DUT (p.Y116C)-Mutation-Induced Thrombocytopenia in Rabbits
Abstract
1. Introduction
2. Results
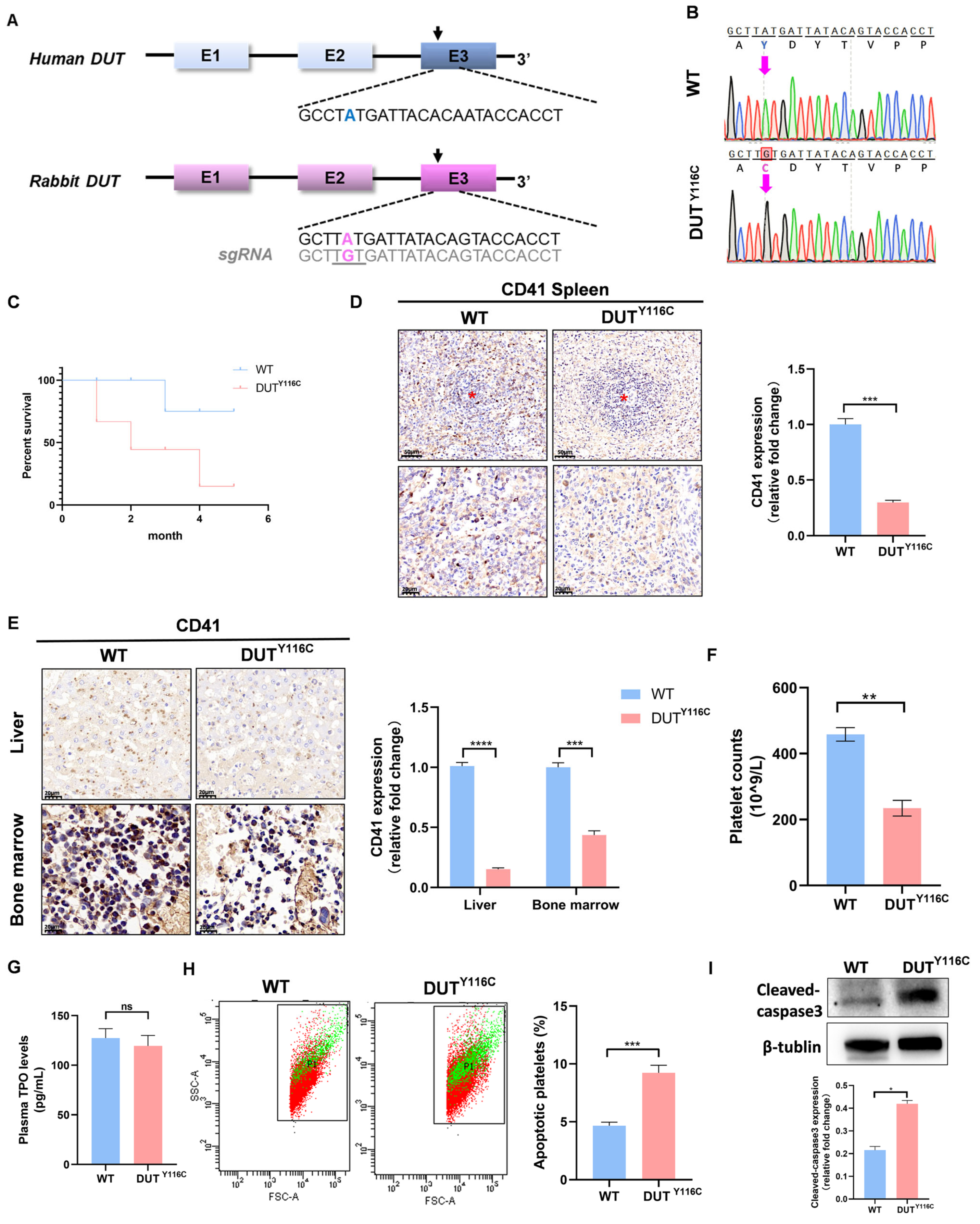
2.1. Generation of DUT (p.Y116C) Rabbits of Thrombocytopenia Using SpRY-ABEmax
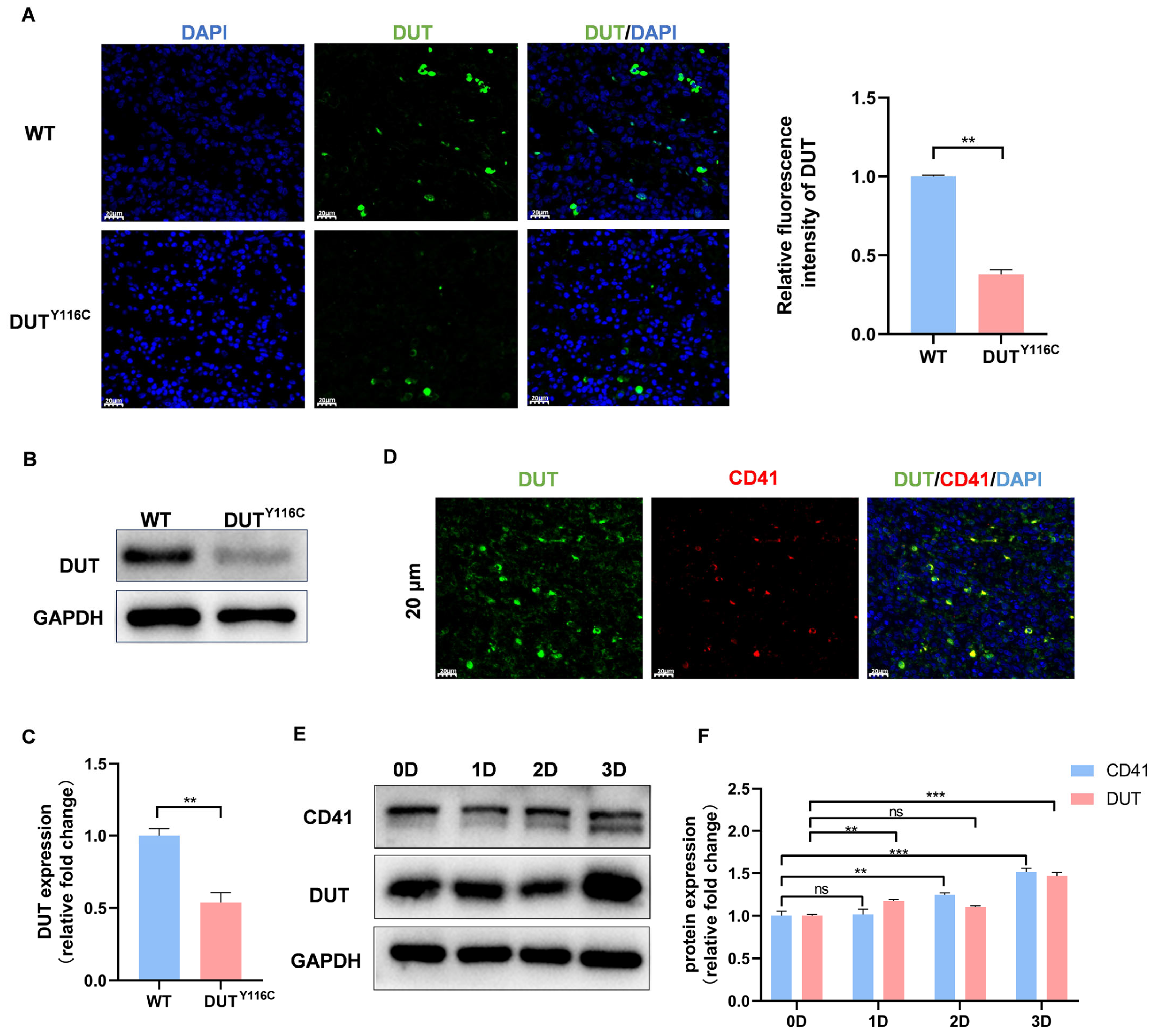
2.2. DUT (p.Y116C) Is a Reduced Mutation and Is Highly Expressed in Megakaryocytes
2.3. DUT Mutations Disruption of Platelet Function in Rabbits
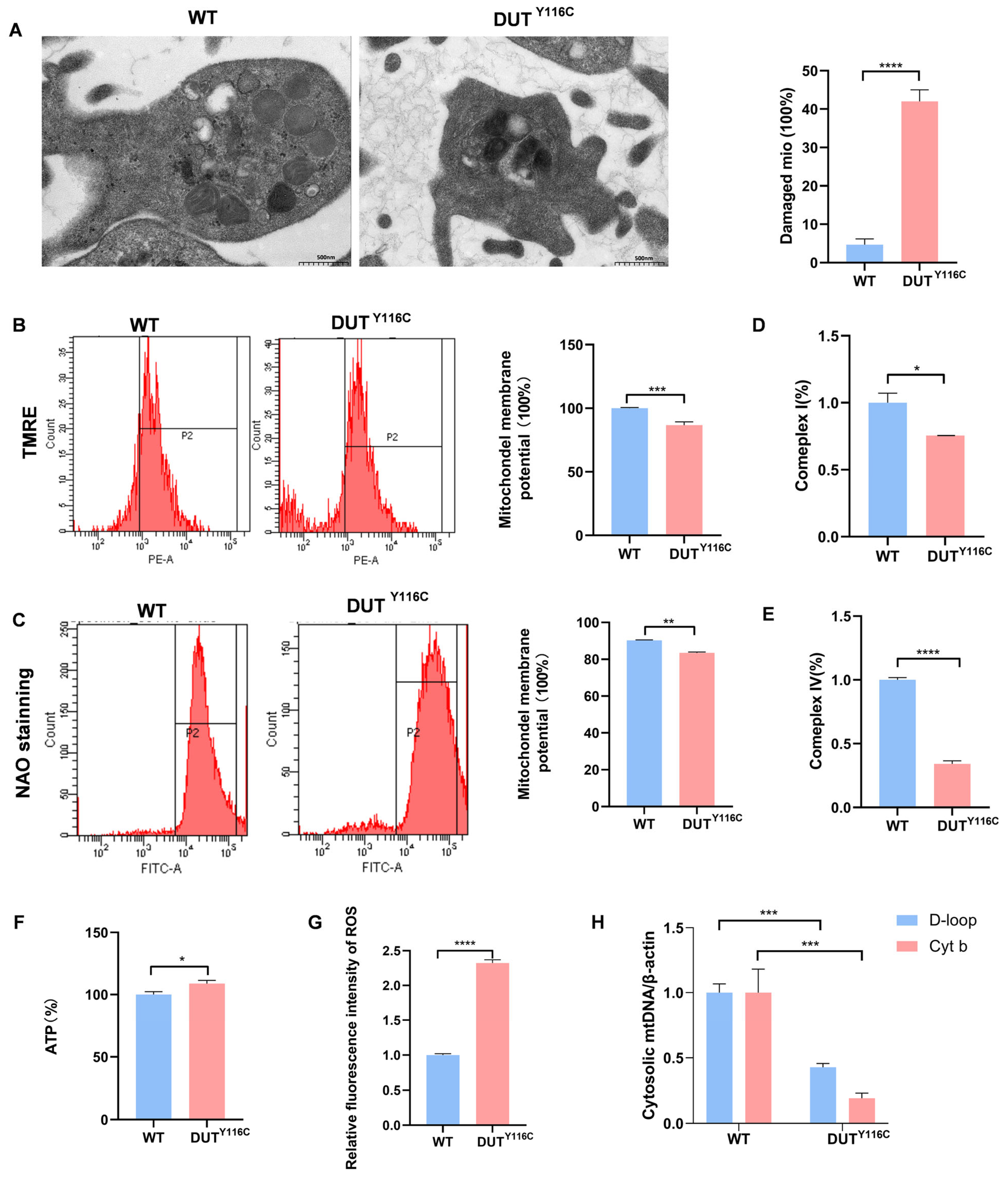
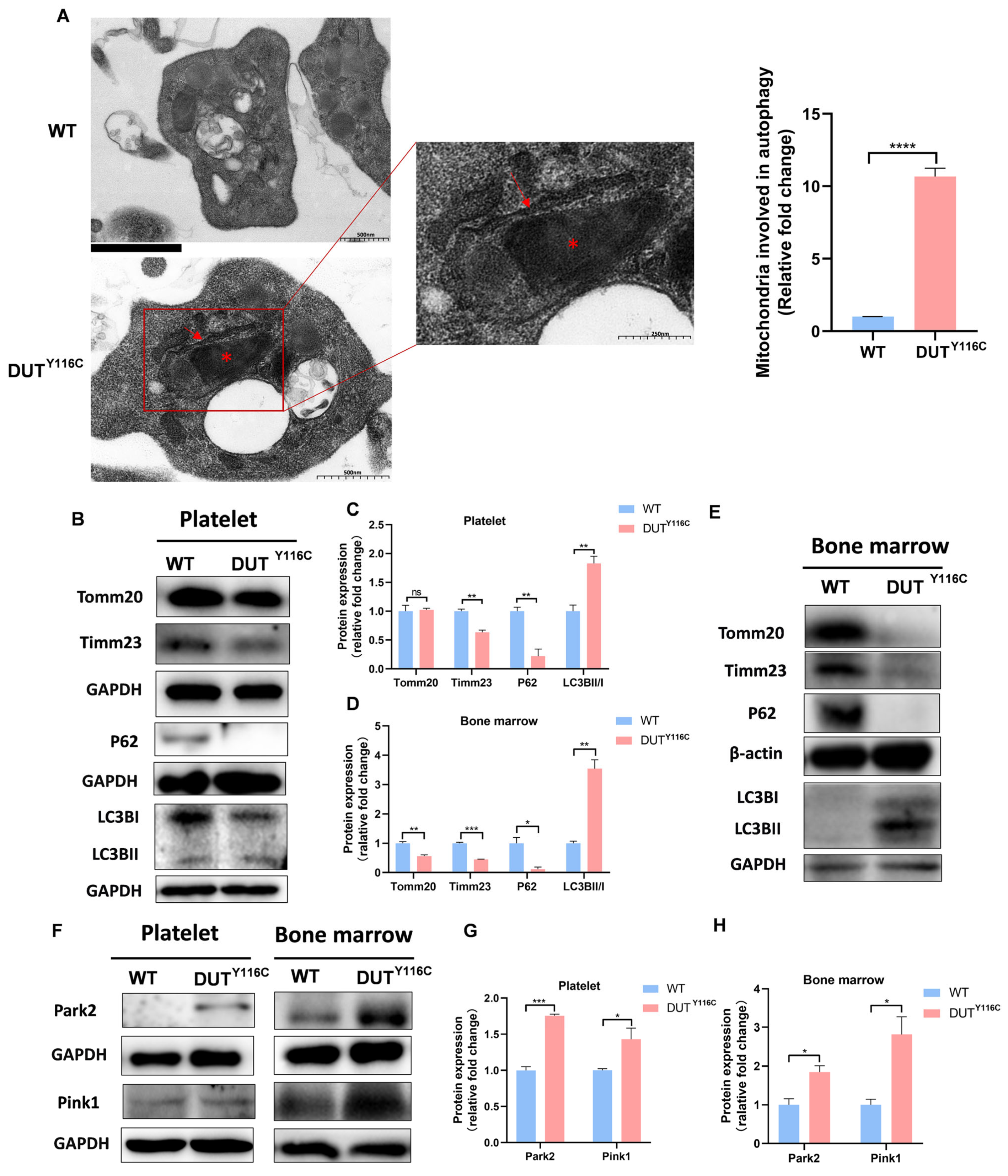
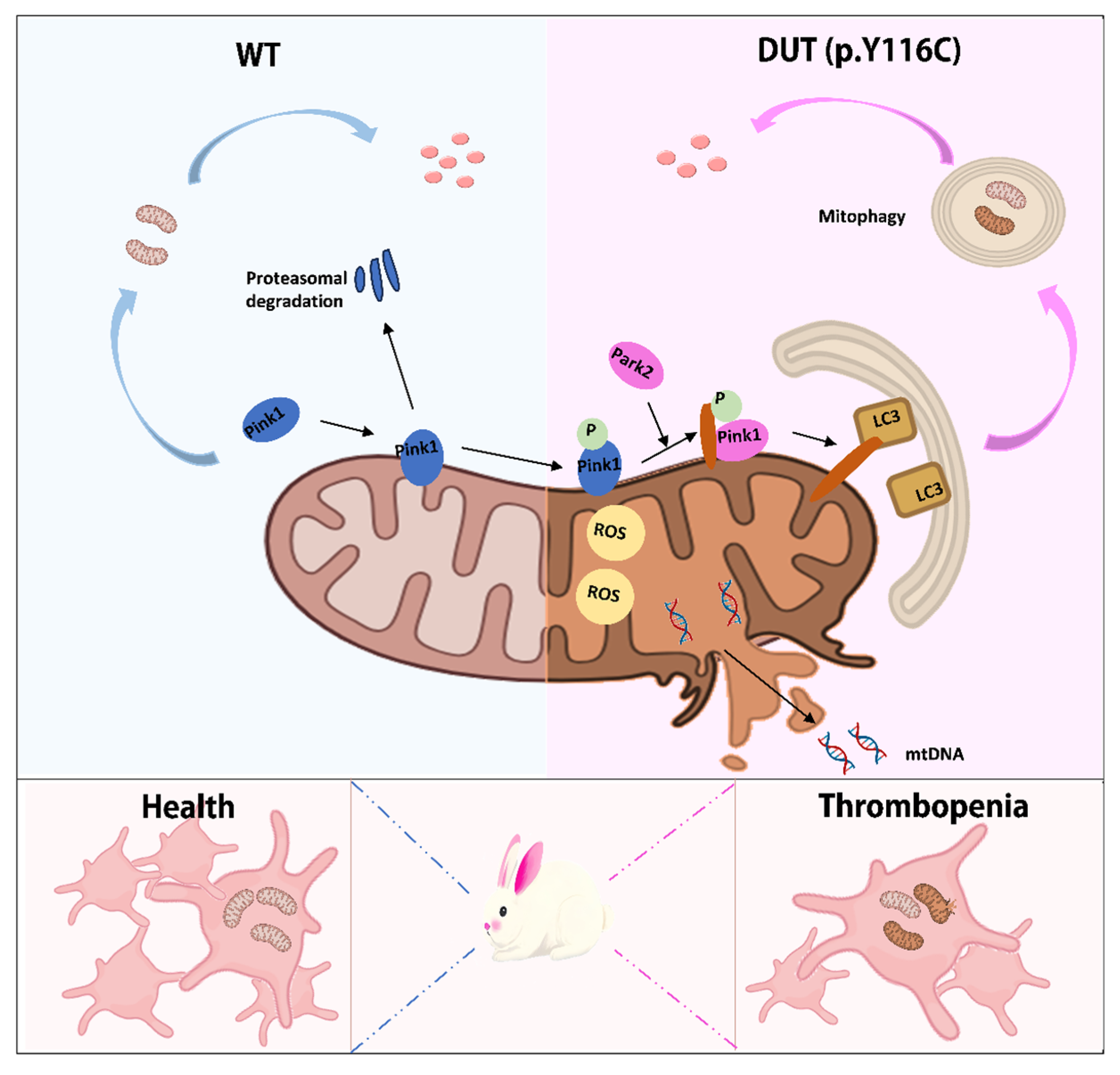
2.4. DUT Mutations Lead to Increased Mitophagy in Rabbits
3. Discussion
4. Materials and Methods
4.1. Animals and Ethics Statement
4.2. Experimental Environment
4.3. Vector Construction and In Vitro Transcription
4.4. Embryo Collection, Microinjection, and Transfer
4.5. Complete Blood Count
4.6. Platelet Preparation
4.7. Western Blots
4.8. Immunohistochemical Staining of Rabbits Femur and Spleen
4.9. Platelet Annexin V Staining
4.10. Detection of TPO in Serum
4.11. Mitophagy Analysis by Electron Microscopy
4.12. Mitochondrial Inner Transmembrane Potential and Mitochondrial Mass Assay
4.13. ATP and ROS Assay
4.14. mtDNA Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, W.; Ma, Q.; Siraj, S.; Ney, P.A.; Liu, J.; Liao, X.; Yuan, Y.; Li, W.; Liu, L.; Chen, Q. Nix-Mediated Mitophagy Regulates Platelet Activation and Life Span. Blood Adv. 2019, 3, 2342–2354. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C. A Mitochondrial Paradigm of Metabolic and Degenerative Diseases, Aging, and Cancer: A Dawn for Evolutionary Medicine. Annu. Rev. Genet. 2005, 39, 359–407. [Google Scholar] [CrossRef]
- Zhang, W.; Ren, H.; Xu, C.; Zhu, C.; Wu, H.; Liu, D.; Wang, J.; Liu, L.; Li, W.; Ma, Q.; et al. Hypoxic Mitophagy Regulates Mitochondrial Quality and Platelet Activation and Determines Severity of I/R Heart Injury. eLife 2016, 5, e21407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Siraj, S.; Zhang, R.; Chen, Q. Mitophagy Receptor FUNDC1 Regulates Mitochondrial Homeostasis and Protects the Heart from I/R Injury. Autophagy 2017, 13, 1080–1081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, C.; Wang, J.; Liu, L.; He, Y.; Chen, Q. Mitophagy in Cardiomyocytes and in Platelets: A Major Mechanism of Cardioprotection Against Ischemia/Reperfusion Injury. Physiology 2018, 33, 86–98. [Google Scholar] [CrossRef]
- Wong, Y.C.; Ysselstein, D.; Krainc, D. Mitochondria–Lysosome Contacts Regulate Mitochondrial Fission via RAB7 GTP Hydrolysis. Nature 2018, 554, 382–386. [Google Scholar] [CrossRef]
- Liu, L.; Sakakibara, K.; Chen, Q.; Okamoto, K. Receptor-Mediated Mitophagy in Yeast and Mammalian Systems. Cell Res. 2014, 24, 787–795. [Google Scholar] [CrossRef]
- Pickrell, A.M.; Youle, R.J. The Roles of PINK1, Parkin, and Mitochondrial Fidelity in Parkinson’s Disease. Neuron 2015, 85, 257–273. [Google Scholar] [CrossRef]
- Yamaguchi, O.; Murakawa, T.; Nishida, K.; Otsu, K. Receptor-Mediated Mitophagy. J. Mol. Cell. Cardiol. 2016, 95, 50–56. [Google Scholar] [CrossRef]
- Ohsumi, Y. Historical Landmarks of Autophagy Research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef]
- Ouseph, M.M.; Huang, Y.; Banerjee, M.; Joshi, S.; MacDonald, L.; Zhong, Y.; Liu, H.; Li, X.; Xiang, B.; Zhang, G.; et al. Autophagy Is Induced upon Platelet Activation and Is Essential for Hemostasis and Thrombosis. Blood 2015, 126, 1224–1233. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Rodriguez-Enriquez, S.; Lemasters, J.J. Selective Degradation of Mitochondria by Mitophagy. Arch. Biochem. Biophys. 2007, 462, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Chiang, W.-C.; Sumpter, R.; Mishra, P.; Levine, B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 2017, 168, 224–238.e10. [Google Scholar] [CrossRef]
- Gong, G.; Song, M.; Csordas, G.; Kelly, D.P.; Matkovich, S.J.; Dorn, G.W. Parkin-Mediated Mitophagy Directs Perinatal Cardiac Metabolic Maturation in Mice. Science 2015, 350, aad2459. [Google Scholar] [CrossRef]
- Murakawa, T.; Yamaguchi, O.; Hashimoto, A.; Hikoso, S.; Takeda, T.; Oka, T.; Yasui, H.; Ueda, H.; Akazawa, Y.; Nakayama, H.; et al. Bcl-2-like Protein 13 Is a Mammalian Atg32 Homologue That Mediates Mitophagy and Mitochondrial Fragmentation. Nat. Commun. 2015, 6, 7527. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, R.S.; Daures, M.; Philippi, A.; Romero, S.; Marselli, L.; Marchetti, P.; Senée, V.; Bacq, D.; Besse, C.; Baz, B.; et al. dUTPase (DUT) Is Mutated in a Novel Monogenic Syndrome With Diabetes and Bone Marrow Failure. Diabetes 2017, 66, 1086–1096. [Google Scholar] [CrossRef]
- Ghawil, M.; Abdulrahman, F.; Hadeed, I.; Doggah, M.; Zarroug, S.; Habeb, A. Further Evidence Supporting the Role of DUT Gene in Diabetes with Bone Marrow Failure Syndrome. Am. J. Med. Genet. A. 2022, 188, 2406–2412. [Google Scholar] [CrossRef] [PubMed]
- Nagy, N.; Hádinger, N.; Tóth, O.; Rácz, G.A.; Pintér, T.; Gál, Z.; Urbán, M.; Gócza, E.; Hiripi, L.; Acsády, L.; et al. Characterization of dUTPase Expression in Mouse Postnatal Development and Adult Neurogenesis. Sci. Rep. 2024, 14, 13139. [Google Scholar] [CrossRef]
- Muñoz-Moreno, E.; Arbat-Plana, A.; Batalle, D.; Soria, G.; Illa, M.; Prats-Galino, A.; Eixarch, E.; Gratacos, E. A Magnetic Resonance Image Based Atlas of the Rabbit Brain for Automatic Parcellation. PLoS ONE 2013, 8, e67418. [Google Scholar] [CrossRef]
- Honda, A.; Ogura, A. Rabbit Models for Biomedical Research Revisited via Genome Editing Approaches. J. Reprod. Dev. 2017, 63, 435–438. [Google Scholar] [CrossRef]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained Genome Targeting with Near-PAMless Engineered CRISPR-Cas9 Variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Qian, Y.; Li, J.; Li, Z.; Lai, L. Highly Efficient A-to-G Base Editing by ABE8.17 in Rabbits. Mol. Ther.-Nucleic Acids 2022, 27, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-J.; Hoffmeister, K.M.; Hu, Z.; Mager, D.E.; Ait-Oudhia, S.; Debrincat, M.A.; Pleines, I.; Josefsson, E.C.; Kile, B.T.; Italiano, J.; et al. Expansion of the Neonatal Platelet Mass Is Achieved via an Extension of Platelet Lifespan. Blood 2014, 123, 3381–3389. [Google Scholar] [CrossRef] [PubMed]
- Szabó, J.E.; Nyíri, K.; Andrási, D.; Matejka, J.; Ozohanics, O.; Vértessy, B. Redox Status of Cysteines Does Not Alter Functional Properties of Human dUTPase but the Y54C Mutation Involved in Monogenic Diabetes Decreases Protein Stability. Sci. Rep. 2021, 11, 19197. [Google Scholar] [CrossRef]
- Smock, K.J.; Perkins, S.L. Thrombocytopenia: An Update. Int. J. Lab. Hematol. 2014, 36, 269–278. [Google Scholar] [CrossRef]
- Song, Y.; Yuan, L.; Wang, Y.; Chen, M.; Deng, J.; Lv, Q.; Sui, T.; Li, Z.; Lai, L. Efficient Dual sgRNA-Directed Large Gene Deletion in Rabbit with CRISPR/Cas9 System. Cell. Mol. Life Sci. 2016, 73, 2959–2968. [Google Scholar] [CrossRef]
- Song, H.; Li, J.; Peng, C.; Liu, D.; Mei, Z.; Yang, Z.; Tian, X.; Zhang, X.; Jing, Q.; Yan, C.; et al. The Role of CREG1 in Megakaryocyte Maturation and Thrombocytopoiesis. Int. J. Biol. Sci. 2023, 19, 3614–3627. [Google Scholar] [CrossRef]
- Chung, Y.; Bishop, C.E.; Treff, N.R.; Walker, S.J.; Sandler, V.M.; Becker, S.; Klimanskaya, I.; Wun, W.-S.; Dunn, R.; Hall, R.M.; et al. Reprogramming of Human Somatic Cells Using Human and Animal Oocytes. Cloning Stem Cells 2009, 11, 213–223. [Google Scholar] [CrossRef]
- Seol, D.; Choe, H.; Zheng, H.; Jang, K.; Ramakrishnan, P.S.; Lim, T.-H.; Martin, J.A. Selection of Reference Genes for Normalization of Quantitative Real-Time PCR in Organ Culture of the Rat and Rabbit Intervertebral Disc. BMC Res. Notes 2011, 4, 162. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, M.; Yang, S.; Liu, R.; Wu, X.; Jiang, L.; Yang, J.; Liu, X.; Wang, G.; Mu, C.; Wang, X.; et al. DUT (p.Y116C)-Mutation-Induced Thrombocytopenia in Rabbits. Int. J. Mol. Sci. 2025, 26, 4169. https://doi.org/10.3390/ijms26094169
Fang M, Yang S, Liu R, Wu X, Jiang L, Yang J, Liu X, Wang G, Mu C, Wang X, et al. DUT (p.Y116C)-Mutation-Induced Thrombocytopenia in Rabbits. International Journal of Molecular Sciences. 2025; 26(9):4169. https://doi.org/10.3390/ijms26094169
Chicago/Turabian StyleFang, Mengmeng, Shujun Yang, Ruonan Liu, Xinyu Wu, Liqiang Jiang, Jie Yang, Xin Liu, Gerong Wang, Chao Mu, Xiuwen Wang, and et al. 2025. "DUT (p.Y116C)-Mutation-Induced Thrombocytopenia in Rabbits" International Journal of Molecular Sciences 26, no. 9: 4169. https://doi.org/10.3390/ijms26094169
APA StyleFang, M., Yang, S., Liu, R., Wu, X., Jiang, L., Yang, J., Liu, X., Wang, G., Mu, C., Wang, X., & Song, Y. (2025). DUT (p.Y116C)-Mutation-Induced Thrombocytopenia in Rabbits. International Journal of Molecular Sciences, 26(9), 4169. https://doi.org/10.3390/ijms26094169



