Molecular Dialog of Ralstonia solanacearum and Plant Hosts with Highlights on Type III Effectors
Abstract
1. Introduction
2. The Complex Evolutionary History of R. solanacearum
3. Main Virulence Factors of R. solanacearum
4. Type III Effectors Secreted by Type III Secretion
5. Basic Characteristics of T3Es
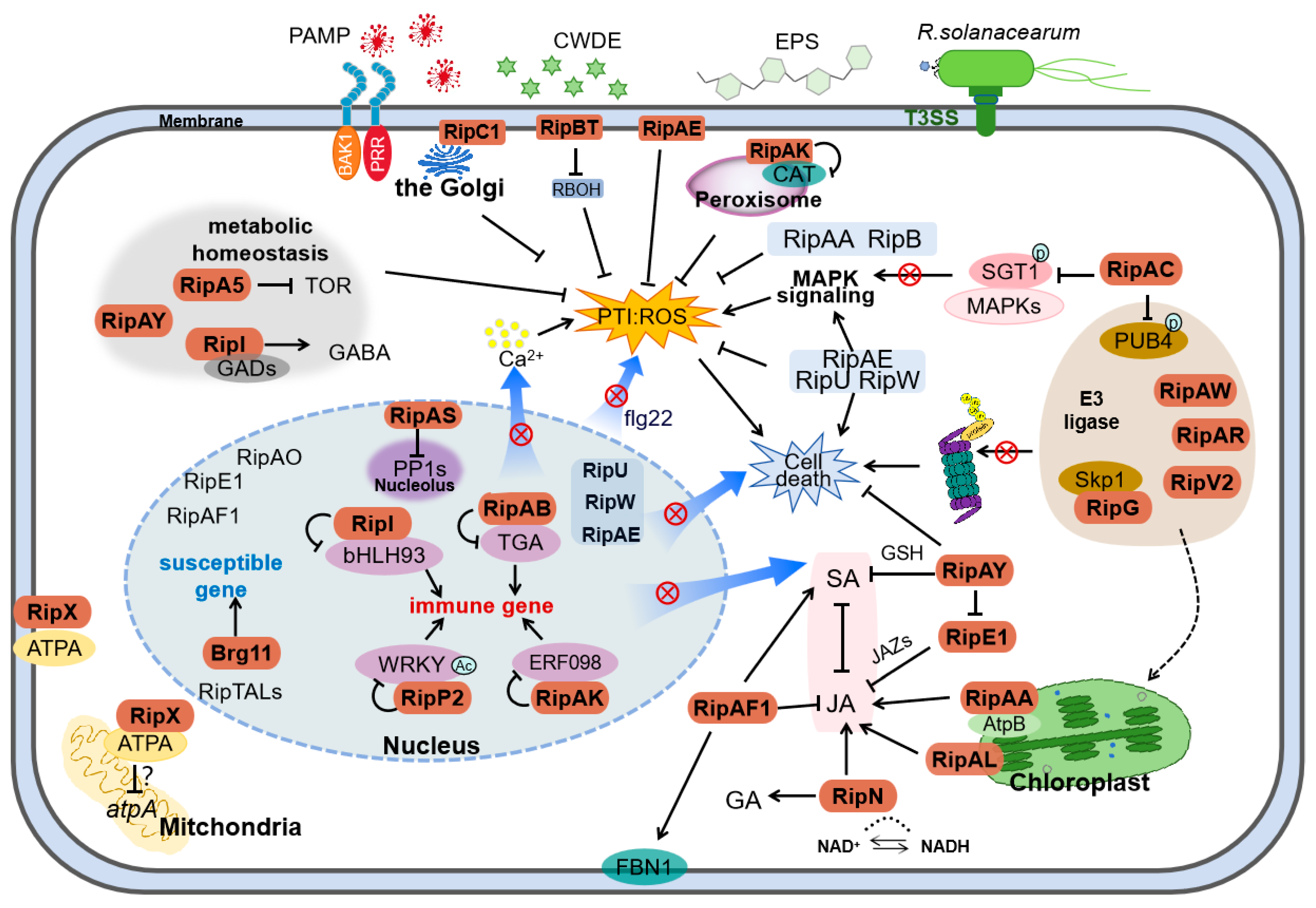
6. Molecular Dialog Between T3Es and Plants
6.1. The Virulence Caused by T3Es
6.1.1. Modulation of Host Plant Protein Metabolism
6.1.2. Subversion of Host Plant Transcription
6.1.3. Manipulation of Plant Hormone Networks
6.1.4. Impact on Host Plant ROS Homeostasis, HR Response, and Beyond
6.2. Host Resistance Mediated by Recognition of T3Es
6.2.1. Avirulence Effectors and Their Roles in HR and ETI-Mediated Host Resistance
6.2.2. Recognition of Type III Effectors by NLRs
7. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Wang, Z.; Luo, W. Ralstonia solanacearum—A soil borne hidden enemy of plants: Research development in management strategies, their action mechanism and challenges. Front. Plant Sci. 2023, 14, 1141902. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, D. The sequevar distribution of Ralstonia solanacearum in tobacco-growing zones of China is structured by elevation. Eur. J. Plant Pathol. 2016, 147, 541–551. [Google Scholar] [CrossRef]
- Ling, L.; Han, X. A Streptomyces sp. NEAU-HV9: Isolation, Identification, and Potential as a Biocontrol Agent against Ralstonia solanacearum of Tomato Plants. Microorganisms 2020, 8, 351. [Google Scholar] [CrossRef]
- Du, H.; Chen, B. Evaluation of Ralstonia solanacearum Infection Dynamics in Resistant and Susceptible Pepper Lines Using Bioluminescence Imaging. Plant Dis. 2017, 101, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Genin, S.; Denny, T.P. Pathogenomics of the Ralstonia solanacearum Species Complex. Annu. Rev. Phytopathol. 2012, 50, 67–89. [Google Scholar] [CrossRef]
- Xu, J.; Pan, Z.C. Genetic diversity of Ralstonia solanacearum strains from China. Eur. J. Plant Pathol. 2009, 125, 641–653. [Google Scholar] [CrossRef]
- Patil, V.U.; Gopal, J. Improvement for Bacterial Wilt Resistance in Potato By Conventional and Biotechnological Approaches. Agric. Res. 2012, 1, 299–316. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Lolle, S.; Stevens, D. Plant NLR-triggered immunity: From receptor activation to downstream signaling. Curr. Opin. Immunol. 2020, 62, 99–105. [Google Scholar] [CrossRef]
- Jacob, F.; Vernaldi, S. Evolution and Conservation of Plant NLR Functions. Front. Immunol. 2013, 4, 297. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Jiang, Y. Manipulation of plant metabolism by pathogen effectors: More than just food. FEMS Microbiol. Rev. 2023, 47, fuad007. [Google Scholar] [CrossRef]
- Hayward, A.C. Characteristics of Pseudomonas solanacearum. J. Appl. Bacteriol. 1964, 27, 265–277. [Google Scholar] [CrossRef]
- Hayward, A.C. Biology and epidemiology of bacterial wilt caused by pseudomonas solanacearum. Annu. Rev. Phytopathol. 1991, 29, 65–87. [Google Scholar] [CrossRef]
- Hayward, A.C. Systematics and Phylogeny of Pseudomonas Solanacearum and Related Bacteria, Bacterial Wilt: The Disease and Its Causative Agent, Pseudomonas Solanacearum; CAB International: Wallingford, UK, 1994; pp. 123–135. [Google Scholar]
- Cook, D.; Sequeira, L. Strain differentiation of Pseudomonas solanacearum by molecular genetic methods. In Bacterial Wilt: The Disease and Its Causative Agent, Pseudomonas Solanacearum; Hayward, A.C., Hartman, G.L., Eds.; CAB International: Wallingford, UK, 1994; pp. 77–93. [Google Scholar]
- Barlow, E.; Cook, D. Genetic Diversity of Pseudomonas solanacearum: Detection of Restriction Fragment Length Polymorphisms with DNA Probes That Specify Virulence and the Hypersensitive Response. Plant-Microbe Interact. 1989, 2, 113. [Google Scholar]
- Fegan, M.; Prior, P. How Complex is the “Ralstonia solanacearum Species Complex”. Bact. Wilt Dis. Ralstonia 2005, 1, 449–461. [Google Scholar]
- Castillo, J.A.; Greenberg, J.T. Evolutionary Dynamics of Ralstonia solanacearum. Appl. Environ. Microbiol. 2007, 73, 1225–1238. [Google Scholar] [CrossRef] [PubMed]
- Safni, I.; Cleenwerck, I. Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: Proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 3087–3103. [Google Scholar]
- Wicker, E.; Lefeuvre, P. Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME 2011, 6, 961–974. [Google Scholar] [CrossRef]
- Sharma, P.; Johnson, M.A. Meta-analysis of the Ralstonia solanacearum species complex (RSSC) based on comparative evolutionary genomics and reverse ecology. Microb. Genom. 2022, 8, 000791. [Google Scholar] [CrossRef]
- Irda, S.; Siti, S. Ecology, Epidemiology and Disease Management ofRalstonia syzygiiin Indonesia. Front. Microbiol. 2018, 9, 419. [Google Scholar]
- Peeters, N.; Guidot, A. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 2013, 14, 651–662. [Google Scholar] [CrossRef]
- Kavitake, D.; Tiwari, S. Antipathogenic potentials of exopolysaccharides produced by lactic acid bacteria and their food and health applications. Food Control 2023, 152, 109850. [Google Scholar] [CrossRef]
- Li, J.; Chu, L.T. Lectins and polysaccharide EPS I have flow-responsive roles in the attachment and biofilm mechanics of plant pathogenic Ralstonia. PLoS Pathog. 2024, 20, e1012358. [Google Scholar]
- Dalsing, B.L.; Allen, C. Nitrate assimilation contributes to Ralstonia solanacearum root attachment, stem colonization, and virulence. J. Bacteriol. 2014, 196, 949–960. [Google Scholar] [CrossRef] [PubMed]
- An, Y.Y.; Zhang, M.X. Advances in understanding the plant-Ralstonia solanacearum interactions. New Crops 2024, 1, 100014. [Google Scholar] [CrossRef]
- Milling, A.; Babujee, L. Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS ONE 2011, 6, e15853. [Google Scholar] [CrossRef]
- Hayashi, K.; Senuma, W. Major exopolysaccharide, EPS I, is associated with the feedback loop in the quorum sensing of Ralstonia solanacearum strain OE1-1. Mol. Plant Pathol. 2019, 20, 1740–1747. [Google Scholar] [CrossRef]
- Denny, T.P. Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 1991, 4, 293–300. [Google Scholar] [CrossRef]
- Tans-Kersten, J.; Huang, H.Y. Ralstonia solanacearum Needs Motility for Invasive Virulence on Tomato. Bacteriology 2001, 183, 3597–3605. [Google Scholar] [CrossRef]
- Yao, J.; Allen, C. Chemotaxis Is Required for Virulence and Competitive Fitness of the Bacterial Wilt Pathogen Ralstonia solanacearum. J. Bacteriol. 2006, 188, 3697–3708. [Google Scholar] [CrossRef] [PubMed]
- Corral, J.; Sebastià, P. Twitching and Swimming Motility Play a Role in Ralstonia solanacearum Pathogenicity. mSphere 2020, 5, 10–1128. [Google Scholar] [CrossRef]
- Juhala, R.J.; Ford, M.E. Genomic sequences of bacteriophages HK97 and HK022: Pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 2000, 299, 27–51. [Google Scholar] [CrossRef] [PubMed]
- Billard-Pomares, T.; Fouteau, S. Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum β-lactamase from an Escherichia coli strain. Antimicrob. Agents Chemother. 2014, 58, 6550–6557. [Google Scholar] [CrossRef]
- Frazão, N.; Sousa, A. Horizontal gene transfer overrides mutation in Escherichia coli colonizing the mammalian gut. Proc. Natl. Acad. Sci. USA 2019, 116, 17906–17915. [Google Scholar] [CrossRef]
- Busby, B.; Kristensen, D.M. Contribution of phage-derived genomic islands to the virulence of facultative bacterial pathogens. Environ. Microbiol. 2013, 15, 307–312. [Google Scholar] [CrossRef]
- Hacker, J.; Carniel, E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Reports 2001, 2, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Greenrod, S.T.E.; Stoycheva, M. Global diversity and distribution of prophages are lineage-specific within the Ralstonia solanacearum species complex. BMC Genomics 2022, 23, 689. [Google Scholar] [CrossRef]
- Ahmad, A.A.; Stulberg, M.J. Molecular and biological characterization of ϕRs551, a filamentous bacteriophage isolated from a race 3 biovar 2 strain of Ralstonia solanacearum. PLoS ONE 2017, 12, e0185034. [Google Scholar] [CrossRef]
- Peyraud, R.; Cottret, L. A Resource Allocation Trade-Off between Virulence and Proliferation Drives Metabolic Versatility in the Plant Pathogen Ralstonia solanacearum. PLoS Pathog. 2016, 12, e1005939. [Google Scholar] [CrossRef]
- Gonçalves, O.S.; Souza, F.D.O. Widespread distribution of prophages signaling the potential for adaptability and pathogenicity evolution of Ralstonia solanacearum species complex. Genomics 2021, 113, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Wanjiru, W.M.; Kang, Z. Importance of Cell Wall Degrading Enzymes Produced by Fusarium graminearum during Infection of Wheat Heads. Plant Pathol. 2002, 108, 803–810. [Google Scholar] [CrossRef]
- Lev, S.; Horwitz, B.A. A Mitogen-activated protein kinase pathway modulates the expression of two cellulase genes in Cochliobolus heterostrophus during plant infection. Plant Cell 2003, 15, 835–844. [Google Scholar] [CrossRef]
- Niture, S.K.; Kumar, A.R. Role of glucose in production and repression of polygalacturonase and pectate lyase from phytopathogenic fungus fusarium moniliforme NCIM 1276. World J. Microbiol. Biotechnol. 2006, 22, 893–899. [Google Scholar] [CrossRef]
- Shi, H.Q.; Liu, Y. Induced defense strategies of plants against Ralstonia solanacearum. Front. Microbiol. 2023, 14, 1059799. [Google Scholar] [CrossRef] [PubMed]
- Gigli-Bisceglia, N.; Engelsdorf, T. Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell. Mol. Life Sci. 2020, 77, 2049–2077. [Google Scholar] [CrossRef]
- Wan, J.; He, M. Cell wall associated immunity in plants. Stress. Biol. 2021, 1, 3. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Demange, N.; Gaspin, C. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 2002, 415, 497–502. [Google Scholar]
- De Chial, M.; Ghysels, B. Identification of type II and type III pyoverdine receptors from Pseudomonas aeruginosa. Microbiology 2003, 149, 821–831. [Google Scholar] [CrossRef]
- Landry, D.; González-Fuente, M. The large, diverse, and robust arsenal of Ralstonia solanacearum type III effectors and their in planta functions. Mol. Plant Pathol. 2020, 21, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- De Ryck, J.; Damme, P.V. From prediction to function: Current practices and challenges towards the functional characterization of type III effectors. Front. Microbiol. 2023, 14, 1113442. [Google Scholar] [CrossRef]
- Sabbagh, C.R.R.; Carrere, S. Pangenomic type III effector database of the plant pathogenic Ralstonia spp. PeerJ 2019, 7, e7346. [Google Scholar] [CrossRef] [PubMed]
- Roux, B.; Bolot, S. Genomics and transcriptomics of Xanthomonas campestris species challenge the concept of core type III effectome. BMC Genom. 2015, 16, 975. [Google Scholar] [CrossRef] [PubMed]
- Dillon, M.M.; Almeida, R.N.D. Molecular Evolution of Pseudomonas syringae Type III Secreted Effector Proteins. Front. Plant Science 2019, 10, 418. [Google Scholar] [CrossRef]
- Peeters, N.; Carrère, S. Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genom. 2013, 14, 859. [Google Scholar] [CrossRef]
- Deslandes, L.; Genin, S. Opening the Ralstonia solanacearum type III effector tool box: Insights into host cell subversion mechanisms. Curr. Opin. Plant Biol. 2014, 20, 110–117. [Google Scholar] [CrossRef]
- Cho, H.; Song, E.S. Prediction of Host-Specific Genes by Pan-Genome Analyses of the Korean Ralstonia solanacearum Species Complex. Front. Microbiol. 2019, 10, 506. [Google Scholar] [CrossRef]
- Cunnac, S.; Occhialini, A. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: Identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 2004, 53, 115–128. [Google Scholar] [CrossRef]
- Delaspre, F.; Peñalver, C.G.N. The Ralstonia solanacearum pathogenicity regulator HrpB induces 3-hydroxy-oxindole synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 15870–15875. [Google Scholar] [CrossRef]
- Occhialini, A.S.; Cunnac, S. Genome-wide analysis of gene expression in Ralstonia solanacearum reveals that the hrpB gene acts as a regulatory switch controlling multiple virulence pathways. Mol. Plant. Microbe Interact. 2005, 18, 938–949. [Google Scholar] [CrossRef]
- Poueymiro, M.; Cazalé, A.C. A Ralstonia solanacearum Type III Effector Directs the Production of the Plant Signal Metabolite Trehalose-6-Phosphate. mBio 2014, 5, e02065-14. [Google Scholar] [CrossRef] [PubMed]
- Mukaihara, T.; Tamura, N. Identification of novel Ralstonia solanacearum type III effector proteins through translocation analysis of hrpB-regulated gene products. Microbiology 2009, 155, 2235–2244. [Google Scholar] [CrossRef]
- Mukaihara, T.; Tamura, N. Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant. Microbe Interact. 2010, 23, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Lonjon, F.; Peeters, N. In Vitro and In Vivo Secretion/Translocation Assays to Identify Novel Ralstonia solanacearum Type 3 Effectors. Methods Mol. Biol. 2018, 1734, 209–222. [Google Scholar]
- Lohou, D.; Turner, M. HpaP modulates type III effector secretion in Ralstonia solanacearum and harbours a substrate specificity switch domain essential for virulence. Mol. Plant Pathol. 2014, 15, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Lonjon, F.; Turner, M. Comparative Secretome Analysis of Ralstonia solanacearum Type 3 Secretion-Associated Mutants Reveals a Fine Control of Effector Delivery, Essential for Bacterial Pathogenicity. Mol. Cell. Proteom. 2016, 15, 598–613. [Google Scholar] [CrossRef]
- Toruño, T.Y.; Stergiopoulos, I. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef]
- Büttner, D. Behind the lines-actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 2016, 40, 894–937. [Google Scholar] [CrossRef]
- Macho, A.P. Subversion of plant cellular functions by bacterial type-III effectors: Beyond suppression of immunity. New Phytol. 2016, 210, 51–57. [Google Scholar] [CrossRef]
- Rohde, J.R.; Breitkreutz, A. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 2007, 1, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Oda, K. Ralstonia solanacearum novel E3 ubiquitin ligase (NEL) effectors RipAW and RipAR suppress pattern-triggered immunity in plants. Microbiology 2017, 163, 992–1002. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.O.; Chen, J. Ubiquitin E3 ligase activity of Ralstonia solanacearum effector RipAW is not essential for induction of plant defense in Nicotiana benthamiana. Front. Microbiol. 2023, 14, 1201444. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Zhou, D. Ralstonia solanacearum type III effector RipV2 encoding a novel E3 ubiquitin ligase (NEL) is required for full virulence by suppressing plant PAMP-triggered immunity. Biochem. Biophys. Res. Commun. 2021, 550, 120–126. [Google Scholar] [CrossRef]
- Dahal, A.; Chen, L. Chloroplastic proteins are targets for the RipG effectors of Ralstonia solanacearum. Int. J. Environ. Technol. Sci. 2018, 5, 147–156. [Google Scholar]
- Tzfira, T.; Vaidya, M. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 2004, 431, 87–92. [Google Scholar] [CrossRef]
- Angot, A.; Peeters, N. Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. USA 2006, 103, 14620–14625. [Google Scholar] [CrossRef]
- Yu, G.; Xian, L. A bacterial effector protein prevents MAPK-mediated phosphorylation of SGT1 to suppress plant immunity. PLoS Pathog. 2020, 16, e1008933. [Google Scholar] [CrossRef]
- Yu, G.; Derkacheva, M. The Arabidopsis E3 ubiquitin ligase PUB4 regulates BIK1 and is targeted by a bacterial type-III effector. EMBO J. 2022, 41, e107257. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y. Arabidopsis PUB2 and PUB4 connect signaling components of pattern-triggered immunity. New Phytol. 2022, 233, 2249–2265. [Google Scholar] [CrossRef]
- Lal, N.K.; Nagalakshmi, U. The Receptor-like Cytoplasmic Kinase BIK1 Localizes to the Nucleus and Regulates Defense Hormone Expression during Plant Innate Immunity. Cell Host Microbe 2018, 23, 485–497.e485. [Google Scholar] [CrossRef]
- Demirjian, C.; Razavi, N. An atypical NLR gene confers bacterial wilt susceptibility in Arabidopsis. Plant Commun. 2023, 4, 100607. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Ichinose, Y. Ralstonia solanacearum Type III Effector RipAC Targets SGT1 to Suppress Effector-Triggered Immunity. Plant Cell Physiol. 2020, 61, 2067–2076. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.W.; Kim, W. Ralstonia solanacearum Type III Effectors with Predicted Nuclear Localization Signal Localize to Various Cell Compartments and Modulate Immune Responses in Nicotiana spp. Plant Pathol. J. 2020, 36, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Huang, M. A Ralstonia solanacearum effector targets TGA transcription factors to subvert salicylic acid signaling. Plant Cell 2022, 34, 1666–1683. [Google Scholar] [CrossRef]
- Zheng, X.; Li, X. A systematic screen of conserved Ralstonia solanacearum effectors reveals the role of RipAB, a nuclear-localized effector that suppresses immune responses in potato. Mol. Plant Pathol. 2019, 20, 547–561. [Google Scholar] [CrossRef]
- Ma, K.W.; Ma, W. YopJ Family Effectors Promote Bacterial Infection through a Unique Acetyltransferase Activity. Microbiol. Mol. Biol. Rev. 2016, 80, 1011–1027. [Google Scholar] [CrossRef]
- Weber, A.N.; Lane, W.S. Serine/threonine acetylation of TGFβ-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 12710–12715. [Google Scholar]
- Mukherjee, S.; Keitany, G. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 2006, 312, 1211–1214. [Google Scholar] [CrossRef]
- Trosky, J.E.; Li, Y. VopA inhibits ATP binding by acetylating the catalytic loop of MAPK kinases. J. Biol. Chem. 2007, 282, 34299–34305. [Google Scholar] [CrossRef]
- Xia, Y.; Zou, R. Secondary-structure switch regulates the substrate binding of a YopJ family acetyltransferase. Nat. Commun. 2021, 12, 5969. [Google Scholar] [CrossRef] [PubMed]
- Roux, C.L.; Huet, G. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 2015, 161, 1074–1088. [Google Scholar]
- Nakano, M.; Mukaihara, T. Comprehensive Identification of PTI Suppressors in Type III Effector Repertoire Reveals that Ralstonia solanacearum Activates Jasmonate Signaling at Two Different Steps. Int. J. Mol. Sci. 2019, 20, 5992. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, T.; Wang, X. The Ralstonia solanacearum effector RipI induces a defence reaction by interacting with the bHLH93 transcription factor in Nicotiana benthamiana. Mol. Plant Pathol. 2020, 21, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- De Lange, O.; Schreiber, T. Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol. 2013, 199, 773–786. [Google Scholar] [CrossRef]
- Wu, D.; von Roepenack-Lahaye, E.; Buntru, M.; de Lange, O.; Schandry, N.; Pérez-Quintero, A.L.; Weinberg, Z.; Lowe-Power, T.M.; Szurek, B.; Michael, A.J.; et al. A Plant Pathogen Type III Effector Protein Subverts Translational Regulation to Boost Host Polyamine Levels. Cell Host Microbe 2019, 26, 638–649.e5. [Google Scholar] [CrossRef]
- Sun, T.W.; Wu, W. Ralstonia solanacearum elicitor RipX Induces Defense Reaction by Suppressing the Mitochondrial atpA Gene in Host Plant. Int. J. Mol. Sci. 2020, 21, 2000. [Google Scholar] [CrossRef]
- Choi, M.S.; Kim, W. Harpins, multifunctional proteins secreted by gram-negative plant-pathogenic bacteria. Mol. Plant. Microbe Interact. 2013, 26, 1115–1122. [Google Scholar] [CrossRef]
- Dong, H.P.; Peng, J. Downstream divergence of the ethylene signaling pathway for harpin-stimulated Arabidopsis growth and insect defense. Plant Physiol. 2004, 136, 3628–3638. [Google Scholar] [CrossRef]
- Ren, X.; Liu, F. Root Growth of Arabidopsis thaliana Is Regulated by Ethylene and Abscisic Acid Signaling Interaction in Response to HrpNEa, a Bacterial Protein of Harpin Group. Plant Mol. Biol. Report. 2008, 26, 225–240. [Google Scholar] [CrossRef]
- Xie, Z.; Chen, Z. Harpin-induced hypersensitive cell death is associated with altered mitochondrial functions in tobacco cells. Mol. Plant-Microbe Interact. 2000, 13, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Ghorbel, M.; Brini, F. Role of jasmonic acid in plants: The molecular point of view. Plant Cell Rep. 2021, 40, 1471–1494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, F. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 2017, 68, 1371–1385. [Google Scholar] [CrossRef]
- Davière, J.M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, P. The Ralstonia solanacearum effector RipN suppresses plant PAMP-triggered immunity, localizes to the endoplasmic reticulum and nucleus, and alters the NADH/NAD+ ratio in Arabidopsis. Mol. Plant Pathol. 2019, 20, 533–546. [Google Scholar] [CrossRef]
- Nguyen, V.N.; Park, A. Substrate specificity characterization for eight putative nudix hydrolases. Evaluation of criteria for substrate identification within the Nudix family. Proteins 2016, 84, 1810–1822. [Google Scholar]
- Srouji, J.R.; Xu, A. The evolution of function within the Nudix homology clan. Proteins 2017, 85, 775–811. [Google Scholar] [CrossRef]
- Dong, Z.; Zheng, X. Comparative transcriptomics and multiple phytohormone profiling reveals the molecular immune response of Arabidopsis thaliana to the pathogen Ralstonia solanacearum type III effector RipN. J. Plant Pathol. 2022, 104, 591–603. [Google Scholar] [CrossRef]
- Wu, W.; Zou, H. Ralstonia solanacearum type III effector RipAF1 mediates plant resistance signaling by ADP-ribosylation of host FBN1. Hortic. Res. 2024, 11, uhae162. [Google Scholar] [CrossRef]
- Youssef, A.; Laizet, Y. Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J. 2010, 61, 436–445. [Google Scholar] [CrossRef]
- Li, J.; Yang, J. Overexpressing OsFBN1 enhances plastoglobule formation, reduces grain-filling percent and jasmonate levels under heat stress in rice. Plant Sci. 2019, 285, 230–238. [Google Scholar] [CrossRef]
- Nakano, M.; Mukaihara, T. Ralstonia solanacearum Type III Effector RipAL Targets Chloroplasts and Induces Jasmonic Acid Production to Suppress Salicylic Acid-Mediated Defense Responses in Plants. Plant Cell Physiol. 2018, 59, 2576–2589. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Yu, W.J. Intra-strain Elicitation and Suppression of Plant Immunity by Ralstonia solanacearum Type-III Effectors in Nicotiana benthamiana. Plant Commun. 2020, 1, 100025. [Google Scholar] [CrossRef]
- Liu, K.; Shi, L. Ralstonia solanacearum effector RipAK suppresses homodimerization of the host transcription factor ERF098 to enhance susceptibility and the sensitivity of pepper plants to dehydration. Plant J. Cell Mol. Biol. 2024, 117, 121–144. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K. Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Chen, J.L. A conserved type III effector RipB is recognized in tobacco and contributes to Ralstonia solanacearum virulence in susceptible host plants. Biochem. Biophys. Res. Commun. 2022, 631, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.C.; Wu, L. Ralstonia solanacearum type III effector RipAA targets chloroplastic AtpB to modulate an incompatible interaction on Nicotiana benthamiana. Front. Microbiol. 2023, 14, 1179824. [Google Scholar] [CrossRef]
- Song, B.T.; Tang, N. A novel effector RipBT contributes to Ralstonia solanacearum virulence on potato. Mol. Plant Pathol. 2023, 24, 947–960. [Google Scholar]
- Cong, S.; Li, J.Z. Diverse interactions of five core type III effectors from Ralstonia solanacearum with plants. Genet. Genom. 2023, 50, 341–352. [Google Scholar] [CrossRef]
- Sun, Y.; Li, P. The Ralstonia solanacearum effector RipAK suppresses plant hypersensitive response by inhibiting the activity of host catalases. Cell Microbiol. 2017, 19, e12736. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, A. A bacterial effector protein uncovers a plant metabolic pathway involved in tolerance to bacterial wilt disease. Mol. Plant 2021, 14, 1281–1296. [Google Scholar] [CrossRef] [PubMed]
- Popa, C.; Li, L. The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Sci. Rep. 2016, 6, 27058. [Google Scholar] [CrossRef]
- Song, Y.; Alyafei, M.S. Contributions of TOR Signaling on Photosynthesis. Int. J. Mol. Sci. 2021, 22, 8959. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Zhang, R. Brassinosteriod Insensitive 2 (BIN2) acts as a downstream effector of the Target of Rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytol. 2017, 213, 233–249. [Google Scholar] [CrossRef]
- Xian, L.; Yu, G. A Bacterial Effector Protein Hijacks Plant Metabolism to Support Pathogen Nutrition. Cell Host Microbe 2020, 28, 548–557.e547. [Google Scholar] [CrossRef]
- Mukaihara, T.; Hatanaka, T. Ralstonia solanacearum Type III Effector RipAY Is a Glutathione-Degrading Enzyme That Is Activated by Plant Cytosolic Thioredoxins and Suppresses Plant Immunity. mBio 2016, 7, e00359-16. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Wang, Y. The Ralstonia solanacearum type III effector RipAY targets plant redox regulators to suppress immune responses. Mol. Plant Pathol. 2016, 19, 129–142. [Google Scholar] [CrossRef]
- Gunawardena, S.R.; Ruis, B.L.; Meyer, J.A.; Kapoor, M.; Conklin, K.F. NOM1 targets protein phosphatase I to the nucleolus. J. Biol. Chem. 2008, 283, 398–404. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, J.; Qin, Q.; Zhang, J.; Chen, Y.; Zhao, L.; He, K.; Hou, S. Type one protein phosphatases (TOPPs) contribute to the plant defense response in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 360–377. [Google Scholar] [CrossRef]
- Wang, B.; Huang, M.; He, W.; Wang, Y.; Yu, L.; Zhou, D.; Meng, C.; Cheng, D.; Qiu, H.; Tan, X.; et al. Protein phosphatase StTOPP6 negatively regulates potato bacterial wilt resistance by modulating MAPK signaling. J. Exp. Bot. 2023, 74, 4208–4224. [Google Scholar] [CrossRef]
- Wang, B.; He, W.; Huang, M.; Feng, J.; Li, Y.; Yu, L.; Wang, Y.; Zhou, D.; Meng, C.; Cheng, D.; et al. Ralstonia solanacearum type III effector RipAS associates with potato type one protein phosphatase StTOPP6 to promote bacterial wilt. Hortic. Res. 2023, 10, uhad087. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.H. The Complementary Genic Systems in Flax and Flax Rust. Adv. Genet. 1956, 8, 29–54. [Google Scholar]
- Dolatabadian, A.; Fernando, W.G.D. Genomic Variations and Mutational Events Associated with Plant-Pathogen Interactions. Biology 2022, 11, 421. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Moon, H. Ralstonia solanacearum Type III Effector RipJ Triggers Bacterial Wilt Resistance in Solanum pimpinellifolium. Mol. Plant-Microbe Interact. 2021, 34, 962–972. [Google Scholar] [CrossRef]
- Su, G.M.; Chu, L.W. Tomato NADPH oxidase SlWfi1 interacts with the effector protein RipBJ of Ralstonia solanacearum to mediate host defence. Plant Cell Environ. 2024, 47, 5007–5020. [Google Scholar] [CrossRef]
- Moon, H.; Pandey, A. Identification of RipAZ1 as an avirulence determinant of Ralstonia solanacearum in Solanum americanum. Mol. Plant Pathol. 2021, 22, 317–333. [Google Scholar] [CrossRef]
- Solé, M.; Popa, C. The awr gene family encodes a novel class of Ralstonia solanacearum type III effectors displaying virulence and avirulence activities. Mol. Plant-Microbe Interact. 2012, 25, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.E.; Wechter, W.P. Relationship between avirulence gene (avrA) diversity in Ralstonia solanacearum and bacterial wilt incidence. Mol. Plant-Microbe Interact. 2004, 17, 1376–1384. [Google Scholar] [CrossRef]
- Liu, Y.; Kanda, A. Distribution of avirulence genes avrA and popP1 in 22 Japanese phylotype I strains of Ralstonia solanacearum. Gen. Appl. Microbiol. 2009, 75, 362–368. [Google Scholar] [CrossRef]
- Poueymiro, M.; Cunnac, S. Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host-range specificity on tobacco. Mol. Plant-Microbe Interact. 2009, 22, 538–550. [Google Scholar] [CrossRef]
- Chen, L.; Dahal, A. Involvement of avirulence genes avrA and popP1 of Japanese Ralstonia solanacearum strains in the pathogenicity to tobacco. Physiol. Mol. Plant Pathol. 2018, 102, 154–162. [Google Scholar] [CrossRef]
- An, Y.; Chen, J. Three amino acid residues are required for the recognition of Ralstonia solanacearum RipTPS in Nicotiana tabacum. Front. Plant Sci. 2022, 13, 1040826. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.R.; Studholme, D.J. Genome-Enabled Phylogeographic Investigation of the Quarantine Pathogen Ralstonia solanacearum Race 3 Biovar 2 and Screening for Sources of Resistance Against Its Core Effectors. Phytopathology 2015, 105, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Tan, X. Comparative genomic analysis of Ralstonia solanacearum reveals candidate avirulence effectors in HA4-1 triggering wild potato immunity. Front. Plant Sci. 2023, 14, 1075042. [Google Scholar] [CrossRef]
- Pensec, F.; Lebeau, A. Towards the Identification of Type III Effectors Associated with Ralstonia solanacearum Virulence on Tomato and Eggplant. Phytopathology 2015, 105, 1529–1544. [Google Scholar] [CrossRef] [PubMed]
- Salgon, S.; Jourda, C. Eggplant Resistance to the Ralstonia solanacearum Species Complex Involves Both Broad-Spectrum and Strain-Specific Quantitative Trait Loci. Front. Plant Sci. 2017, 8, 828. [Google Scholar] [CrossRef]
- Morel, A.; Guinard, J. The eggplant AG91-25 recognizes the Type III-secreted effector RipAX2 to trigger resistance to bacterial wilt (Ralstonia solanacearum species complex). Mol. Plant Pathol. 2018, 19, 2459–2472. [Google Scholar] [CrossRef]
- Nahar, K.; Matsumoto, I. Ralstonia solanacearum type III secretion system effector Rip36 induces a hypersensitive response in the nonhost wild eggplant Solanum torvum. Mol. Plant Pathol. 2013, 15, 297–303. [Google Scholar] [CrossRef]
- Lebeau, A.; Daunay, M.C. Bacterial wilt resistance in tomato, pepper, and eggplant: Genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 2010, 101, 154–165. [Google Scholar] [CrossRef]
- Chen, L.; Lei, N. Contribution of RipS type III effector family of Ralstonia solanacearum Japanese strain OE1-1 to disease development in eggplant. J. Gen. Plant Pathol. 2021, 87, 77–82. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2014, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Ngou, B.P.M.; Ahn, H.K. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Franchi, L.; Warner, N. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009, 227, 106–128. [Google Scholar] [CrossRef] [PubMed]
- Maruta, N.; Burdett, H. Structural basis of NLR activation and innate immune signalling in plants. Immunogenetics 2022, 74, 5–26. [Google Scholar] [CrossRef]
- Ijaz, S.; Haq, I.U. Genome-wide identification, and gene expression analysis of NBS-LRR domain containing R genes in Chenopodium quinoa for unveiling the dynamic contribution in plant immunity against Cercospora cf. chenopodii. Physiol. Mol. Biol. Plants 2024, 30, 1129–1144. [Google Scholar] [CrossRef]
- Monteiro, F.; Nishimura, M.T. Structural, Functional, and Genomic Diversity of Plant NLR Proteins: An Evolved Resource for Rational Engineering of Plant Immunity. Annu. Rev. Phytopathol. 2018, 56, 243–267. [Google Scholar] [CrossRef]
- Sarris, P.; Duxbury, Z. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Segonzac, C. Autoimmunity and effector recognition in Arabidopsis thaliana can be uncoupled by mutations in the RRS1-R immune receptor. New Phytol. 2019, 222, 954–965. [Google Scholar]
- Sohn, K.H.; Segonzac, C. The Nuclear Immune Receptor RPS4 Is Required for RRS1SLH1-Dependent Constitutive Defense Activation in Arabidopsis thaliana. PLoS Genet. 2014, 10, e1004655. [Google Scholar] [CrossRef]
- Guo, H.; Ahn, H.K. Phosphorylation-Regulated Activation of the Arabidopsis RRS1-R/RPS4 Immune Receptor Complex Reveals Two Distinct Effector Recognition Mechanisms. Cell Host Microbe 2020, 27, 769–781.e766. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J. Molecular basis for the interference of the Arabidopsis WRKY54-mediated immune response by two sequence-unrelated bacterial effectors. Plant J. 2024, 118, 839–855. [Google Scholar] [CrossRef] [PubMed]
- Mukhi, N.; Brown, H. Perception of structurally distinct effectors by the integrated WRKY domain of a plant immune receptor. Proc. Natl. Acad. Sci. USA 2021, 118, e2113996118. [Google Scholar] [CrossRef]
- Kim, B.; Yu, W. A plasma membrane nucleotide-binding leucine-rich repeat receptor mediates the recognition of the Ralstonia pseudosolanacearum effector RipY in Nicotiana benthamiana. Plant Commun. 2023, 4, 100640. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Kloek, A.P. The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol. Plant-Microbe Interact. 2000, 13, 1312–1321. [Google Scholar] [CrossRef]
- Axtell, M.J.; Chisholm, S.T. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 2003, 49, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Molina, C.M.; Mainiero, S. The Ptr1 Locus of Solanum lycopersicoides Confers Resistance to Race 1 Strains of Pseudomonas syringae pv. tomato and to Ralstonia pseudosolanacearum by Recognizing the Type III Effectors AvrRpt2 and RipBN. Mol. Plant-Microbe Interact. 2019, 32, 949–960. [Google Scholar] [CrossRef]
- Bent, A.F.; Kunkel, B.N. RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 1994, 265, 1856–1860. [Google Scholar] [CrossRef]
- Mackey, D.; Belkhadir, Y. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef]
- Ahn, Y.J.; Kim, H. Ptr1 and ZAR1 immune receptors confer overlapping and distinct bacterial pathogen effector specificities. New Phytol. 2023, 239, 1935–1953. [Google Scholar] [CrossRef]
- Tsakiri, D.; Kotsaridis, K. The core effector RipE1 of Ralstonia solanacearum interacts with and cleaves Exo70B1 and is recognized by the Ptr1 immune receptor. bioRxiv 2023, 2022, 506019. [Google Scholar]
- Schultink, A.; Qi, T. Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 2017, 92, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Mukaihara, T. The type III effector RipB from Ralstonia solanacearum RS1000 acts as a major avirulence factor in Nicotiana benthamiana and other Nicotiana species. Mol. Plant Pathol. 2019, 20, 1237–1251. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Cai, W.; Zhang, L.; Zhu, Z.; Okita, T.W.; Tian, L. Molecular Dialog of Ralstonia solanacearum and Plant Hosts with Highlights on Type III Effectors. Int. J. Mol. Sci. 2025, 26, 3686. https://doi.org/10.3390/ijms26083686
Hu X, Cai W, Zhang L, Zhu Z, Okita TW, Tian L. Molecular Dialog of Ralstonia solanacearum and Plant Hosts with Highlights on Type III Effectors. International Journal of Molecular Sciences. 2025; 26(8):3686. https://doi.org/10.3390/ijms26083686
Chicago/Turabian StyleHu, Xinyu, Weiwei Cai, Laining Zhang, Zhujun Zhu, Thomas W. Okita, and Li Tian. 2025. "Molecular Dialog of Ralstonia solanacearum and Plant Hosts with Highlights on Type III Effectors" International Journal of Molecular Sciences 26, no. 8: 3686. https://doi.org/10.3390/ijms26083686
APA StyleHu, X., Cai, W., Zhang, L., Zhu, Z., Okita, T. W., & Tian, L. (2025). Molecular Dialog of Ralstonia solanacearum and Plant Hosts with Highlights on Type III Effectors. International Journal of Molecular Sciences, 26(8), 3686. https://doi.org/10.3390/ijms26083686







