Cytokine Receptor-like Factor 3 (CRLF3) and Its Emerging Roles in Neurobiology, Hematopoiesis and Related Human Diseases
Abstract
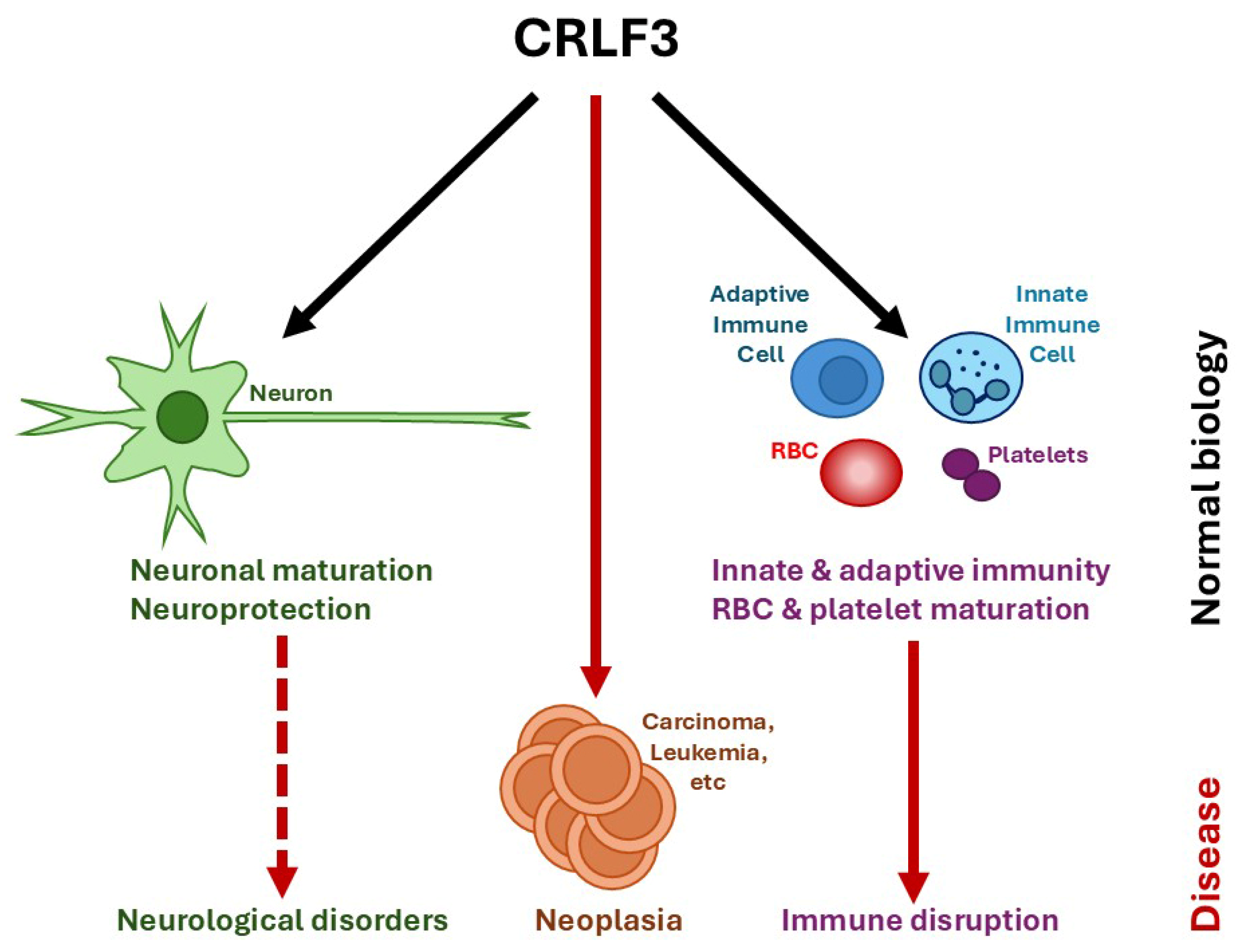
:1. Introduction
2. Biological Roles of CRLF3
2.1. Neuronal Cells
2.2. Hematopoietic Cells
2.3. Other Tissues
3. Functional Context of CRLF3
3.1. Structural Information
3.2. Functional Modalities
3.2.1. Classical Cytokine Receptor Signaling
3.2.2. Microtubule Stability via Hippo/Rho Signaling
3.2.3. Stress Signaling
3.3. Evolutionary Considerations
4. Involvement of CRLF3 in Human Diseases
4.1. Neurological Disorders
4.2. Hematological Disorders
4.3. Other Diseases
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liongue, C.; Ward, A.C. Evolution of class I cytokine receptors. BMC Evol. Biol. 2007, 7, 120. [Google Scholar] [CrossRef] [PubMed]
- Wyder, S.; Kriventseva, E.V.; Schroder, R.; Kadowaki, T.; Zdobnov, E.M. Quantification of ortholog losses in insects and vertebrates. Genome Biol. 2007, 8, R242. [Google Scholar] [CrossRef] [PubMed]
- Hahn, N.; Buschgens, L.; Schwedhelm-Domeyer, N.; Bank, S.; Geurten, B.R.H.; Neugebauer, P.; Massih, B.; Gopfert, M.C.; Heinrich, R. The orphan cytokine receptor CRLF3 emerged with the origin of the nervous system and is a neuroprotective erythropoietin receptor in locusts. Front. Mol. Neurosci. 2019, 12, 251. [Google Scholar] [CrossRef] [PubMed]
- Knorr, D.Y.; Hartung, D.; Schneider, K.; Hintz, L.; Pies, H.S.; Heinrich, R. Locust hemolymph conveys erythropoietin-like cytoprotection via activation of the cytokine receptor CRLF3. Front. Physiol. 2021, 12, 648245. [Google Scholar] [CrossRef]
- Wegscheid, M.L.; Anastasaki, C.; Hartigan, K.A.; Cobb, O.M.; Papke, J.B.; Traber, J.N.; Morris, S.M.; Gutmann, D.H. Patient-derived iPSC-cerebral organoid modeling of the 17q11.2 microdeletion syndrome establishes CRLF3 as a critical regulator of neurogenesis. Cell Rep. 2021, 36, 109315. [Google Scholar] [CrossRef]
- Bennett, C.; Lawrence, M.; Guerrero, J.A.; Stritt, S.; Waller, A.K.; Yan, Y.; Mifsud, R.W.; Ballester-Beltran, J.; Baig, A.A.; Mueller, A.; et al. CRLF3 plays a key role in the final stage of platelet genesis and is a potential therapeutic target for thrombocythaemia. Blood 2022, 139, 2227–2239. [Google Scholar] [CrossRef]
- Taznin, T.; Perera, K.; Gibert, Y.; Ward, A.C.; Liongue, C. Cytokine receptor-like factor 3 (CRLF3) contributes to early zebrafish hematopoiesis. Front. Immunol. 2022, 13, 910428. [Google Scholar]
- Castellucci, L.C.; Almeida, L.; Cherlin, S.; Fakiola, M.; Francis, R.W.; Carvalho, E.M.; Santos da Hora, A.; do Lago, T.S.; Figueiredo, A.B.; Cavalcanti, C.M.; et al. A genome-wide association study identifies SERPINB10, CRLF3, STX7, LAMP3, IFNG-AS1, and KRT80 as risk loci contributing to cutaneous Leishmaniasis in Brazil. Clin. Infect. Dis. 2021, 72, e515–e525. [Google Scholar] [CrossRef]
- Huang, Z.; Huang, L.; Huang, C.; Zhang, X.; Zhang, X. Expressional and prognostic value of CRLF3 in liver hepatocellular carcinoma patients via integrated bioinformatics analyses and experiments. Res. Sq. 2025. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Muramatsu, K.; Kunii, M.; Yoshimura, S.; Yamada, M.; Sato, T.; Ishida, Y.; Harada, R.; Harada, A. Uncovering genes required for neuronal morphology by morphology-based gene trap screening with a revertible retrovirus vector. FASEB J. 2012, 26, 4662–4674. [Google Scholar] [CrossRef]
- Wilson, A.F.; Barakat, R.; Mu, R.; Karush, L.L.; Gao, Y.; Hartigan, K.A.; Chen, J.K.; Shu, H.; Turner, T.N.; Maloney, S.E.; et al. A common single nucleotide variant in the cytokine receptor-like factor-3 (CRLF3) gene causes neuronal deficits in human and mouse cells. Hum. Mol. Genet. 2023, 32, 3342–3352. [Google Scholar] [CrossRef] [PubMed]
- Bult, C.J.; Blake, J.A.; Smith, C.L.; Kadin, J.A.; Richardson, J.E.; Mouse Genome Database, G. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019, 47, D801–D806. [Google Scholar] [CrossRef] [PubMed]
- Hahn, N.; Knorr, D.Y.; Liebig, J.; Wustefeld, L.; Peters, K.; Buscher, M.; Bucher, G.; Ehrenreich, H.; Heinrich, R. The insect ortholog of the human orphan cytokine receptor CRLF3 is a neuroprotective erythropoietin receptor. Front. Mol. Neurosci. 2017, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Knorr, D.Y.; Rodriguez Polo, I.; Pies, H.S.; Schwedhelm-Domeyer, N.; Pauls, S.; Behr, R.; Heinrich, R. The cytokine receptor CRLF3 is a human neuroprotective EV-3 (Epo) receptor. Front. Mol. Neurosci. 2023, 16, 1154509. [Google Scholar] [CrossRef]
- Dickinson, M.E.; Flenniken, A.M.; Ji, X.; Teboul, L.; Wong, M.D.; White, J.K.; Meehan, T.F.; Weninger, W.J.; Westerberg, H.; Adissu, H.; et al. High-throughput discovery of novel developmental phenotypes. Nature 2016, 537, 508–514. [Google Scholar] [CrossRef]
- Polewko-Klim, A.; Lesinski, W.; Golinska, A.K.; Mnich, K.; Siwek, M.; Rudnicki, W.R. Sensitivity analysis based on the random forest machine learning algorithm identifies candidate genes for regulation of innate and adaptive immune response of chicken. Poult. Sci. 2020, 99, 6341–6354. [Google Scholar] [CrossRef]
- Yan, X.; Zheng, W.; Geng, S.; Zhou, M.; Xu, T. Cytokine receptor-like factor 3 negatively regulates antiviral immunity by promoting the degradation of TBK1 in teleost fish. J. Virol. 2023, 97, e0179222. [Google Scholar] [CrossRef]
- Smith, C.M.; Hayamizu, T.F.; Finger, J.H.; Bello, S.M.; McCright, I.J.; Xu, J.; Baldarelli, R.M.; Beal, J.S.; Campbell, J.; Corbani, L.E.; et al. The mouse gene expression database (GXD): 2019 update. Nucleic Acids Res. 2019, 47, D774–D779. [Google Scholar] [CrossRef]
- Karimi, K.; Fortriede, J.D.; Lotay, V.S.; Burns, K.A.; Wang, D.Z.; Fisher, M.E.; Pells, T.J.; James-Zorn, C.; Wang, Y.; Ponferrada, V.G. Xenbase: A genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 2017, 46, D861–D868. [Google Scholar] [CrossRef]
- Karlsson, M.; Zhang, C.; Mear, L.; Zhong, W.; Digre, A.; Katona, B.; Sjostedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Miljus, N.; Massih, B.; Weis, M.A.; Rison, J.V.; Bonnas, C.B.; Sillaber, I.; Ehrenreich, H.; Geurten, B.R.; Heinrich, R. Neuroprotection and endocytosis: Erythropoietin receptors in insect nervous systems. J. Neurochem. 2017, 141, 63–74. [Google Scholar] [PubMed]
- Knorr, D.Y.; Schneider, K.; Buschgens, L.; Forster, J.; Georges, N.S.; Geurten, B.R.H.; Heinrich, R. Protection of insect neurons by erythropoietin/CRLF3-mediated regulation of pro-apoptotic acetylcholinesterase. Sci. Rep. 2022, 12, 18565. [Google Scholar]
- Carmona, S.J.; Teichmann, S.A.; Ferreira, L.; Macaulay, I.C.; Stubbington, M.J.; Cvejic, A.; Gfeller, D. Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 2017, 27, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Iyer, S.; Lobbardi, R.; Moore, J.C.; Chen, H.; Lareau, C.; Hebert, C.; Shaw, M.L.; Neftel, C.; Suva, M.L.; et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med. 2017, 214, 2875–2887. [Google Scholar]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar]
- Zhao, Y.; Duan, S.S.; Che, C.Y.; Zhang, L.; Zhang, Y.; Wang, S.H.; Liu, L. The expression of human cytokine receptor-like factor 3 in mammalian and E. coli cells. Xi Bao Yu Fen. Zi Mian Yi Xue Za Zhi 2008, 24, 27–29. [Google Scholar]
- Jenne, D.E.; Tinschert, S.; Dorschner, M.O.; Hameister, H.; Stephens, K.; Kehrer-Sawatzki, H. Complete physical map and gene content of the human NF1 tumor suppressor region in human and mouse. Genes Chromosomes Cancer 2003, 37, 111–120. [Google Scholar]
- Yang, F.; Xu, Y.P.; Li, J.; Duan, S.S.; Fu, Y.J.; Zhang, Y.; Zhao, Y.; Qiao, W.T.; Chen, Q.M.; Geng, Y.Q.; et al. Cloning and characterization of a novel intracellular protein p48.2 that negatively regulates cell cycle progression. Int. J. Biochem. Cell Biol. 2009, 41, 2240–2250. [Google Scholar]
- Boulay, J.L.; O’Shea, J.J.; Paul, W.E. Molecular phylogeny within type I cytokines and their cognate receptors. Immunity 2003, 19, 159–163. [Google Scholar]
- Hilton, D.J.; Watowich, S.S.; Katz, L.; Lodish, H.F. Saturation mutagenesis of the WSXWS motif of the erythropoietin receptor. J. Biol. Chem. 1996, 271, 4699–4708. [Google Scholar] [CrossRef]
- Dagil, R.; Knudsen, M.J.; Olsen, J.G.; O’Shea, C.; Franzmann, M.; Goffin, V.; Teilum, K.; Breinholt, J.; Kragelund, B.B. The WSXWS motif in cytokine receptors is a molecular switch involved in receptor activation: Insight from structures of the prolactin receptor. Structure 2012, 20, 270–282. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar]
- D’Cruz, A.A.; Babon, J.J.; Norton, R.S.; Nicola, N.A.; Nicholson, S.E. Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci. 2013, 22, 1–10. [Google Scholar]
- Robb, L. Cytokine receptors and hematopoietic differentiation. Oncogene 2007, 26, 6715–6723. [Google Scholar]
- Liongue, C.; Sertori, R.; Ward, A.C. Evolution of cytokine receptor signaling. J. Immunol. 2016, 197, 11–18. [Google Scholar] [CrossRef]
- O’Sullivan, L.A.; Liongue, C.; Lewis, R.S.; Stephenson, S.E.M.; Ward, A.C. Cytokine receptor signaling through the Jak/Stat/Socs pathway in disease. Mol. Immunol. 2007, 44, 2497–2506. [Google Scholar]
- Miljus, N.; Heibeck, S.; Jarrar, M.; Micke, M.; Ostrowski, D.; Ehrenreich, H.; Heinrich, R. Erythropoietin-mediated protection of insect brain neurons involves JAK and STAT but not PI3K transduction pathways. Neuroscience 2014, 258, 218–227. [Google Scholar]
- Hitchcock, I.S.; Hafer, M.; Sangkhae, V.; Tucker, J.A. The thrombopoietin receptor: Revisiting the master regulator of platelet production. Platelets 2021, 32, 770–778. [Google Scholar]
- Tichil, I.; Mitre, I.; Zdrenghea, M.T.; Bojan, A.S.; Tomuleasa, C.I.; Cenariu, D. A review of key regulators of steady-state and ineffective erythropoiesis. J. Clin. Med. 2024, 13, 2585. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Estienne, M.; Bessoles, S.; Echchakir, H.; Pederzoli-Ribeil, M.; Chiron, A.; Aldaz-Carroll, L.; Leducq, V.; Zhang, Y.; Souyri, M.; et al. Erythropoietin is a major regulator of thrombopoiesis in thrombopoietin-dependent and -independent contexts. Exp. Hematol. 2020, 88, 15–27. [Google Scholar] [CrossRef]
- Borriello, F.; Galdiero, M.R.; Varricchi, G.; Loffredo, S.; Spadaro, G.; Marone, G. Innate immune modulation by GM-CSF and IL-3 in health and disease. Int. J. Mol. Sci. 2019, 20, 834. [Google Scholar] [CrossRef] [PubMed]
- Boulay, J.L.; Du Pasquier, L.; Cooper, M.D. Cytokine receptor diversity in the lamprey predicts the minimal essential cytokine networks of vertebrates. J. Immunol. 2022, 209, 1013–1020. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, D.; Heinrich, R. Alternative erythropoietin receptors in the nervous system. J. Clin. Med. 2018, 7, 24. [Google Scholar] [CrossRef]
- Wakhloo, D.; Scharkowski, F.; Curto, Y.; Javed Butt, U.; Bansal, V.; Steixner-Kumar, A.A.; Wustefeld, L.; Rajput, A.; Arinrad, S.; Zillmann, M.R.; et al. Functional hypoxia drives neuroplasticity and neurogenesis via brain erythropoietin. Nat. Commun. 2020, 11, 1313. [Google Scholar] [CrossRef]
- Thompson, A.M.; Farmer, K.; Rowe, E.M.; Hayley, S. Erythropoietin modulates striatal antioxidant signalling to reduce neurodegeneration in a toxicant model of Parkinson’s disease. Mol. Cell Neurosci. 2020, 109, 103554. [Google Scholar] [CrossRef]
- Rey, F.; Ottolenghi, S.; Giallongo, T.; Balsari, A.; Martinelli, C.; Rey, R.; Allevi, R.; Giulio, A.M.D.; Zuccotti, G.V.; Mazzucchelli, S.; et al. Mitochondrial metabolism as target of the neuroprotective role of erythropoietin in Parkinson’s disease. Antioxidants 2021, 10, 121. [Google Scholar] [CrossRef]
- Brines, M.; Cerami, A. Emerging biological roles for erythropoietin in the nervous system. Nat. Rev. Neurosci. 2005, 6, 484–494. [Google Scholar] [CrossRef]
- Leist, M.; Ghezzi, P.; Grasso, G.; Bianchi, R.; Villa, P.; Fratelli, M.; Savino, C.; Bianchi, M.; Nielsen, J.; Gerwien, J.; et al. Derivatives of erythropoietin that are tissue protective but not erythropoietic. Science 2004, 305, 239–242. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, Y.; Chen, Y. Spatiotemporal regulation of Rho GTPases in neuronal migration. Cells 2019, 8, 568. [Google Scholar] [CrossRef]
- Phillips, J.E.; Zheng, Y.; Pan, D. Assembling a Hippo: The evolutionary emergence of an animal developmental signaling pathway. Trends Biochem. Sci. 2024, 49, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Couzens, A.L.; Knight, J.D.; Kean, M.J.; Teo, G.; Weiss, A.; Dunham, W.H.; Lin, Z.Y.; Bagshaw, R.D.; Sicheri, F.; Pawson, T.; et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal 2013, 6, rs15. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Couzens, A.L.; Kean, M.J.; Mao, D.Y.; Guettler, S.; Kurinov, I.; Gingras, A.C.; Sicheri, F. Regulation of protein interactions by mps one binder (MOB1) phosphorylation. Mol. Cell Proteom. 2017, 16, 1111–1125. [Google Scholar] [CrossRef] [PubMed]
- Masters, S.L.; Palmer, K.R.; Stevenson, W.S.; Metcalf, D.; Viney, E.M.; Sprigg, N.S.; Alexander, W.S.; Nicola, N.A.; Nicholson, S.E. Genetic deletion of murine SPRY domain-containing SOCS box protein 2 (SSB-2) results in very mild thrombocytopenia. Mol. Cell Biol. 2005, 25, 5639–5647. [Google Scholar] [CrossRef] [PubMed]
- Pilli, M.; Arko-Mensah, J.; Ponpuak, M.; Roberts, E.; Master, S.; Mandell, M.A.; Dupont, N.; Ornatowski, W.; Jiang, S.; Bradfute, S.B.; et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012, 37, 223–234. [Google Scholar] [CrossRef]
- Zhu, H.; Zhou, Z.; Wang, D.; Liu, W.; Zhu, H. Hippo pathway genes developed varied exon numbers and coevolved functional domains in metazoans for species specific growth control. BMC Evol. Biol. 2013, 13, 76. [Google Scholar] [CrossRef]
- Martin, J.F.; D’Avino, P.P. A theory of rapid evolutionary change explaining the de novo appearance of megakaryocytes and platelets in mammals. J. Cell Sci. 2022, 135, jcs260286. [Google Scholar] [CrossRef]
- Davidson, C.J.; Tuddenham, E.G.; McVey, J.H. 450 million years of hemostasis. J. Thromb. Haemost. 2003, 1, 1487–1494. [Google Scholar] [CrossRef]
- Jenne, D.E.; Tinschert, S.; Reimann, H.; Lasinger, W.; Thiel, G.; Hameister, H.; Kehrer-Sawatzki, H. Molecular characterization and gene content of breakpoint boundaries in patients with neurofibromatosis type 1 with 17q11.2 microdeletions. Am. J. Hum. Genet. 2001, 69, 516–527. [Google Scholar] [CrossRef]
- Kehrer-Sawatzki, H.; Cooper, D.N. Classification of NF1 microdeletions and its importance for establishing genotype/phenotype correlations in patients with NF1 microdeletions. Hum. Genet. 2021, 140, 1635–1649. [Google Scholar] [CrossRef]
- Sharma, A.; Dey, P. A machine learning approach to unmask novel gene signatures and prediction of Alzheimer’s disease within different brain regions. Genomics 2021, 113, 1778–1789. [Google Scholar] [CrossRef]
- Cirulli, E.T.; Lasseigne, B.N.; Petrovski, S.; Sapp, P.C.; Dion, P.A.; Leblond, C.S.; Couthouis, J.; Lu, Y.F.; Wang, Q.; Krueger, B.J.; et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 2015, 347, 1436–1441. [Google Scholar] [PubMed]
- Nijs, M.; Van Damme, P. The genetics of amyotrophic lateral sclerosis. Curr. Opin. Neurol. 2024, 37, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Ota, V.K.; Oliveira, A.M.; Bugiga, A.V.G.; Conceicao, H.B.; Galante, P.A.F.; Asprino, P.F.; Schafer, J.L.; Hoffmann, M.S.; Bressan, R.; Brietzke, E.; et al. Impact of life adversity and gene expression on psychiatric symptoms in children and adolescents: Findings from the Brazilian high risk cohort study. Front. Psychiatry 2025, 16, 1505421. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.T.; Hannon, E.; Levine, M.E.; Crimmins, E.M.; Lunnon, K.; Mill, J.; Geschwind, D.H.; Horvath, S. Genetic architecture of epigenetic and neuronal ageing rates in human brain regions. Nat. Commun. 2017, 8, 15353. [Google Scholar]
- Maghsoudloo, M.; Azimzadeh Jamalkandi, S.; Najafi, A.; Masoudi-Nejad, A. An efficient hybrid feature selection method to identify potential biomarkers in common chronic lung inflammatory diseases. Genomics 2020, 112, 3284–3293. [Google Scholar]
- Cancer Genome Atlas Research, N.; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar]
- Ponten, F.; Jirstrom, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar]
- Dang, C.; Gottschling, M.; Manning, K.; O’Currain, E.; Schneider, S.; Sterry, W.; Stockfleth, E.; Nindl, I. Identification of dysregulated genes in cutaneous squamous cell carcinoma. Oncol. Rep. 2006, 16, 513–519. [Google Scholar]
- Ahmad, W.; Ijaz, B.; Hassan, S. Gene expression profiling of HCV genotype 3a initial liver fibrosis and cirrhosis patients using microarray. J. Transl. Med. 2012, 10, 41. [Google Scholar] [CrossRef]
- Luo, R.; Chong, W.; Wei, Q.; Zhang, Z.; Wang, C.; Ye, Z.; Abu-Khalaf, M.M.; Silver, D.P.; Stapp, R.T.; Jiang, W.; et al. Whole-exome sequencing identifies somatic mutations and intratumor heterogeneity in inflammatory breast cancer. NPJ Breast Cancer 2021, 7, 72. [Google Scholar]
- Padella, A.; Simonetti, G.; Paciello, G.; Giotopoulos, G.; Baldazzi, C.; Righi, S.; Ghetti, M.; Stengel, A.; Guadagnuolo, V.; De Tommaso, R.; et al. Novel and rare fusion transcripts involving transcription factors and tumor suppressor genes in acute myeloid leukemia. Cancers 2019, 11, 1951. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [PubMed]
- Elfman, J.; Goins, L.; Heller, T.; Singh, S.; Wang, Y.H.; Li, H. Discovery of a polymorphic gene fusion via bottom-up chimeric RNA prediction. Nucleic Acids Res. 2024, 52, 4409–4421. [Google Scholar] [PubMed]

| Phenotype | Species | System | Study | Refs | |
|---|---|---|---|---|---|
| Neuronal | Neuronal maturation defect | Rat | Neuronal cell line | Crlf3 KO (gene trap) | [10] |
| Mouse | Whole animal | Crlf3L389P | [11] | ||
| Human | iPSC-cerebral organoid | CRLF3L389P + CRLF3 KD | [5] | ||
| Behavioral defects & tremors | Mouse | Whole animal | Crlf3 KO | [12] | |
| Neuroprotection | Red flour beetle | Brain-derived neurons | crlf3 KD (RNAi) | [13] | |
| Migratory locust | Brain-derived neurons | crlf3 KD (RNAi) | [4] | ||
| Human | iPSC-derived neurons | CRLF3 KO | [14] | ||
| Hematopoietic/immune | Platelet maturation defect | Mouse | Whole animal | Crlf3 KO | [6] |
| RBC maturation defect | Mouse | Whole animal | Crlf3 KO | [12,15] | |
| HSC decrease | Zebrafish | Whole animal | crlf3 KO | [7] | |
| Myeloid cell decrease | Zebrafish | Whole animal | crlf3 KO | [7] | |
| Lymphoid cell increase | Human | Blood samples | CRLF3 SNV association | [6] | |
| Adaptive immunity | Chicken | Whole animal | crlf3 SNV association | [16] | |
| Innate immunity | Chicken | Whole animal | crlf3 SNV association | [16] | |
| Type I IFN responses | Brown croaker | Kidney cells | crlf3 KD + OE | [17] | |
| Leishmaniasis risk | Human | Case/control data | CRLF3 SNV association | [8] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liongue, C.; Ward, A.C. Cytokine Receptor-like Factor 3 (CRLF3) and Its Emerging Roles in Neurobiology, Hematopoiesis and Related Human Diseases. Int. J. Mol. Sci. 2025, 26, 3498. https://doi.org/10.3390/ijms26083498
Liongue C, Ward AC. Cytokine Receptor-like Factor 3 (CRLF3) and Its Emerging Roles in Neurobiology, Hematopoiesis and Related Human Diseases. International Journal of Molecular Sciences. 2025; 26(8):3498. https://doi.org/10.3390/ijms26083498
Chicago/Turabian StyleLiongue, Clifford, and Alister C. Ward. 2025. "Cytokine Receptor-like Factor 3 (CRLF3) and Its Emerging Roles in Neurobiology, Hematopoiesis and Related Human Diseases" International Journal of Molecular Sciences 26, no. 8: 3498. https://doi.org/10.3390/ijms26083498
APA StyleLiongue, C., & Ward, A. C. (2025). Cytokine Receptor-like Factor 3 (CRLF3) and Its Emerging Roles in Neurobiology, Hematopoiesis and Related Human Diseases. International Journal of Molecular Sciences, 26(8), 3498. https://doi.org/10.3390/ijms26083498







