Virus-Induced Genome Editing (VIGE): One Step Away from an Agricultural Revolution
Abstract
1. Importance of Transgene-Free Technologies in Agriculture
2. Plant Viruses: Structure and Mechanism of Action
3. Plant Viruses as Vectors
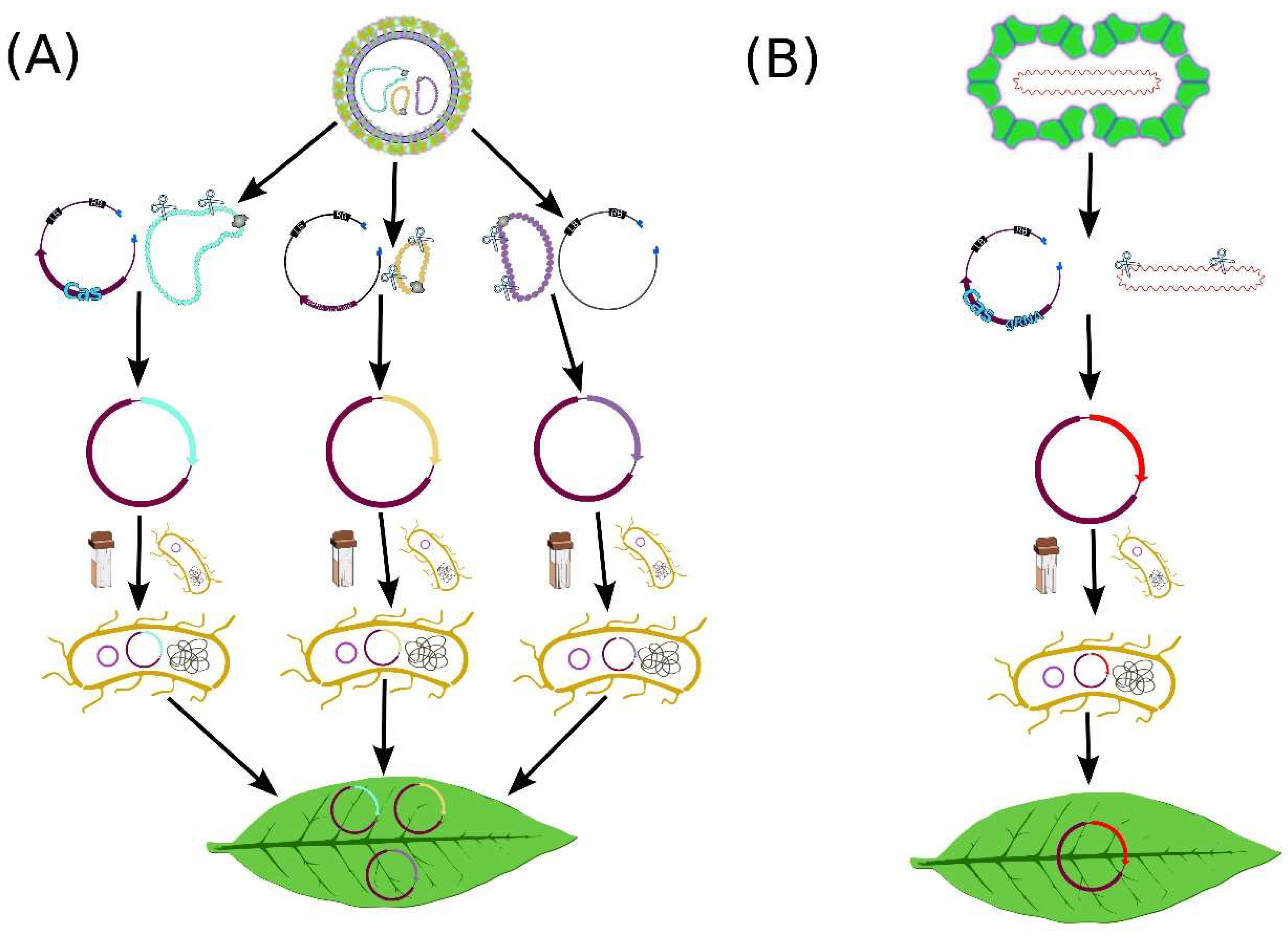
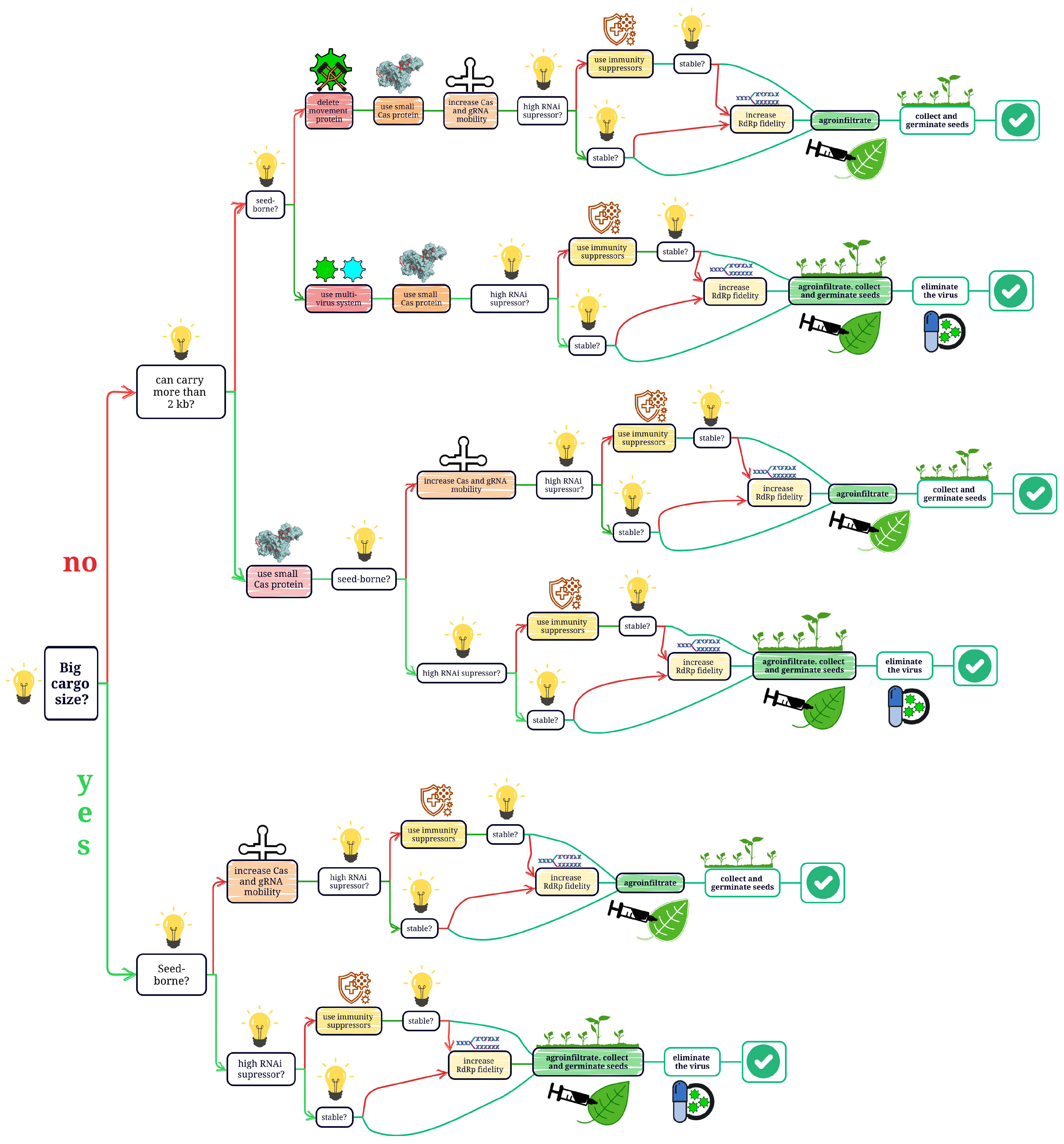
4. Recent Advances in VIGE
4.1. Increasing Cargo Capacity
4.2. Increasing Stability of Viral Vectors
4.3. Decreasing the Size of Cas Protein
4.4. Increasing Mobility of CRISPR/Cas System
4.5. Decreasing Host Immune Response
4.6. Multivirus Vector Systems
4.7. Regulating the Temperature Conditions
4.8. Providing Seed Transmission
4.9. Elimination of Viruses
4.10. Application of VIGE in Crops
4.11. Targeting Economically Valuable Traits
5. Conclusions
Funding
Conflicts of Interest
References
- Hwang, H.-H.; Yu, M.; Lai, E.-M. Agrobacterium-Mediated Plant Transformation: Biology and Applications. Arab. Book 2017, 15, e0186. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kumar, A.; Patel, R.; Kumar, V. Genetically Modified Crop Regulations: Scope and Opportunity Using the CRISPR-Cas9 Genome Editing Approach. Mol. Biol. Rep. 2021, 48, 4851–4863. [Google Scholar] [CrossRef]
- Mikhaylova, E.V. Crop genome editing–regulations and policies. In Genome Editing and Global Food Security: Molecular Engineering Technologies for Sustainable Agriculture, 1st ed.; Routledge: London, UK, 2023; ISBN 978-1-00-338210-2. [Google Scholar] [CrossRef]
- EU Rethinks Genome Editing. Nat. Plants 2023, 9, 1169–1170. [CrossRef] [PubMed]
- Waltz, E. GABA-Enriched Tomato Is First CRISPR-Edited Food to Enter Market. Nat. Biotechnol. 2022, 40, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Permyakova, N.V.; Deineko, E.V. Crop Improvement: Comparison of Transgenesis and Gene Editing. Horticulturae 2024, 10, 57. [Google Scholar] [CrossRef]
- Mikhaylova, E.V.; Khusnutdinov, E.A.; Chemeris, A.V.; Kuluev, B.R. Available Toolkits for CRISPR/CAS Genome Editing in Plants. Russ. J. Plant Physiol. 2022, 69, 3. [Google Scholar] [CrossRef]
- Hull, R.; Harper, G.; Lockhart, B. Viral Sequences Integrated into Plant Genomes. Trends Plant Sci. 2000, 5, 362–365. [Google Scholar] [CrossRef]
- Callaway, A.; Giesman-Cookmeyer, D.; Gillock, E.T.; Sit, T.L.; Lommel, S.A. THE MULTIFUNCTIONAL CAPSID PROTEINS OF PLANT RNA VIRUSES. Annu. Rev. Phytopathol. 2001, 39, 419–460. [Google Scholar] [CrossRef]
- Wu, B.; Grigull, J.; Ore, M.O.; Morin, S.; White, K.A. Global Organization of a Positive-Strand RNA Virus Genome. PLoS Pathog. 2013, 9, e1003363. [Google Scholar] [CrossRef]
- Oliver, J.E.; Whitfield, A.E. The Genus Tospovirus: Emerging Bunyaviruses That Threaten Food Security. Annu. Rev. Virol. 2016, 3, 101–124. [Google Scholar] [CrossRef]
- Porta, C.; Lomonossoff, G.P. Viruses as Vectors for the Expression of Foreign Sequences in Plants. Biotechnol. Genet. Eng. Rev. 2002, 19, 245–292. [Google Scholar] [CrossRef]
- Shafiq, M.; Qurashi, F.; Mushtaq, S.; Hussain, M.; Hameed, A.; Saleem Haider, M. DNA Plant Viruses: Biochemistry, Replication, and Molecular Genetics. In Applied Plant Virology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 169–182. ISBN 978-0-12-818654-1. [Google Scholar] [CrossRef]
- Malathi, V.G.; Renuka Devi, P. ssDNA Viruses: Key Players in Global Virome. VirusDisease 2019, 30, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhou, T. Unraveling the Mechanisms of Virus-Induced Symptom Development in Plants. Plants 2023, 12, 2830. [Google Scholar] [CrossRef]
- Stafford, C.A.; Walker, G.P.; Ullman, D.E. Hitching a Ride: Vector Feeding and Virus Transmission. Commun. Integr. Biol. 2012, 5, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Prakash, V.; Singh, A.; Singh, A.K.; Dalmay, T.; Chakraborty, S. Tobacco RNA-Dependent RNA Polymerase 1 Affects the Expression of Defence-Related Genes in Nicotiana Benthamiana upon Tomato Leaf Curl Gujarat Virus Infection. Planta 2020, 252, 11. [Google Scholar] [CrossRef]
- Carrington, J.C.; Kasschau, K.D.; Johansen, L.K. Activation and Suppression of RNA Silencing by Plant Viruses. Virology 2001, 281, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Escalante, C.; Sanz-Saez, A.; Jacobson, A.; Otulak-Kozieł, K.; Kozieł, E.; Balkcom, K.S.; Zhao, C.; Conner, K. Plant Virus Transmission during Seed Development and Implications to Plant Defense System. Front. Plant Sci. 2024, 15, 1385456. [Google Scholar] [CrossRef]
- Pagán, I. Transmission through Seeds: The Unknown Life of Plant Viruses. PLoS Pathog. 2022, 18, e1010707. [Google Scholar] [CrossRef]
- Sandra, N.; Mandal, B. Emerging Evidence of Seed Transmission of Begomoviruses: Implications in Global Circulation and Disease Outbreak. Front. Plant Sci. 2024, 15, 1376284. [Google Scholar] [CrossRef]
- Belaffif, M.B.; Brown, M.C.; Marcial, B.; Baysal, C.; Swaminathan, K. New Strategies to Advance Plant Transformation. Curr. Opin. Biotechnol. 2025, 91, 103241. [Google Scholar] [CrossRef]
- Gleba, Y.; Marillonnet, S.; Klimyuk, V. Engineering Viral Expression Vectors for Plants: The ‘Full Virus’ and the ‘Deconstructed Virus’ Strategies. Curr. Opin. Plant Biol. 2004, 7, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lomonossoff, G.P. Agroinfection as a Rapid Method for Propagating Cowpea Mosaic Virus-Based Constructs. J. Virol. Methods 2002, 105, 343–348. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, C.; Sun, K.; Deng, Y.; Li, Z. Engineered Biocontainable RNA Virus Vectors for Non-Transgenic Genome Editing across Crop Species and Genotypes. Mol. Plant 2023, 16, 616–631. [Google Scholar] [CrossRef]
- Yang, S.-J.; Carter, S.A.; Cole, A.B.; Cheng, N.-H.; Nelson, R.S. A Natural Variant of a Host RNA-Dependent RNA Polymerase Is Associated with Increased Susceptibility to Viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 2004, 101, 6297–6302. [Google Scholar] [CrossRef]
- Zulfiqar, S.; Farooq, M.A.; Zhao, T.; Wang, P.; Tabusam, J.; Wang, Y.; Xuan, S.; Zhao, J.; Chen, X.; Shen, S.; et al. Virus-Induced Gene Silencing (VIGS): A Powerful Tool for Crop Improvement and Its Advancement towards Epigenetics. Int. J. Mol. Sci. 2023, 24, 5608. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Li, X.; Fan, B.; Zhu, C.; Chen, Z. Maximizing the Production of Recombinant Proteins in Plants: From Transcription to Protein Stability. Int. J. Mol. Sci. 2022, 23, 13516. [Google Scholar] [CrossRef] [PubMed]
- Gleba, Y.Y.; Tusé, D.; Giritch, A. Plant Viral Vectors for Delivery by Agrobacterium. In Plant Viral Vectors; Palmer, K., Gleba, Y., Eds.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2013; Volume 375, pp. 155–192. ISBN 978-3-642-40828-1. [Google Scholar] [CrossRef]
- Anand, A.; Uppalapati, S.R.; Ryu, C.-M.; Allen, S.N.; Kang, L.; Tang, Y.; Mysore, K.S. Salicylic Acid and Systemic Acquired Resistance Play a Role in Attenuating Crown Gall Disease Caused by Agrobacterium tumefaciens. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef]
- Azizi-Dargahlou, S.; Pouresmaeil, M. Agrobacterium Tumefaciens-Mediated Plant Transformation: A Review. Mol. Biotechnol. 2024, 66, 1563–1580. [Google Scholar] [CrossRef]
- Gelvin, S.B. Plant DNA Repair and Agrobacterium T−DNA Integration. Int. J. Mol. Sci. 2021, 22, 8458. [Google Scholar] [CrossRef]
- Monroy-Borrego, A.G.; Steinmetz, N.F. Three Methods for Inoculation of Viral Vectors into Plants. Front. Plant Sci. 2022, 13, 963756. [Google Scholar] [CrossRef]
- Lacroix, B.; Citovsky, V. Biolistic Approach for Transient Gene Expression Studies in Plants. In Biolistic DNA Delivery in Plants; Rustgi, S., Luo, H., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; Volume 2124, pp. 125–139. ISBN 978-1-07-160355-0. [Google Scholar] [CrossRef]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-Frequency, Precise Modification of the Tomato Genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [PubMed]
- Butler, N.M.; Baltes, N.J.; Voytas, D.F.; Douches, D.S. Geminivirus-Mediated Genome Editing in Potato (Solanum tuberosum L.) Using Sequence-Specific Nucleases. Front. Plant Sci. 2016, 7, 1045. [Google Scholar] [CrossRef]
- Li, B.; Fu, C.; Zhou, J.; Hui, F.; Wang, Q.; Wang, F.; Wang, G.; Xu, Z.; Che, L.; Yuan, D.; et al. Highly Efficient Genome Editing Using Geminivirus-Based CRISPR/Cas9 System in Cotton Plant. Cells 2022, 11, 2902. [Google Scholar] [CrossRef] [PubMed]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency Gene Targeting in Hexaploid Wheat Using DNA Replicons and CRISPR /Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lu, Y.; Botella, J.R.; Mao, Y.; Hua, K.; Zhu, J. Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol. Plant 2017, 10, 1007–1010. [Google Scholar] [CrossRef]
- Palmer, K.E.; Rybicki, E.P. Investigation of the Potential of Maize Streak Virus to Act as an Infectious Gene Vector in Maize Plants. Arch. Virol. 2001, 146, 1089–1104. [Google Scholar] [CrossRef]
- Yin, K.; Han, T.; Liu, G.; Chen, T.; Wang, Y.; Yu, A.Y.L.; Liu, Y. A Geminivirus-Based Guide RNA Delivery System for CRISPR/Cas9 Mediated Plant Genome Editing. Sci. Rep. 2015, 5, 14926. [Google Scholar] [CrossRef]
- Tuo, D.; Yao, Y.; Yan, P.; Chen, X.; Qu, F.; Xue, W.; Liu, J.; Kong, H.; Guo, J.; Cui, H.; et al. Development of Cassava Common Mosaic Virus-Based Vector for Protein Expression and Gene Editing in Cassava. Plant Methods 2023, 19, 78. [Google Scholar] [CrossRef]
- Wyant, P.S.; Kober, S.; Schwierzok, A.; Kocher, C.; Schäfer, B.; Jeske, H.; Wege, C. Cloned Tomato Golden Mosaic Virus Back in Tomatoes. Virus Res. 2012, 167, 397–403. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Elmer, J.S.; Rogers, S.G. Transient Expression of Heterologous RNAs Using Tomato Golden Mosaic Virus. Nucleic Acids Res. 1988, 16, 10511–10528. [Google Scholar] [CrossRef]
- Del Rosario Abraham-Juárez, M.; Del Carmen Rocha-Granados, M.; López, M.G.; Rivera-Bustamante, R.F.; Ochoa-Alejo, N. Virus-Induced Silencing of Comt, pAmt and Kas Genes Results in a Reduction of Capsaicinoid Accumulation in Chili Pepper Fruits. Planta 2008, 227, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Zhou, X. A Modified Viral Satellite DNA That Suppresses Gene Expression in Plants. Plant J. 2004, 38, 850–860. [Google Scholar] [CrossRef]
- Ishibashi, K.; Sukegawa, S.; Endo, M.; Hara, N.; Nureki, O.; Saika, H.; Toki, S. Systemic Delivery of Engineered Compact AsCas12f by a Positive-Strand RNA Virus Vector Enables Highly Efficient Targeted Mutagenesis in Plants. Front. Plant Sci. 2024, 15, 1454554. [Google Scholar] [CrossRef]
- Kim, N.-S.; Lee, K.-R.; Lee, J.; Kil, E.-J.; Lee, J.; Lee, S.-K. High Production of Recombinant Protein Using Geminivirus-Based Deconstructed Vectors in Nicotiana Benthamiana. Front. Plant Sci. 2024, 15, 1407240. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, N.K.; Chakraborty, S. Chilli Leaf Curl Virus-Based Vector for Phloem-Specific Silencing of Endogenous Genes and Overexpression of Foreign Genes. Appl. Microbiol. Biotechnol. 2017, 101, 2121–2129. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Li, Y.; Dai, P.; Liu, C.; Zhao, Y.; You, Y.; Qu, Y.; Chen, Q.; Liu, X. Efficient Virus-Mediated Genome Editing in Cotton Using the CRISPR/Cas9 System. Front. Plant Sci. 2022, 13, 1032799. [Google Scholar] [CrossRef]
- Lei, J.; Dai, P.; Li, Y.; Zhang, W.; Zhou, G.; Liu, C.; Liu, X. Heritable Gene Editing Using FT Mobile Guide RNAs and DNA Viruses. Plant Methods 2021, 17, 20. [Google Scholar] [CrossRef]
- Jeyabharathy, C.; Shakila, H.; Usha, R. Development of a VIGS Vector Based on the β-Satellite DNA Associated with Bhendi Yellow Vein Mosaic Virus. Virus Res. 2015, 195, 73–78. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, X.; Sun, H.; Liang, Q.; Wang, W.; Zhang, C.; Bian, X.; Cao, Q.; Li, Q.; Xie, Y.; et al. Improving CRISPR-Cas-mediated RNA Targeting and Gene Editing Using SPLCV Replicon-based Expression Vectors in Nicotiana benthamiana. Plant Biotechnol. J. 2020, 18, 1993–1995. [Google Scholar] [CrossRef]
- Zhao, C.; Lou, H.; Liu, Q.; Pei, S.; Liao, Q.; Li, Z. Efficient and Transformation-free Genome Editing in Pepper Enabled by RNA Virus-mediated Delivery of CRISPR/Cas9. J. Integr. Plant Biol. 2024, 66, 2079–2082. [Google Scholar] [CrossRef]
- Lee, H.; Baik, J.E.; Kim, K.-N. Development of an Efficient and Heritable Virus-Induced Genome Editing System in Solanum lycopersicum. Hortic. Res. 2024, 12, uhae364. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.H.; Ko, Y.; Lee, J.M. Enhancing Virus-Mediated Genome Editing for Cultivated Tomato Through Low Temperature. Plant Cell Rep. 2025, 44, 22. [Google Scholar] [CrossRef]
- Yoshida, T.; Ishikawa, M.; Toki, S.; Ishibashi, K. Heritable Tissue-Culture-Free Gene Editing in Nicotiana Benthamiana through Viral Delivery of SpCas9 and sgRNA. Plant Cell Physiol. 2024, 65, 1743–1750. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, D.; Dai, Y.; Liu, C. Efficient Tobacco Rattle Virus-induced Gene Editing in Tomato Mediated by the CRISPR/Cas9 System. Biotechnol. J. 2024, 19, 2400204. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Eid, A.; Ali, S.; Mahfouz, M.M. Pea Early-Browning Virus-Mediated Genome Editing via the CRISPR/Cas9 System in Nicotiana Benthamiana and Arabidopsis. Virus Res. 2018, 244, 333–337. [Google Scholar] [CrossRef]
- Ali, Z.; Abul-faraj, A.; Li, L.; Ghosh, N.; Piatek, M.; Mahjoub, A.; Aouida, M.; Piatek, A.; Baltes, N.J.; Voytas, D.F.; et al. Efficient Virus-Mediated Genome Editing in Plants Using the CRISPR/Cas9 System. Mol. Plant 2015, 8, 1288–1291. [Google Scholar] [CrossRef] [PubMed]
- Ellison, E.E.; Nagalakshmi, U.; Gamo, M.E.; Huang, P.; Dinesh-Kumar, S.; Voytas, D.F. Multiplexed Heritable Gene Editing Using RNA Viruses and Mobile Single Guide RNAs. Nat. Plants 2020, 6, 620–624. [Google Scholar] [CrossRef]
- Tavares-Esashika, M.L.; Campos, R.N.S.; Blawid, R.; Da Luz, L.L.; Inoue-Nagata, A.K.; Nagata, T. Characterization of an Infectious Clone of Pepper Ringspot Virus and Its Use as a Viral Vector. Arch. Virol. 2020, 165, 367–375. [Google Scholar] [CrossRef]
- Tavares-Esashika, M.L.; Campos, R.N.S.; Maeda, M.H.K.; Koyama, L.H.H.; Hamann, P.R.V.; Noronha, E.F.; Nagata, T. Development of a Heterologous Gene Expression Vector in Plants Based on an Infectious Clone of a Tobravirus, Pepper Ringspot Virus. Ann. Appl. Biol. 2022, 181, 107–116. [Google Scholar] [CrossRef]
- Selma, S.; Gianoglio, S.; Uranga, M.; Vázquez-Vilar, M.; Espinosa-Ruiz, A.; Drapal, M.; Fraser, P.D.; Daròs, J.; Orzáez, D. Potato Virus X -delivered CRISPR Activation Programs Lead to Strong Endogenous Gene Induction and Transient Metabolic Reprogramming in Nicotiana benthamiana. Plant J. 2022, 111, 1550–1564. [Google Scholar] [CrossRef]
- Ariga, H.; Toki, S.; Ishibashi, K. Potato Virus X Vector-Mediated DNA-Free Genome Editing in Plants. Plant Cell Physiol. 2020, 61, 1946–1953. [Google Scholar] [CrossRef]
- Bouton, C.; King, R.C.; Chen, H.; Azhakanandam, K.; Bieri, S.; Hammond-Kosack, K.E.; Kanyuka, K. Foxtail Mosaic Virus: A Viral Vector for Protein Expression in Cereals. Plant Physiol. 2018, 177, 1352–1367. [Google Scholar] [CrossRef] [PubMed]
- Baysal, C.; Kausch, A.P.; Cody, J.P.; Altpeter, F.; Voytas, D.F. Rapid and Efficient in Planta Genome Editing in Sorghum Using Foxtail Mosaic Virus-mediated sgRNA Delivery. Plant J. 2025, 121, e17196. [Google Scholar] [CrossRef]
- Mei, Y.; Beernink, B.M.; Ellison, E.E.; Konečná, E.; Neelakandan, A.K.; Voytas, D.F.; Whitham, S.A. Protein Expression and Gene Editing in Monocots Using Foxtail Mosaic Virus Vectors. Plant Direct 2019, 3, e00181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kang, L.; Zhang, Q.; Meng, Q.; Pan, Y.; Yu, Z.; Shi, N.; Jackson, S.; Zhang, X.; Wang, H.; et al. An RNAi Suppressor Activates in Planta Virus–Mediated Gene Editing. Funct. Integr. Genom. 2020, 20, 471–477. [Google Scholar] [CrossRef]
- Ruiz-Ramón, F.; Sempere, R.N.; Méndez-López, E.; Sánchez-Pina, M.A.; Aranda, M.A. Second Generation of Pepino Mosaic Virus Vectors: Improved Stability in Tomato and a Wide Range of Reporter Genes. Plant Methods 2019, 15, 58. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.; Wang, Y.; Xu, L.; Qin, T.; Xia, K.; Zhang, P.; Ma, Y.; Zhang, K.; Wang, L.; Dong, J.; et al. Establishing VIGS and CRISPR/Cas9 Techniques to Verify RsPDS Function in Radish. J. Integr. Agric. 2024, 23, 1557–1567. [Google Scholar] [CrossRef]
- Pflieger, S.; Blanchet, S.; Camborde, L.; Drugeon, G.; Rousseau, A.; Noizet, M.; Planchais, S.; Jupin, I. Efficient Virus-induced Gene Silencing in Arabidopsis Using a ‘One-step’ TYMV-derived Vector. Plant J. 2008, 56, 678–690. [Google Scholar] [CrossRef]
- Li, T.; Hu, J.; Sun, Y.; Li, B.; Zhang, D.; Li, W.; Liu, J.; Li, D.; Gao, C.; Zhang, Y.; et al. Highly Efficient Heritable Genome Editing in Wheat Using an RNA Virus and Bypassing Tissue Culture. Mol. Plant 2021, 14, 1787–1798. [Google Scholar] [CrossRef]
- Hu, J.; Li, S.; Li, Z.; Li, H.; Song, W.; Zhao, H.; Lai, J.; Xia, L.; Li, D.; Zhang, Y. A Barley Stripe Mosaic Virus-based Guide RNA Delivery System for Targeted Mutagenesis in Wheat and Maize. Mol. Plant Pathol. 2019, 20, 1463–1474. [Google Scholar] [CrossRef]
- Tamilselvan-Nattar-Amutha, S.; Hiekel, S.; Hartmann, F.; Lorenz, J.; Dabhi, R.V.; Dreissig, S.; Hensel, G.; Kumlehn, J.; Heckmann, S. Barley Stripe Mosaic Virus-Mediated Somatic and Heritable Gene Editing in Barley (Hordeum vulgare L.). Front. Plant Sci. 2023, 14, 1201446. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, C.; Liu, J.; Guo, Z.; Zhang, Z.; Han, C.; Wang, Y. Development of Beet Necrotic Yellow Vein Virus-based Vectors for Multiple-gene Expression and Guide RNA Delivery in Plant Genome Editing. Plant Biotechnol. J. 2019, 17, 1302–1315. [Google Scholar] [CrossRef]
- Kaya, H.; Ishibashi, K.; Toki, S. A Split Staphylococcus Aureus Cas9 as a Compact Genome-Editing Tool in Plants. Plant Cell Physiol. 2017, 58, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Yoshikawa, M.; Ariga, H.; Endo, M.; Toki, S.; Ishibashi, K. A Removable Virus Vector Suitable for Plant Genome Editing. Plant J. 2017, 91, 558–561. [Google Scholar] [CrossRef] [PubMed]
- Cody, W.B.; Scholthof, H.B.; Mirkov, T.E. Multiplexed Gene Editing and Protein Overexpression Using a Tobacco Mosaic Virus Viral Vector. Plant Physiol. 2017, 175, 23–35. [Google Scholar] [CrossRef]
- Gao, Q.; Xu, W.; Yan, T.; Fang, X.; Cao, Q.; Zhang, Z.; Ding, Z.; Wang, Y.; Wang, X. Rescue of a Plant Cytorhabdovirus as Versatile Expression Platforms for Planthopper and Cereal Genomic Studies. New Phytol. 2019, 223, 2120–2133. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, X.; Liu, H.; Li, Z. Highly Efficient DNA-Free Plant Genome Editing Using Virally Delivered CRISPR–Cas9. Nat. Plants 2020, 6, 773–779. [Google Scholar] [CrossRef]
- Xiang, H.; Chen, B.; Wang, S.; Zeng, W.; Jiang, J.; Kong, W.; Huang, H.; Mi, Q.; Ni, S.; Gao, Q.; et al. Development of an RNA Virus Vector for Non-Transgenic Genome Editing in Tobacco and Generation of Berberine Bridge Enzyme-like Mutants with Reduced Nicotine Content. Abiotech 2024, 5, 449–464. [Google Scholar] [CrossRef]
- Cañizares, M.C.; Lomonossoff, G.P.; Nicholson, L. Development of Cowpea Mosaic Virus-Based Vectors for the Production of Vaccines in Plants. Expert Rev. Vaccines 2005, 4, 687–697. [Google Scholar] [CrossRef]
- Luo, Y.; Na, R.; Nowak, J.S.; Qiu, Y.; Lu, Q.S.; Yang, C.; Marsolais, F.; Tian, L. Development of a Csy4-Processed Guide RNA Delivery System with Soybean-Infecting Virus ALSV for Genome Editing. BMC Plant Biol. 2021, 21, 419. [Google Scholar] [CrossRef]
- Melgar, A.E.; Palacios, M.B.; Tosar, L.J.M.; Zelada, A.M. A Novel and Efficient Apple Latent Spherical Virus-Based Gene Silencing Method for Functional Genomic Studies in Chenopodium Quinoa. Sci. Hortic. 2024, 333, 113258. [Google Scholar] [CrossRef]
- Zhao, F.; Lim, S.; Igori, D.; Yoo, R.H.; Kwon, S.-Y.; Moon, J.S. Development of Tobacco Ringspot Virus-Based Vectors for Foreign Gene Expression and Virus-Induced Gene Silencing in a Variety of Plants. Virology 2016, 492, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Adhab, M.; Zhang, Y.; Schoelz, J. Transient Expression of Cauliflower Mosaic Virus (CaMV) P6-GFP Complements a Defective CaMV Replicon to Facilitate Viral Gene Expression, Replication and Virion Formation. Virology 2023, 587, 109854. [Google Scholar] [CrossRef]
- Kant, R.; Dasgupta, I. Phenotyping of VIGS-Mediated Gene Silencing in Rice Using a Vector Derived from a DNA Virus. Plant Cell Rep. 2017, 36, 1159–1170. [Google Scholar] [CrossRef]
- Zhang, B.; Shi, J.-A.; Chen, J.-B.; Li, D.-D.; Li, J.-J.; Guo, H.-L.; Zong, J.-Q.; Wang, Y.; Guo, A.-G.; Liu, J.-X. Efficient Virus-Induced Gene Silencing in Cynodon Dactylon and Zoysia Japonica Using Rice Tungro Bacilliform Virus Vectors. Sci. Hortic. 2016, 207, 97–103. [Google Scholar] [CrossRef]
- Otagaki, S.; Arai, M.; Takahashi, A.; Goto, K.; Hong, J.-S.; Masuta, C.; Kanazawa, A. Rapid Induction of Transcriptional and Post-Transcriptional Gene Silencing Using a Novel Cucumber Mosaic Virus Vector. Plant Biotechnol. 2006, 23, 259–265. [Google Scholar] [CrossRef]
- Tzean, Y.; Lee, M.-C.; Jan, H.-H.; Chiu, Y.-S.; Tu, T.-C.; Hou, B.-H.; Chen, H.-M.; Chou, C.-N.; Yeh, H.-H. Cucumber Mosaic Virus-Induced Gene Silencing in Banana. Sci. Rep. 2019, 9, 11553. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.S.; Mannas, S.W.; Bishop, B.A.; Rao, X.; Lecoultre, M.; Kwon, S.; Nelson, R.S. An Improved Brome Mosaic Virus Silencing Vector: Greater Insert Stability and More Extensive VIGS. Plant Physiol. 2018, 176, 496–510. [Google Scholar] [CrossRef]
- Wang, Y.; Chai, C.; Khatabi, B.; Scheible, W.-R.; Udvardi, M.K.; Saha, M.C.; Kang, Y.; Nelson, R.S. An Efficient Brome Mosaic Virus-Based Gene Silencing Protocol for Hexaploid Wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 685187. [Google Scholar] [CrossRef]
- Choi, I.; Stenger, D.C.; Morris, T.J.; French, R. A Plant Virus Vector for Systemic Expression of Foreign Genes in Cereals. Plant J. 2000, 23, 547–555. [Google Scholar] [CrossRef]
- Fernández-Fernández, M.R.; Mouriño, M.; Rivera, J.; Rodríguez, F.; Plana-Durán, J.; García, J.A. Protection of Rabbits against Rabbit Hemorrhagic Disease Virus by Immunization with the VP60 Protein Expressed in Plants with a Potyvirus-Based Vector. Virology 2001, 280, 283–291. [Google Scholar] [CrossRef]
- Masuta, C.; Yamana, T.; Tacahashi, Y.; Uyeda, I.; Sato, M.; Ueda, S.; Matsumura, T. Development of Clover Yellow Vein Virus as an Efficient, Stable Gene-expression System for Legume Species. Plant J. 2000, 23, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Uranga, M.; Vazquez-Vilar, M.; Orzáez, D.; Daròs, J.-A. CRISPR-Cas12a Genome Editing at the Whole-Plant Level Using Two Compatible RNA Virus Vectors. Cris. J. 2021, 4, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, S.; McMechan, A.J.; Bartels, M.; Hein, G.L.; Graybosch, R.A. In Vitro Transcripts of Wild-Type and Fluorescent Protein-Tagged Triticum mosaic virus (Family Potyviridae) Are Biologically Active in Wheat. Phytopathology 2015, 105, 1496–1505. [Google Scholar] [CrossRef]
- DeMell, A.; Mendoza, M.R.; Scholthof, H.B. A Tomato Bushy Stunt Virus–Based Vector for Simultaneous Editing and Sensing to Survey the Host Antiviral RNA Silencing Machinery. PNAS Nexus 2023, 3, pgad436. [Google Scholar] [CrossRef]
- Roumagnac, P.; Lett, J.-M.; Fiallo-Olivé, E.; Navas-Castillo, J.; Zerbini, F.M.; Martin, D.P.; Varsani, A. Establishment of Five New Genera in the Family Geminiviridae: Citlodavirus, Maldovirus, Mulcrilevirus, Opunvirus, and Topilevirus. Arch. Virol. 2022, 167, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Boezen, D.; Zwart, M.P. Metagenomic Studies of Viruses in Weeds and Wild Plants: A Powerful Approach to Characterise Variable Virus Communities. Viruses 2021, 13, 1939. [Google Scholar] [CrossRef]
- Shen, Y.; Ye, T.; Li, Z.; Kimutai, T.H.; Song, H.; Dong, X.; Wan, J. Exploiting Viral Vectors to Deliver Genome Editing Reagents in Plants. Abiotech 2024, 5, 247–261. [Google Scholar] [CrossRef]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA Replicons for Plant Genome Engineering. Plant Cell 2014, 26, 151–163. [Google Scholar] [CrossRef]
- Gleba, Y.; Klimyuk, V.; Marillonnet, S. Viral Vectors for the Expression of Proteins in Plants. Curr. Opin. Biotechnol. 2007, 18, 134–141. [Google Scholar] [CrossRef]
- Khakhar, A.; Voytas, D.F. RNA Viral Vectors for Accelerating Plant Synthetic Biology. Front. Plant Sci. 2021, 12, 668580. [Google Scholar] [CrossRef]
- Shi, J.; Perryman, J.M.; Yang, X.; Liu, X.; Musser, D.M.; Boehr, A.K.; Moustafa, I.M.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Rational Control of Poliovirus RNA-Dependent RNA Polymerase Fidelity by Modulating Motif-D Loop Conformational Dynamics. Biochemistry 2019, 58, 3735–3743. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhong, Z.; Wu, Z.; Pausch, P.; Al-Shayeb, B.; Amerasekera, J.; Doudna, J.A.; Jacobsen, S.E. Genome Editing in Plants Using the Compact Editor CasΦ. Proc. Natl. Acad. Sci. USA 2023, 120, e2216822120. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Cao, X.; Hu, L.; Hu, D.; Li, D.; Sun, Y. Engineering an Optimized Hypercompact CRISPR /Cas12j-8 System for Efficient Genome Editing in Plants. Plant Biotechnol. J. 2025, 23, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Previtera, D.A.; Wang, Y.; Botella, J.R. Geminiviral-Induced Genome Editing Using Miniature CRISPR/Cas12j (CasΦ) and Cas12f Variants in Plants. Plant Cell Rep. 2024, 43, 71. [Google Scholar] [CrossRef]
- Awan, M.J.A.; Amin, I.; Mansoor, S. Mini CRISPR-Cas12f1: A New Genome Editing Tool. Trends Plant Sci. 2022, 27, 110–112. [Google Scholar] [CrossRef]
- Takeda, S.N.; Nakagawa, R.; Okazaki, S.; Hirano, H.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Yamashita, K.; Nishimasu, H.; Nureki, O. Structure of the Miniature Type V-F CRISPR-Cas Effector Enzyme. Mol. Cell 2021, 81, 558–570.e3. [Google Scholar] [CrossRef]
- Tang, Z.; Chen, S.; Chen, A.; He, B.; Zhou, Y.; Chai, G.; Guo, F.; Huang, J. CasPDB: An Integrated and Annotated Database for Cas Proteins from Bacteria and Archaea. Database 2019, 2019, baz093. [Google Scholar] [CrossRef]
- Kehr, J.; Kragler, F. Long Distance RNA Movement. New Phytol. 2018, 218, 29–40. [Google Scholar] [CrossRef]
- Beernink, B.M.; Lappe, R.R.; Bredow, M.; Whitham, S.A. Impacts of RNA Mobility Signals on Virus Induced Somatic and Germline Gene Editing. Front. Genome Ed. 2022, 4, 925088. [Google Scholar] [CrossRef]
- Zhang, W.; Thieme, C.J.; Kollwig, G.; Apelt, F.; Yang, L.; Winter, N.; Andresen, N.; Walther, D.; Kragler, F. tRNA-Related Sequences Trigger Systemic mRNA Transport in Plants. Plant Cell 2016, 28, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Vong, B.; Picard, C.L.; Feng, S.; Tam, J.M.; Jacobsen, S.E. A Viral Guide RNA Delivery System for CRISPR-Based Transcriptional Activation and Heritable Targeted DNA Demethylation in Arabidopsis Thaliana. PLoS Genet. 2020, 16, e1008983. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Xuan, S.; Prichard, L.E.; Donahue, L.I.; Pan, C.; Nagalakshmi, U.; Ellison, E.E.; Starker, C.G.; Dinesh-Kumar, S.P.; Qi, Y.; et al. Heritable Base-Editing in Arabidopsis Using RNA Viral Vectors. Plant Physiol. 2022, 189, 1920–1924. [Google Scholar] [CrossRef]
- Anikina, I.; Kamarova, A.; Issayeva, K.; Issakhanova, S.; Mustafayeva, N.; Insebayeva, M.; Mukhamedzhanova, A.; Khan, S.M.; Ahmad, Z.; Lho, L.H.; et al. Plant Protection from Virus: A Review of Different Approaches. Front. Plant Sci. 2023, 14, 1163270. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Parks, G.; Endres, M.W.; Baulcombe, D.; Bowman, L.H.; Pruss, G.J.; Vance, V.B. The Amplicon-plus System for High-Level Expression of Transgenes in Plants. Nat. Biotechnol. 2002, 20, 622–625. [Google Scholar] [CrossRef]
- Sánchez-Tovar, M.R.; Rivera-Bustamante, R.F.; Saavedra-Trejo, D.L.; Guevara-González, R.G.; Torres-Pacheco, I. Mixed Plant Viral Infections: Complementation, Interference and Their Effects, a Review. Agronomy 2025, 15, 620. [Google Scholar] [CrossRef]
- Zhao, F.; Li, Y.; Chen, L.; Zhu, L.; Ren, H.; Lin, H.; Xi, D. Temperature Dependent Defence of Nicotiana Tabacum against Cucumber Mosaic Virus and Recovery Occurs with the Formation of Dark Green Islands. J. Plant Biol. 2016, 59, 293–301. [Google Scholar] [CrossRef]
- Naeem, M.; Zaman, W.; Saqib, S.; Shahzad, A.; Rahman, S.U.; Ahmad, N. CRISPR/Cas-Mediated Genome Editing for Efficient Tomato Breeding: Past Achievements and Future Directions. S. Afr. J. Bot. 2024, 172, 277–288. [Google Scholar] [CrossRef]
- Liu, D.; Ellison, E.E.; Myers, E.A.; Donahue, L.I.; Xuan, S.; Swanson, R.; Qi, S.; Prichard, L.E.; Starker, C.G.; Voytas, D.F. Heritable Gene Editing in Tomato through Viral Delivery of Isopentenyl Transferase and Single-Guide RNAs to Latent Axillary Meristematic Cells. Proc. Natl. Acad. Sci. USA 2024, 121, e2406486121. [Google Scholar] [CrossRef]
- Khusnutdinov, E.; Sukhareva, A.; Panfilova, M.; Mikhaylova, E. Anthocyanin Biosynthesis Genes as Model Genes for Genome Editing in Plants. Int. J. Mol. Sci. 2021, 22, 8752. [Google Scholar] [CrossRef]
- Kong, W.; Wang, M.; Huang, L.; Wu, F.; Tao, J.; Mo, B.; Yu, Y. A High-Efficient and Naked-Eye Visible CRISPR/Cas9 System in Arabidopsis. Planta 2023, 257, 30. [Google Scholar] [CrossRef] [PubMed]
| VIGS | Transient Protein Production | Agrobacterium-Mediated Genome Editing | RNP-Mediated Genome Editing | VIGE | |
|---|---|---|---|---|---|
| Use of virus | Yes | Yes | No | No | Yes |
| Goal | Silence gene expression | Short-term production of recombinant proteins | Easy selection of editing events | DNA-free genome editing | Transgene-free, in planta genome editing |
| Mechanism | A fragment of the target gene, delivered by viral vector, triggers the immune response | Viral vectors ensure high expression levels of recombinant proteins | The same as transgenesis | Delivery of RNPs by protoplast electroporation or particle bombardment | Transient expression from viral vector |
| Genome Integration | No | No | Yes | No | No |
| Tissue culture | No | No | Yes (except Arabidopsis) | Yes | Not tissue-culture-free yet |
| Heritability | No | No | Yes | Yes | Potentially yes |
| Key Applications | Functional genomics, pathway validation | Vaccine/antibody production | Genome editing in varieties susceptible to agrobacterium-mediated transformation | Genome editing in varieties with well-developed protoplast isolation and regeneration protocols | Genome editing of varieties infected by certain viruses |
| Genus | Type and Shape | Genome | Transmission | Virus | Hosts | VIGE | References |
|---|---|---|---|---|---|---|---|
| Geminivirus, Mastrevirus | Twinned icosahedral circular monopartite | ssDNA, 2.5–3.0 kb | Leafhoppers | Bean yellow dwarf virus (BeYDV); | French bean (Phaseolus vulgaris), tobacco, tomato, potato, Datura stramonium, Arabidopsis thaliana | + | [35,36,37] |
| Wheat dwarf virus (WDV) | Wheat, barley, oats | + | [38,39] | ||||
| Maize streak virus (MSV) | Maize, other grasses | - | [40] | ||||
| Geminivirus, Begomovirus | Twinned icosahedral circular bipartite | ssDNA, each 2.6–2.8 kb | Whiteflies | Cabbage leaf curl virus (CaLCuV) | Cabbage, other crucifers | + | [41] |
| African cassava mosaic virus (ACMV) | Euphorbiaceae family | + | [42] | ||||
| Tomato golden mosaic virus (TGMV) | Tomato, other solanaceous plants | - | [43,44] | ||||
| Pepper huasteco yellow vein virus (PHYVV) | Peppers, other solanaceous plants | - | [45] | ||||
| Tomato yellow leaf curl China virus (TYLCCNV) | Tomato, other solanaceous plants | - | [46,47] | ||||
| Honeysuckle yellow vein virus (HYVV) | Tomato, other solanaceous plants | - | [48] | ||||
| Chilli leaf curl viruses (ChLCV) | Chili peppers, other solanaceous plants | - | [49] | ||||
| Cotton leaf crumple virus (CLCrV) | Cotton, other malvaceous plants | + | [50,51] | ||||
| Bhendi yellow vein mosaic virus (BYVMV) | Okra (bhendi), other malvaceous plants | - | [52] | ||||
| Monopartite | ssDNA, 2.6–2.8 kb | Whiteflies, seeds | Sweet potato leaf curl virus (SPLCV) | Ipomoea | + | [53] | |
| Geminivirus, Curtovirus | Twinned icosahedral circular monopartite | ssDNA, ~3.0 kb | Leafhoppers | Beet mild curly top virus (BMCTV) | Beets, other chenopods | - | [48] |
| Bunyavirus, Tospovirus | Spherical or pleomorphic tripartite | ssRNA (-), parts: L (8.9 kb), M (4.8 kb), and S (2.9 kb) | Thrips, seeds (in some hosts) | Tomato spotted wilt virus (TSWV) | Over 1090 dicotyledonous and monocotyledonous species | + | [25,54] |
| Tobravirus, Virgaviridae | Rod-shaped bipartite | ssRNA (+); RNA1, ~6.8 kb; RNA2, ~1.9–4.3 kb | Nematodes, soil, seeds | Tobacco rattle virus (TRV) | Tobacco, potato, over 400 species from 50 families | + | [55,56,57,58] |
| ssRNA (+); RNA1, ~7.1 kb; RNA2, ~3.5 kb | Pea early-browning virus (PEBV) | 30 legume species, N. benthamiana, A. thaliana | + | [59,60,61] | |||
| ssRNA (+); RNA1, ~6.8 kb; RNA2, ~1.7 kb | Pepper ringspot virus (PRSV) | Pepper, tomato, artichoke, potato | - | [62,63] | |||
| Potexvirus, Alphaflexiviridae | Flexuous rod monopartite | ssRNA (+), ~6.4 kb | Mechanical | Potato virus X (PVX) | Mostly limited to Solanaceae | + | [64,65] |
| Foxtail mosaic virus (FoMV) | 56 monocot species and 35 dicot species | + | [66,67,68,69] | ||||
| Cassava common mosaic virus (CsCMV) | Cassava | + | [42] | ||||
| Pepino mosaic virus (PepMV) | Mostly limited to Solanaceae | - | [70] | ||||
| Tymovirus | Monopartite icosahedral | ssRNA (+), ~6.3 kb | Beetles, seeds | Turnip yellow mosaic virus (TYMV) | Brassicaeae | - | [71,72] |
| Hordeivirus, Virgaviridae | Rod-shaped tripartite | ssRNA (+), total ~12.7 kb | Mechanical, seeds, soil | Barley stripe mosaic virus (BSMV) | Barley, wheat, other gramineous plants | + | [73,74,75] |
| Benyvirus, Flexiviridae | Filamentous multipartite | ssRNA (+), total ~15–16 kb | Polymyxa betae (soil) | Beet necrotic yellow vein virus (BNYVV) | Chenopodiaceae, Amaranthaceae, Caryophyllaceae | + | [76] |
| Tobamovirus, Virgaviridae | Rod-shaped monopartite | ssRNA (+), 6.4 kb | Mechanical | Tomato mosaic virus (ToMV) | Over 200 species | + | [77,78] |
| Tobacco mosaic virus (TMV) | Over 125 species | + | [79] | ||||
| Cytorhabdovirus, Rhabdoviridae | Bullet-shaped monopartite | ssRNA (-), 12.7 kb | Planthoppers | Barley yellow striate mosaic virus (BYSMV) | Poaceae | + | [80] |
| Nucleorhabdovirus, Rhabdoviridae | Bullet-shaped monopartite | ssRNA (-), ~13 kb | Aphids | Sonchus yellow net virus (SYNV) | Compositae, Solanaceae, and Chenopodiaceae families | + | [81] |
| ssRNA (-), ~14 kb | Leafhoppers, mechanical | Eggplant mottled dwarf virus (EMDV) | Wide host range (Solanaceae, Cucurbitaceae, Chenopodiaceae, Amaranthaceae, Malvaceae, Hydrangeaceae, Caprifoliaceae, Geraniaceae, Pittosporaceae) | + | [82] | ||
| Comovirus, Secoviridae | Icosahedral bipartite | ssRNA (+); RNA1, ~5.8 kb; RNA2, ~3.5 kb | Leaf-feeding beetles, mechanical, seeds | Cowpea mosaic virus (CPMV) | Fabaceae | - | [24,83] |
| ssRNA (+); RNA1, ~6.8 kb; RNA2, ~3.3 kb | Nematodes, seeds | Apple latent spherical virus (ALSV) | Caryophyllaceae, Chenopodiaceae, Cryptomeria, Fabaceae, Cucurbitaceae, Gentianaceae, Pinus, Rosaceae, Rutaceae, Solanaceae, Arabidopsis | + | [57,84,85] | ||
| Nepovirus, Secoviridae | Icosahedral bipartite | ssRNA (+), 4.8 and 7.2 kb | Nematodes, thrips, seeds, mechanical | Tobacco ringspot virus (TRSV) | Broad host range (Solanaceae, Fabaceae, Cucurbitaceae, woody plants) | + | [57,86] |
| Caulimovirus, Caulimoviridae | Icosahedral monopartite | dsDNA, 8 kb | Aphids, seeds | Cauliflower mosaic virus (CaMV) | Brassicaceae | - | [87] |
| Caulimovirus, Tungrovirus | Bacilliform circular monopartite | dsDNA, ~8.3 kb | Leafhoppers | Rice tungro bacilliform virus (RTBV) | Rice | - | [88,89] |
| Cucumovirus, Bromoviridae | Icosahedral tripartite | ssRNA (+), total ~8.6 kb | Aphids, seeds | Cucumber mosaic virus (CMV) | Over 1300 species | - | [90,91] |
| Bromovirus, Bromoviridae | Icosahedral tripartite | ssRNA (+), total ~8.6 kb | Mechanical, spotted cucumber, beetles, nematodes | Brome mosaic virus (BMV) | Monocotyledonous cereal crops | - | [92,93] |
| Potyvirus, Tritimovirus | Filamentous monopartite | ssRNA (+), ~9.4 kb | Mites, mechanical, low rates of seed transmission | Wheat streak mosaic virus (WSMV) | Poaceae | - | [94] |
| Potyvirus | Filamentous monopartite | ssRNA (+), ~9.8 kb | Aphids, mechanical | Plum pox virus (PPV) | Prunus | - | [95] |
| ssRNA (+), ~9.6 kb | Clover yellow vein virus (CYVV) | Fabaceae | - | [96] | |||
| ssRNA (+), ~9.5 kb | Tobacco etch virus (TEV) | Solanaceae | + | [97] | |||
| Potyvirus, Poacevirus | Filamentous monopartite | ssRNA (+), ~9.5 kb | Mites, mechanical | Triticum mosaic virus (TriMV) | 12 species of grasses | - | [98] |
| Tombusvirus | Icosahedral monopartite | ssRNA (+), ~4.8 kb | Mechanical, soil | Tomato bushy stunt virus (TBSV) | Over 120 plant species across 20 families | + | [99] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikhaylova, E. Virus-Induced Genome Editing (VIGE): One Step Away from an Agricultural Revolution. Int. J. Mol. Sci. 2025, 26, 4599. https://doi.org/10.3390/ijms26104599
Mikhaylova E. Virus-Induced Genome Editing (VIGE): One Step Away from an Agricultural Revolution. International Journal of Molecular Sciences. 2025; 26(10):4599. https://doi.org/10.3390/ijms26104599
Chicago/Turabian StyleMikhaylova, Elena. 2025. "Virus-Induced Genome Editing (VIGE): One Step Away from an Agricultural Revolution" International Journal of Molecular Sciences 26, no. 10: 4599. https://doi.org/10.3390/ijms26104599
APA StyleMikhaylova, E. (2025). Virus-Induced Genome Editing (VIGE): One Step Away from an Agricultural Revolution. International Journal of Molecular Sciences, 26(10), 4599. https://doi.org/10.3390/ijms26104599







