transcription factor family and its response to water stress in sunflower (Helianthus annuus). Cell Stress. Chaperones 2023, 28, 943–958. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, Y.; Xiao, Y.; Zhang, X.; Du, B.; Turupu, M.; Wang, C.; Yao, Q.; Gai, S.; Huang, J.; et al. Genome-Wide Identification of the SQUAMOSA Promoter-Binding Protein-like (SPL) Transcription Factor Family in Sweet Cherry Fruit. Int. J. Mol. Sci. 2023, 24, 2880. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wu, J.; Jing, X.; Khan, F.S.; Chen, Y.; Chen, Z.; Zhang, H.; Wei, Y. Genome-Wide Identification of Litchi SPL Gene Family and Expression Analysis in Pericarp Anthocyanin Biosynthesis. Horticulturae 2024, 10, 762. [Google Scholar] [CrossRef]
- Jerome Jeyakumar, J.M.; Ali, A.; Wang, W.M.; Thiruvengadam, M. Characterizing the Role of the miR156-SPL Network in Plant Development and Stress Response. Plants 2020, 9, 1206. [Google Scholar] [CrossRef]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H. The miR156/SPL Module, a Regulatory Hub and Versatile Toolbox, Gears up Crops for Enhanced Agronomic Traits. Mol. Plant 2015, 8, 677–688. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Wang, X.; Yang, R.; Wu, Z.; Wang, H.; Wang, L.; Hu, Z.; Guo, S.; Zhang, H.; et al. MiR156 regulates anthocyanin biosynthesis through SPL targets and other microRNAs in poplar. Hortic. Res. 2020, 7, 118. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.-Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.-G.; Shan, J.-X.; Shi, M.; Gao, J.-P.; Lin, H.-X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Salinas, M.; Hohmann, S.; Berndtgen, R.; Huijser, P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, Q.; Zhai, X.; Tu, Z.; Wang, J.; Wang, M.; Li, H. Genome-wide investigation of SQUAMOSA promoter binding protein-like genes in Liriodendron and functional characterization of LcSPL2. AoB Plants 2024, 16, plae008. [Google Scholar] [CrossRef]
- Li, Y.; Han, S.; Sun, X.; Khan, N.U.; Zhong, Q.; Zhang, Z.; Zhang, H.; Ming, F.; Li, Z.; Li, J. Variations in OsSPL10 confer drought tolerance by directly regulating OsNAC2 expression and ROS production in rice. J. Integr. Plant Biol. 2023, 65, 918–933. [Google Scholar] [CrossRef]
- Li, S.; Cheng, Z.; Li, Z.; Dong, S.; Yu, X.; Zhao, P.; Liao, W.; Yu, X.; Peng, M. MeSPL9 attenuates drought resistance by regulating JA signaling and protectant metabolite contents in cassava. Theor. Appl. Genet. 2022, 135, 817–832. [Google Scholar] [CrossRef]
- Zhao, Y.; He, J.; Liu, M.; Miao, J.; Ma, C.; Feng, Y.; Qian, J.; Li, H.; Bi, H.; Liu, W. The SPL transcription factor TaSPL6 negatively regulates drought stress response in wheat. Plant Physiol. Biochem. 2024, 206, 108264. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Wang, L.; Cui, J.; Jin, B.; Zhao, J.; Xu, H.; Lu, Z.; Li, W.; Li, X.; Li, L.; Liang, E.; et al. Multifeature analyses of vascular cambial cells reveal longevity mechanisms in old Ginkgo biloba trees. Proc. Natl. Acad. Sci. USA 2020, 117, 2201–2210. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wang, L.; Pang, S.; Jia, Z.; Wang, L.; Li, W.; Jin, B. UV-B promotes flavonoid synthesis in Ginkgo biloba leaves. Ind. Crops Prod. 2020, 151, 112483. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Wang, G.; Cui, P.; Wu, S.; Ai, C.; Hu, N.; Li, A.; He, B.; Shao, X.; et al. The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution. Nat. Plants 2021, 7, 748–756. [Google Scholar] [CrossRef]
- Ye, J.; Cheng, S.; Zhou, X.; Chen, Z.; Kim, S.U.; Tan, J.; Zheng, J.; Xu, F.; Zhang, W.; Liao, Y.; et al. A global survey of full-length transcriptome of Ginkgo biloba reveals transcript variants involved in flavonoid biosynthesis. Ind. Crops Prod. 2019, 139, 111547. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, J.; Zhou, Q.; Xin, Y.; Wang, G.; Xu, L.-A. De novo transcriptome analysis revealed genes involved in flavonoid biosynthesis, transport and regulation in Ginkgo biloba. Ind. Crops Prod. 2018, 124, 226–235. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, J.; Wang, T.; Cao, F.; Wang, G. Metabolomic and transcriptomic analyses of mutant yellow leaves provide insights into pigment synthesis and metabolism in Ginkgo biloba. BMC Genom. 2020, 21, 858. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Wang, B.; He, G.; Wang, Y.; Tang, X.; Wang, S.; Ma, Y.; Fu, C.; Chai, G.; Zhou, G. Metabolomics Integrated with Transcriptomics Reveals Redirection of the Phenylpropanoids Metabolic Flux in Ginkgo biloba. J. Agric. Food Chem. 2019, 67, 3284–3291. [Google Scholar] [CrossRef]
- Ming, M.; Zhang, J.; Tang, J.; Zhang, J.; Fu, F.; Cao, F. Transcriptome Profiling Revealed ABA Signaling Pathway-Related Genes and Major Transcription Factors Involved in the Response to Water Shock and Rehydration in Ginkgo biloba. Forests 2024, 15, 1690. [Google Scholar] [CrossRef]
- Ming, M.; Zhang, J.; Zhang, J.; Tang, J.; Fu, F.; Cao, F. Transcriptome Profiling Identifies Plant Hormone Signaling Pathway-Related Genes and Transcription Factors in the Drought and Re-Watering Response of Ginkgo biloba. Plants 2024, 13, 2685. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Z.; Liu, D.; Zhang, K.; Li, A.; Mao, L. SQUAMOSA Promoter-Binding Protein-Like Transcription Factors: Star Players for Plant Growth and Development. J. Integr. Plant Biol. 2010, 52, 946–951. [Google Scholar] [CrossRef]
- Qin, S.W.; Bao, L.H.; He, Z.G.; Li, C.L.; La, H.G.; Zhao, L.F. Identification and regulatory network analysis of SPL family transcription factors in Populus euphratica Oliv. heteromorphic leaves. Sci. Rep. 2022, 12, 2856. [Google Scholar] [CrossRef]
- Zhang, S.-D.; Ling, L.-Z.; Yi, T.-S. Evolution and divergence of SBP-box genes in land plants. Bmc Genom. 2015, 16, 787. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Y.; Lin, E.-P.; Huang, H.-H.; Niu, M.-Y.; Tong, Z.-K.; Zhang, J.-H. Molecular Characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Gene Family in Betula luminifera. Front. Plant Sci. 2018, 9, 608. [Google Scholar] [CrossRef]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef]
- Liu, S.; Gu, X.; Jiang, Y.; Wang, L.; Xiao, N.; Chen, Y.; Jin, B.; Wang, L.; Li, W. UV-B promotes flavonoid biosynthesis in Ginkgo biloba by inducing the GbHY5-GbMYB1-GbFLS module. Hortic. Res. 2023, 10, uhad118. [Google Scholar] [CrossRef]
- Sun, Y.J.; Fu, M.; Wang, L.; Bai, Y.X.; Fang, X.L.; Wang, Q.; He, Y.; Zeng, H.L. OsSPLs Regulate Male Fertility in Response to Different Temperatures by Flavonoid Biosynthesis and Tapetum PCD in PTGMS Rice. Int. J. Mol. Sci. 2022, 23, 3744. [Google Scholar] [CrossRef]
- Xue, G.; Wu, W.; Fan, Y.; Ma, C.; Xiong, R.; Bai, Q.; Yao, X.; Weng, W.; Cheng, J.; Ruan, J. Genome-wide identification, evolution, and role of SPL gene family in beet (Beta vulgaris L.) under cold stress. BMC Genom. 2024, 25, 101. [Google Scholar] [CrossRef]
- Lai, D.; Fan, Y.; Xue, G.; He, A.; Yang, H.; He, C.; Li, Y.; Ruan, J.; Yan, J.; Cheng, J. Genome-wide identification and characterization of the SPL gene family and its expression in the various developmental stages and stress conditions in foxtail millet (Setaria italica). Bmc Genom. 2022, 23, 389. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Bai, S.; Ma, J.; Zhang, L.; Shao, F.; Zhang, K.; Yang, Y.; Sun, T.; Huang, J.; Zhou, Y.; et al. The genome of Populus alba x Populus tremula var. glandulosa clone 84K. DNA Res. 2019, 26, 423–431. [Google Scholar] [CrossRef] [PubMed]
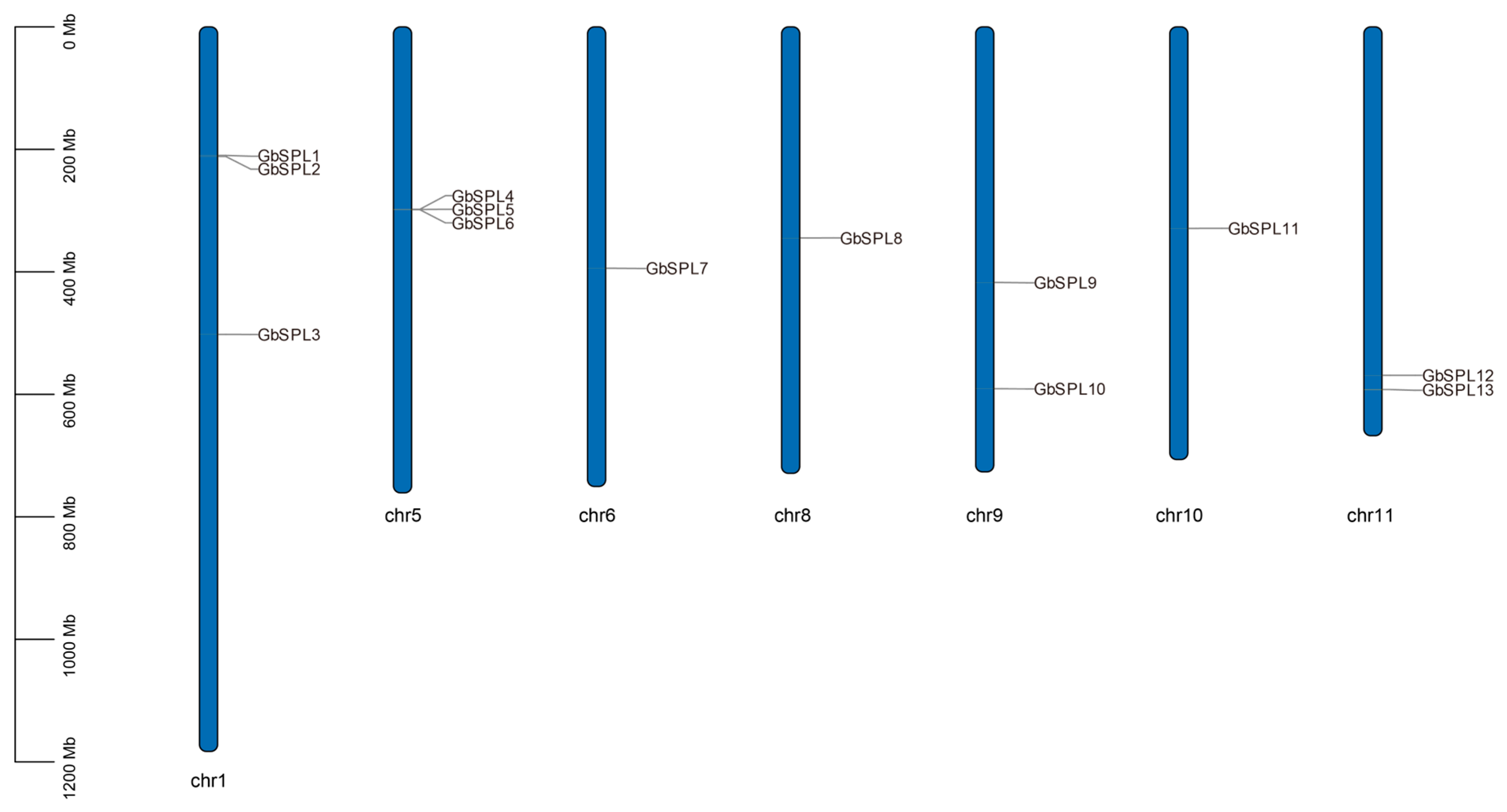
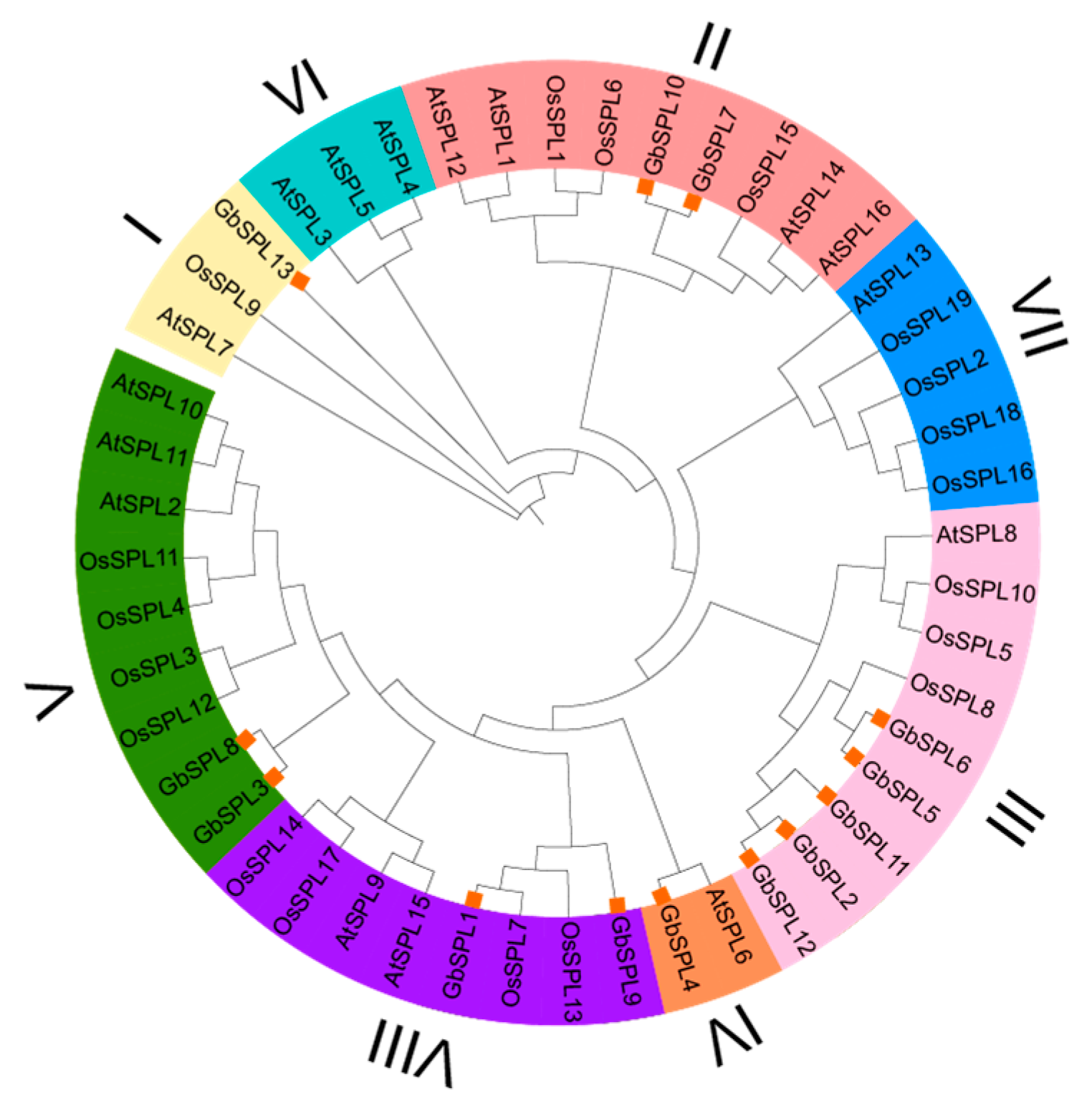
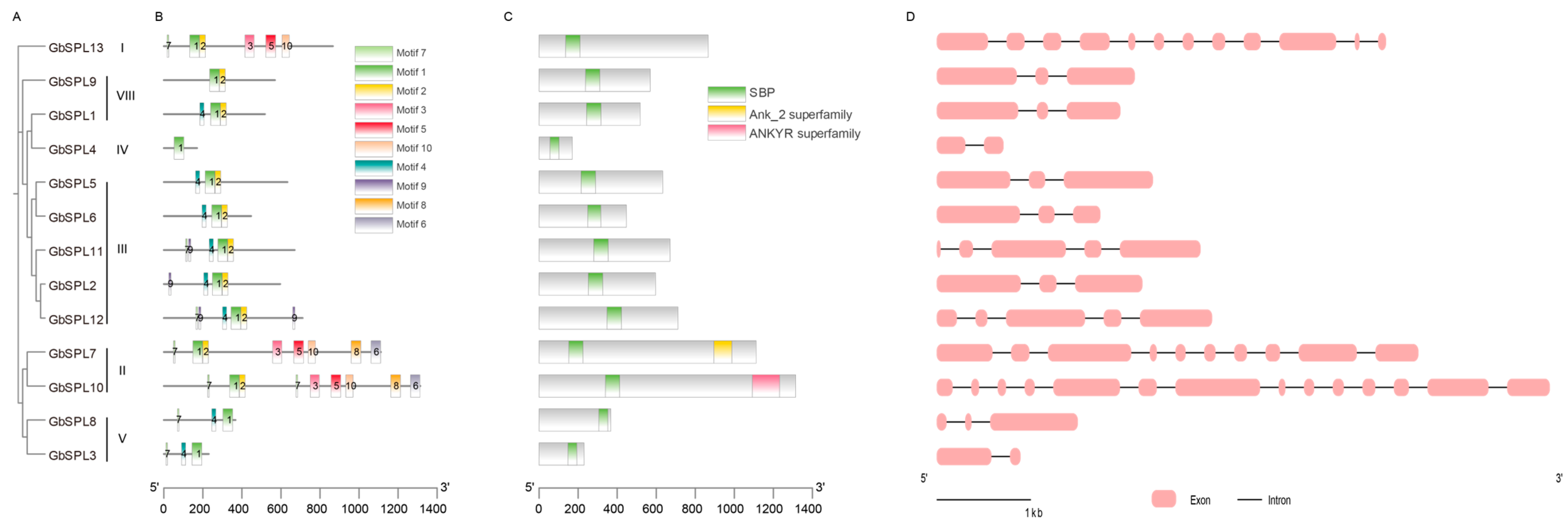

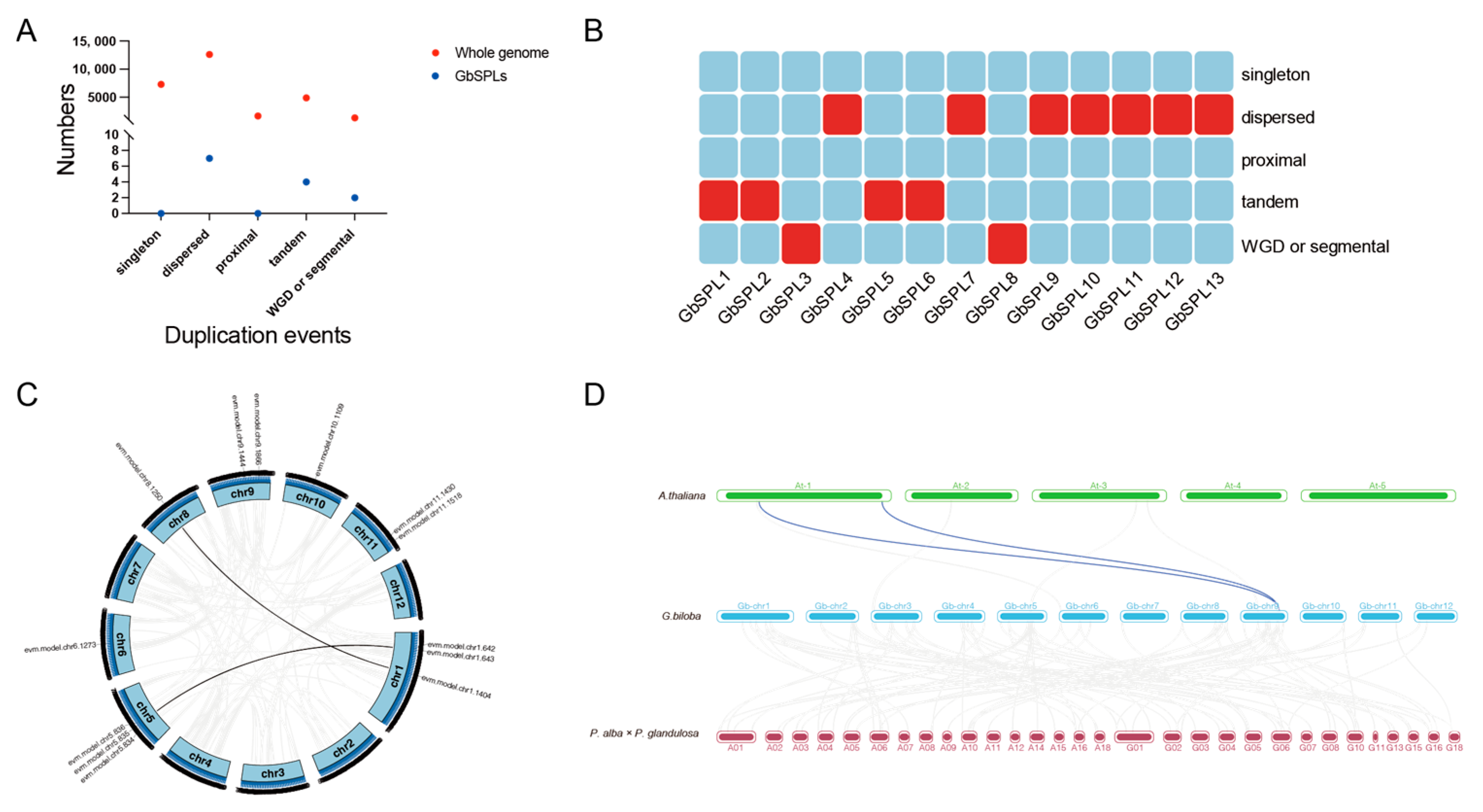

| Gene Name | Gene ID | Peptide (aa) | Molecular Weight MW (Da) | Theoretical pI | Grand Average of Hydropathicity (GRAVY) | Instability Index | Aliphatic Index | Subcellular Localization Predictions | miR156 Target |
|---|---|---|---|---|---|---|---|---|---|
| GbSPL1 | evm.model.chr1.642 | 518 | 57,582.8 | 7.36 | −0.776 | 58.94 | 58.90 | nuclear | YES |
| GbSPL2 | evm.model.chr1.643 | 596 | 64,192.75 | 7.10 | −0.657 | 55.27 | 65.30 | nuclear | NO |
| GbSPL3 | evm.model.chr1.1404 | 230 | 25,387.53 | 8.98 | −0.750 | 55.07 | 50.91 | nuclear | NO |
| GbSPL4 | evm.model.chr5.834 | 170 | 18,774.35 | 6.52 | −0.514 | 51.55 | 79.24 | nuclear | NO |
| GbSPL5 | evm.model.chr5.835 | 633 | 70,406.88 | 7.61 | −0.765 | 65.35 | 60.39 | nuclear | NO |
| GbSPL6 | evm.model.chr5.836 | 447 | 49,707.51 | 8.55 | −0.654 | 65.54 | 64.41 | nuclear | NO |
| GbSPL7 | evm.model.chr6.1273 | 1112 | 123,372.1 | 8.56 | −0.499 | 55.69 | 75.50 | nuclear | NO |
| GbSPL8 | evm.model.chr8.1250 | 367 | 40,125.93 | 7.52 | −0.622 | 37.22 | 58.96 | nuclear | NO |
| GbSPL9 | evm.model.chr9.1444 | 569 | 63,284.01 | 9.27 | −0.707 | 53.76 | 66.40 | nuclear | YES |
| GbSPL10 | evm.model.chr9.1866 | 1314 | 145,251.18 | 8.69 | −0.414 | 58.05 | 78.76 | chloroplast | NO |
| GbSPL11 | evm.model.chr10.1109 | 670 | 72,574.69 | 8.92 | −0.741 | 59.74 | 60.70 | nuclear | NO |
| GbSPL12 | evm.model.chr11.1430 | 711 | 77,686.16 | 8.58 | −0.693 | 56.53 | 58.30 | nuclear | NO |
| GbSPL13 | evm.model.chr11.1518 | 866 | 97,933.52 | 5.57 | −0.523 | 50.55 | 76.74 | nuclear | NO |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ming, M.; Yi, M.; Qin, C.; Yan, L.; Sun, Y.; Zhang, J.; Cao, F.; Fu, F. Genome-Wide Identification of Ginkgo biloba SPL Gene Family and Expression Analysis in Flavonoid Biosynthesis and Water Stress. Int. J. Mol. Sci. 2025, 26, 4932. https://doi.org/10.3390/ijms26104932
Ming M, Yi M, Qin C, Yan L, Sun Y, Zhang J, Cao F, Fu F. Genome-Wide Identification of Ginkgo biloba SPL Gene Family and Expression Analysis in Flavonoid Biosynthesis and Water Stress. International Journal of Molecular Sciences. 2025; 26(10):4932. https://doi.org/10.3390/ijms26104932
Chicago/Turabian StyleMing, Meiling, Mulin Yi, Chunyue Qin, Luyao Yan, Yuhan Sun, Juan Zhang, Fuliang Cao, and Fangfang Fu. 2025. "Genome-Wide Identification of Ginkgo biloba SPL Gene Family and Expression Analysis in Flavonoid Biosynthesis and Water Stress" International Journal of Molecular Sciences 26, no. 10: 4932. https://doi.org/10.3390/ijms26104932
APA StyleMing, M., Yi, M., Qin, C., Yan, L., Sun, Y., Zhang, J., Cao, F., & Fu, F. (2025). Genome-Wide Identification of Ginkgo biloba SPL Gene Family and Expression Analysis in Flavonoid Biosynthesis and Water Stress. International Journal of Molecular Sciences, 26(10), 4932. https://doi.org/10.3390/ijms26104932







