Limited Diversity of Thermal Adaptation to a Critical Temperature in Zymomonas mobilis: Evidence from Multiple-Parallel Laboratory Evolution Experiments
Abstract
1. Introduction
2. Results
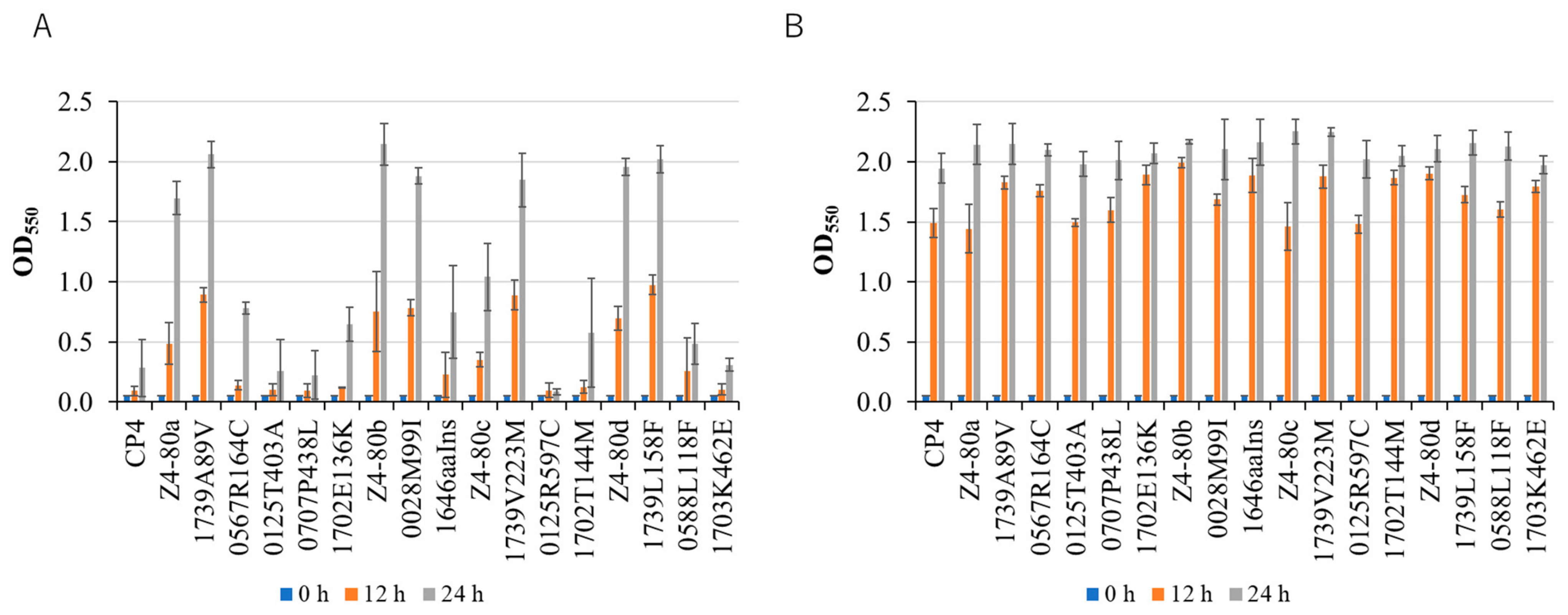
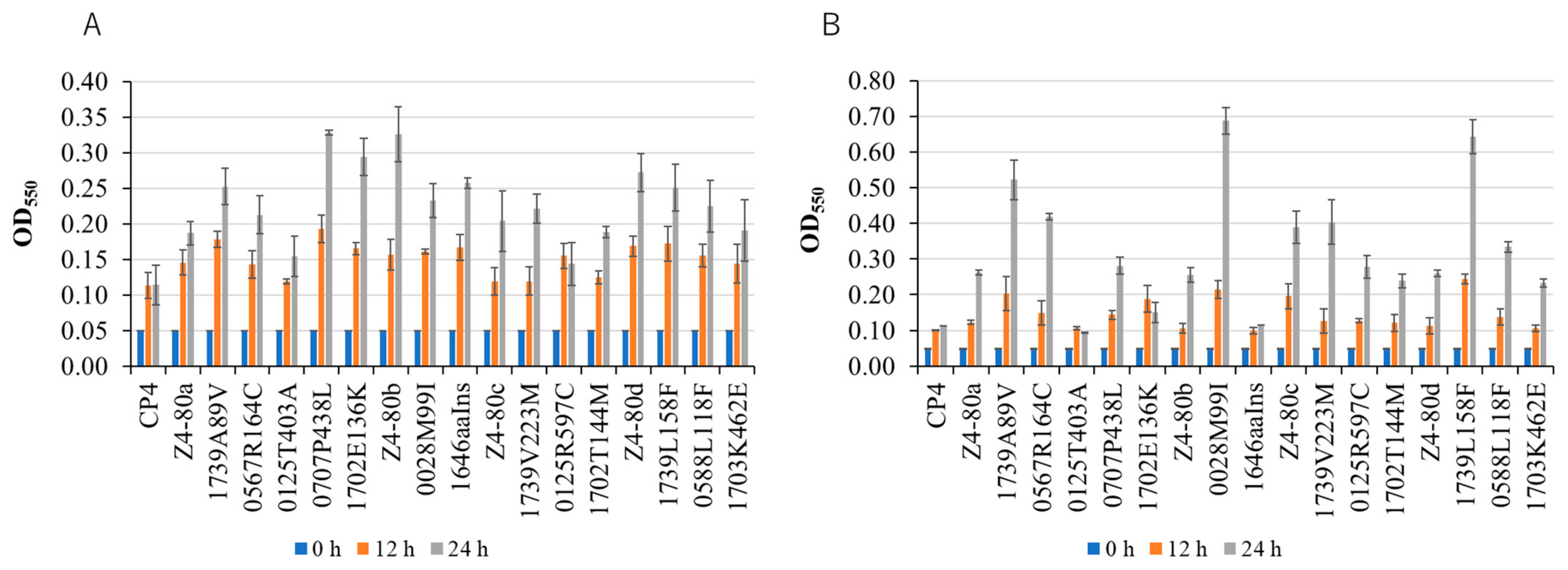
2.1. Identification of Mutations That Caused an Increase in Thermal Resistance
2.2. RNA-Seq Analysis of Thermoadapted Mutants and Single-Mutation Mutants
2.3. Factors for Acquired Thermal Resistance
2.4. Minor but Important Contribution of Transporters to Thermal Adaptation
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Construction of Single-Mutation Mutants
4.3. Two-Step Cultivation Assay
4.4. Characterization of Thermoadapted Mutants and Single-Mutation Mutants
4.5. Cell Preparation for RNA-Seq
4.6. RNA-Seq Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RNA-seq | RNA sequencing |
| CHT | Critical high temperature |
| DEGs | Differentially expressed genes |
| GO | Gene Ontology |
| LPS | Lipopolysaccharide |
| IMP | Inner membrane protein |
| PAP | Periplasmic adapter protein |
| OMF | Outer membrane factor |
| MFS | Major facilitator superfamily |
| ROS | Reactive oxygen species |
| LB | Luria–Bertani |
| cDNA | Complementary DNA |
| TPM | Transcripts per million |
| TMM | Trimmed mean of M-values |
| GLM | Generalized linear models |
References
- Soemphol, W.; Deeraksa, A.; Matsutani, M.; Yakushi, T.; Toyama, H.; Adachi, O.; Yamada, M.; Matsushita, K. Global analysis of the genes involved in the thermotolerance mechanism of thermotolerant Acetobacter tropicalis SKU1100. Biosci. Biotechnol. Biochem. 2011, 75, 1921–1928. [Google Scholar] [PubMed]
- Charoensuk, K.; Sukurada, T.; Tokiyama, A.; Murata, M.; Kosaka, T.; Thanonkeo, P.; Yamada, M. Thermotolerant genes essential for survival at a critical high temperature in thermotolerant ethanologenic Zymomonas mobilis TISTR 548. Biotechnol. Biofuels 2017, 10, 204. [Google Scholar]
- Murata, M.; Ishii, A.; Fujimoto, H.; Nishimura, K.; Kosaka, T.; Mori, H.; Yamada, M. Update of thermotolerant genes essential for survival at a critical high temperature in Escherichia coli. PLoS ONE 2018, 13, e0189487. [Google Scholar]
- Tan, Y.S.; Zhang, R.K.; Liu, Z.H.; Li, B.Z.; Yuan, Y.J. Microbial adaptation to enhance stress tolerance. Front. Microbiol. 2022, 13, 888746. [Google Scholar]
- Wang, G.; Li, Q.; Zhang, Z.; Yin, X.; Wang, B.; Yang, X. Recent progress in adaptive laboratory evolution of industrial microorganisms. J. Ind. Microbiol. Biotechnol. 2023, 50, kuac023. [Google Scholar]
- Hirasawa, T.; Maeda, T. Adaptive laboratory evolution of microorganisms: Methodology and application for bioproduction. Microorganisms 2023, 11, 92. [Google Scholar]
- Tenaillon, O.; Rodriguez-Verdugo, A.; Gaut, R.L.; McDonald, P.; Bennett, A.F.; Long, A.D.; Gaut, B.S. The molecular diversity of adaptive convergence. Science 2012, 335, 457–461. [Google Scholar]
- Sandberg, T.E.; Pedersen, M.; LaCroix, R.A.; Ebrahim, A.; Bonde, M.; Herrgard, M.J.; Palsson, B.O.; Sommer, M.; Feist, A.M. Evolution of Escherichia coli to 42 °C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol. Biol. Evol. 2014, 31, 2647–2662. [Google Scholar]
- Tenaillon, O.; Barrick, J.E.; Ribeck, N.; Deatherage, D.E.; Blanchard, J.L.; Dasgupta, A.; Wu, G.C.; Wielgoss, S.; Cruveiller, S.; Médigue, C.; et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 2016, 536, 165–170. [Google Scholar]
- Deatherage, D.E.; Kepner, J.L.; Bennett, A.F.; Lenski, R.E.; Barrick, J.E. Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc. Natl. Acad. Sci. USA 2017, 114, E1904–E1912. [Google Scholar]
- Kishimoto, T.; Lijima, L.; Tatsumi, M.; Ono, N.; Oyake, A.; Hashimoto, T.; Matsuo, M.; Okubo, M.; Suzuki, S.; Mori, K.; et al. Transition from positive to neutral in mutation fixation along with continuing rising fitness in thermal adaptive evolution. PLoS Genet. 2010, 6, e1001164. [Google Scholar]
- Rudolph, B.; Gebendorfer, K.M.; Buchner, J.; Winter, J. Evolution of Escherichia coli for growth at high temperatures. J Biol Chem. 2010, 285, 19029–19034. [Google Scholar] [PubMed]
- Blaby, I.K.; Lyons, B.J.; Wroclawska-Hughes, E.; Philips, G.C.F.; Pyle, T.P.; Chamberlin, S.G.; Benner, S.A.; Lyons, T.J.; de Crécy-Lagard, V.; de Crécy, E. Experimental evolution of a facultative thermophile from a mesophilic ancestor. Appl. Environ. Microbiol. 2012, 78, 144–155. [Google Scholar] [PubMed]
- Kosaka, T.; Nakajima, Y.; Ishii, A.; Yamashita, M.; Yoshida, S.; Murata, M.; Kat, K.; Shiromaru, Y.; Kato, S.; Kanasaki, Y.; et al. Capacity for survival in global warming: Adaptation of mesophiles to the temperature upper limit. PLoS ONE 2019, 14, e0218985. [Google Scholar]
- Chen, J.; Shen, J.; Hellgren, L.H.; Jensen, P.R.; Solem, C. Adaptation of Lactococcus lactis to high growth temperature leads to a dramatic increase in acidification rate. Sci. Rep. 2015, 5, 14199. [Google Scholar]
- García-Ríos, E.; Lairón-Peris, M.; Muñiz-Calvo, S.; Heras, J.M.; Ortiz-Julien, A.; Poirot, P.; Rozès, N.; Querol, A.; Guillamón, J.M. Thermo-adaptive evolution to generate improved Saccharomyces cerevisiae strains for cocoa pulp fermentations. Int. J. Food Microbiol. 2021, 342, 109077. [Google Scholar]
- Liu, M.; Cheng, H. Discovery and functional evaluation of heat tolerance genes in the nonconventional yeast Yarrowia lipolytica. Fermentation 2023, 9, 509. [Google Scholar] [CrossRef]
- Li, W.; Yang, J.; Chen, Y.; Xu, N.; Liu, J.; Wang, J. Thermo-adaptive evolution of Corynebacterium glutamicum reveals the regulatory functions of fasR and hrcA in heat tolerance. Microb. Cell Fact. 2024, 23, 294. [Google Scholar]
- Breland, E.J.; Eberly, A.R. An overview of two-component signal transduction systems implicated in extra-intestinal pathogenic E. coli infections. Front. Cell. Infect. Microbiol. 2017, 7, 162. [Google Scholar]
- Choudhary, K.S.; Kleinmanns, J.A.; Decker, K.; Sastry, A.V.; Gao, Y.; Szubin, R.; Seif, Y.; Palsson, B.O. Elucidation of regulatory modes for five two-component systems in Escherichia coli reveals novel relationships. mSystems 2020, 5, e00980-20. [Google Scholar]
- Almassy, R.J.; Janson, C.A.; Kan, C.C.; Hostomska, Z. Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase. Proc. Natl. Acad. Sci. USA 1992, 89, 6114–6118. [Google Scholar]
- Kulis-Horn, R.K.; Persicke, M.; Kalinowski, J. Histidine biosynthesis, its regulation and biotechnological application in Corynebacterium glutamicum. Microb. Biotechnol. 2013, 7, 5–25. [Google Scholar]
- Badola, P.; Sanders, C.R. Escherichia coli diacylglycerol kinase is an evolutionarily optimized membrane enzyme and catalyzes direct phosphoryl transfer. J. Biol. Chem. 1997, 272, 24176–24182. [Google Scholar]
- Purcell, A.B.; Voss, B.J.; Trent, M.S. Diacylglycerol kinase A is essential for polymyxin pesistance provided by EptA, MCR-1, and other lipid A phosphoethanolamine transferases. J. Bacteriol. 2022, 204, e0049821. [Google Scholar]
- Dahiya, V.; Chaudhuri, T.K. Chaperones GroEL/GroES accelerate the refolding of a multidomain protein through modulating on-pathway intermediates. J. Biol. Chem. 2014, 289, 286–298. [Google Scholar]
- Illingworth, M.; Ellis, H.; Chen, L. Creating the functional single-ring GroEL-GroES chaperonin systems via modulating GroEL-GroES Interaction. Sci. Rep. 2017, 7, 9710. [Google Scholar]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell wall hydrolases in bacteria: Insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol. 2019, 10, 418687. [Google Scholar]
- Sakunda, A.; Murata, M.; Keisuke, K.; Kosaka, T.; Sootsuwan, K.; Thanonkeo, P.; Yamada, M. Improvement of thermotolerance of Zymomonas mobilis by genes for reactive oxygen species-scavenging enzymes and heat-shock proteins. Front. Microbiol. 2020, 10, 501735. [Google Scholar]
- Mueller, E.A.; Levin, P.A. Bacterial cell wall quality control during environmental stress. mBio 2020, 11, e02456-20. [Google Scholar]
- Tascón, I.; Sousa, J.S.; Corey, R.A.; Mills, D.J.; Griwatz, D.; Aumüller, N.; Mikusevic, V.; Stansfeld, P.J.; Vonck, J.; Hänelt, I. Structural basis of proton-coupled potassium transport in the KUP family. Nat. Commun. 2020, 11, 626. [Google Scholar]
- Kosaka, T.; Nishioka, A.; Sakurada, T.; Miura, K.; Sakunda, A.; Yamada, M. Enhancement of thermal resistance by metal ions in the thermotolerant Zymomonas mobilis TISTR 548. Front. Microbiol. 2020, 11, 503372. [Google Scholar]
- Symmons, M.F.; Marshall, R.L.; Bavro, V.N. Architecture and roles of periplasmic adaptor proteins in tripartite efflux assemblies. Front. Microbiol. 2015, 6, 145258. [Google Scholar]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015, 7, 14. [Google Scholar]
- Sanchez-Vazquez, P.; Dewey, C.N.; Kitten, N.; Ross, W.; Gourse, R.L. Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. USA 2019, 116, 8310–8319. [Google Scholar]
- Spira, B.; Ospino, K. Diversity in E.coli (p)ppGpp levels and its consequences. Front. Microbiol. 2020, 11, 1759. [Google Scholar]
- Matsumoto, N.; Osumi, N.; Matsutani, M.; Phathanathavorn, T.; Kataoka, N.; Theeragool, G.; Yakushi, T.; Shiraishi, Y.; Matsushita, K. Thermal adaptation of acetic acid bacteria for practical high-temperature vinegar fermentation. Biosci. Biotechnol. Biochem. 2021, 85, 1243–1251. [Google Scholar]
- Matsumoto, N.; Matsutani, M.; Tanimoto, Y.; Nakanishi, R.; Tanaka, S.; Kanesaki, Y.; Theeragool, G.; Kataoka, N.; Yakushi, T.; Matsushita, K. Implication of amino acid metabolism and cell surface integrity for the thermotolerance mechanism in the thermally adapted acetic acid bacterium Acetobacter pasteurianus TH-3. J. Bacteriol. 2023, 205, e0010123. [Google Scholar]
- Schulte, M.; Frick, K.; Gnandt, E.; Jurkovic, S.; Burschel, S.; Labatzke, R.; Aierstock, K.; Fiegen, D.; Wohlwend, D.; Gerhardt, S.; et al. A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I. Nat. Commun. 2019, 10, 2551. [Google Scholar]
- Matsumoto, N.; Matsutani, M.; Azuma, Y.; Kataoka, N.; Yakushi, T.; Matsushita, K. In vitro thermal adaptation of mesophilic Acetobacter pasteurianus NBRC 3283 generates thermotolerant strains with evolutionary trade-offs. Biosci. Biotechnol. Biochem. 2020, 84, 832–841. [Google Scholar]
- Shin, J.; Noireaux, V. Efficient cell-free expression with the endogenous E. Coli RNA polymerase and sigma factor 70. J. Biol. Eng. 2010, 4, 8. [Google Scholar]
- Simon, R.; Reifer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram-negative bacteria. Bio/Technol. 1983, 1, 784–791. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Katzen, K.; Becker, A.; Ielmini, M.V.; Oddo, C.G.; Ielpi, L. New mobilizable vectors suitable for gene replacement in Gram-negative bacteria and their use in mapping of the 3′ end of the Xanthomonas campestris pv. campestris gum operon. Appl. Environ. Microbiol. 1999, 65, 278–282. [Google Scholar]
- Kim, I.S.; Kim, Y.S.; Kim, H.; Jin, I.; Yoon, H.S. Saccharomyces cerevisiae KNU5377 stress response during high-temperature ethanol fermentation. Mol. Cells 2013, 35, 210–218. [Google Scholar]
- Tomczak, A.; Mortensen, J.M.; Winnenburg, R.; Liu, C.; Alessi, D.T.; Swamy, V.; Vallania, F.; Lofgren, S.; Haynes, W.; Shah, N.H.; et al. Interpretation of biological experiments changes with evolution of the Gene Ontology and its annotations. Sci. Rep. 2018, 8, 5115. [Google Scholar]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar]

| Classification | Locus Tag | Old Locus Tag | Gene Product | Z4-80a | Z4-80b | Z4-80c | Z4-80d |
|---|---|---|---|---|---|---|---|
| Transcriptional regulation | ZCP4_RS00160 | ZCP4_0028 | Two-component system, sensor histidine kinase for signal transduction | M99I | |||
| ZCP4_RS02830 | ZCP4_0567 | RpoB, DNA-directed RNA polymerase subunit beta | R164C | R164C | |||
| Membrane stabilization | ZCP4_RS08695 | ZCP4_1739 | DgkA, Diacylglycerol kinase | A89V | V223M | L158F | |
| General metabolism | ZCP4_RS02940 | ZCP4_0588 | PurN: Formyltetrahydrofolate-dependent phosphoribosylglycinamide formyltransferase | L118F | |||
| ZCP4_RS08220 | ZCP4_1646 | HisA, 1-(5-phosphoribosyl)-5-[(5- phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase | IYDGSL insertion | ||||
| Transporter | ZCP4_RS00655 | ZCP4_0125 | Kup, Potassium transporter | T403A | R597C | ||
| ZCP4_RS03540 | ZCP4_0707 | Nitrate/sulfonate/bicarbonate ABC transporter, ATP-binding protein | P438L | ||||
| ZCP4_RS08505 | ZCP4_1702 | RND family efflux transporter, MFP subunit(HlyD family secretion protein) | E136K | T144M | |||
| ZCP4_RS08510 | ZCP4_1703 | Efflux transporter, outer membrane factor lipoprotein, NodT family (TolC family protein) | K462E |
| Locus Tag | Gene Product | Mutant | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0028M99I | 0567R164C | 0588L118F | 1646aaIns | 1739A89V | 1739V223M | 1739L158F | Z4-80a | Z4-80b | Z4-80c | Z4-80d | ||
| Gene Expression * | ||||||||||||
| (Z4-80a, Z4-80c, Z4-80d) | ||||||||||||
| ZCP4_RS08695 | DgkA, diacylglycerol kinase family protein | ↓↓ | ↓↓ | ↓ | ↓ | ↑↑ | ↑↑ | - | ↑↑ | ↓ | ↑↑ | ↑ |
| (Z4-80a, Z4-80c) | ||||||||||||
| ZCP4_RS02830 | RpoB, DNA-directed RNA polymerase subunit beta | - | ↑ | - | - | - | - | - | ↑ | - | ↑↑ | - |
| ZCP4_RS00655 | Kup, potassium transporter | - | - | - | - | - | - | - | ↑ | ↓ | - | - |
| ZCP4_RS08505 | RND family efflux transporter, MFP subunit (HlyD family secretion protein) | ↑ | ↑ | - | - | - | - | - | ↑ | - | ↑↑ | ↑ |
| (Z4-80a) | ||||||||||||
| ZCP4_RS03540 | Nitrate/sulfonate/bicarbonate ABC transporter ATP-binding protein | - | ↓ | ↓ | ↓ | - | - | - | ↓↓ | - | ↓↓ | - |
| (Z4-80b) | ||||||||||||
| ZCP4_RS00160 | HAMP domain-containing sensor histidine kinase (two component system) | - | - | - | - | ↑ | - | - | ↓ | ↓ | - | ↓ |
| ZCP4_RS08220 | HisA, 1-(5-phosphoribosyl)-5-[(5- phosphoribosylamino)methylideneamino]imidazole-4- carboxamide isomerase | ↑ | - | - | - | - | - | - | - | - | ↑↑ | - |
| (Z4-80d) | ||||||||||||
| ZCP4_RS02940 | PurN, phosphoribosylglycinamide formyltransferase | - | - | - | - | - | - | - | - | - | ↓ | - |
| ZCP4_RS08510 | TolC family protein (efflux transporter, outer membrane factor lipoprotein) | ↑ | ↑↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑↑ | - | ↑↑ | ↑ |
| Locus Tag | Gene Product | Mutant | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0028M99I | 0567R164C | 0588L118F | 1646aaIns | 1739A89V | 1739 V223M | 1739 L158F | Z4-80a | Z4-80b | Z4-80c | Z4-80d | ||
| Gene Expression * | ||||||||||||
| ZCP4_RS08515 | DgkB: diacylglycerol kinase family lipid kinase | ↓↓ | ↓↓ | ↓↓ | - | - | - | - | ↓↓ | - | ↓↓ | ↓ |
| ZCP4_RS06260 | GroEL: Chaperonin GroEL | - | ↑↑ | - | ↑↑ | - | - | ↑ | ↑↑ | ↑↑ | ↑ | - |
| ZCP4_RS06265 | GroES: Co-chaperone GroES | - | ↑↑ | - | ↑↑ | - | - | - | ↑↑ | ↑↑ | ↑ | - |
| ZCP4_RS04150 | Cell wall hydrolase | - | - | ↑↑ | - | - | - | - | - | - | ↑↑ | ↑↑ |
| Locus Tag | Gene Product | Mutant | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0028M99I | 0567R164C | 0588L118F | 1646aaIns | 1739A89V | 1739V223M | 1739L158F | Z4-80a | Z4-80b | Z4-80c | Z4-80d | Description | ||
| Gene Expression * | |||||||||||||
| (Operon) | |||||||||||||
| ZCP4_RS03540 | Nitrate/sulfonate/bicarbonate ABC transporter ATP-binding protein | - | ↓ | ↓ | ↓ | - | - | - | ↓↓ | - | ↓↓ | - | Mutation in Z4-80a |
| ZCP4_RS03545 | ABC transporter permease subunit | - | ↓ | ↓ | ↓ | ↓ | - | - | ↓↓ | ↓ | ↓↓ | - | |
| (Operon) | |||||||||||||
| ZCP4_RS08495 | FUSC family protein | ↑ | ↑ | - | - | - | - | - | ↑ | - | ↑↑ | ↑ | |
| ZCP4_RS08500 | DUF1656 domain-containing protein | ↑ | ↑↑ | - | - | - | - | - | ↑↑ | - | ↑↑ | ↑↑ | |
| ZCP4_RS08505 | HlyD family secretion protein (efflux transporter, MFP subunit) | ↑ | ↑ | - | - | - | - | - | ↑ | - | ↑↑ | ↑ | Mutation in Z4-80a and Z4-80c |
| ZCP4_RS08510 | TolC family protein (efflux transporter, outer membrane factor lipoprotein) | ↑ | ↑↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑↑ | - | ↑↑ | ↑ | Mutation in Z4-80d |
| (Operon) | |||||||||||||
| ZCP4_RS02520 | HlyD family efflux transporter periplasmic adaptor subunit | ↑↑ | ↑↑ | ↑ | ↑↑ | - | ↑ | ↑ | ↑↑ | ↑↑ | ↑↑ | ↑↑ | |
| ZCP4_RS02525 | ribosome-associated ATPase/putative transporter RbbA | ↑↑ | ↑ | ↑ | ↑ | - | ↑ | - | ↑ | ↑ | ↑↑ | ↑ | |
| ZCP4_RS02530 | ABC transporter permease | - | ↑ | - | ↑ | - | - | - | ↑ | ↑ | ↑ | - | |
| ZCP4_RS02535 | TolC family protein | - | ↑ | - | ↑ | - | - | - | ↑ | ↑ | ↑ | - | |
| (Operon) | |||||||||||||
| ZCP4_RS07805 | ABC transporter permease | ↑↑ | - | ↑↑ | - | ↑ | ↑ | - | - | - | ↑↑ | ↑↑ | |
| ZCP4_RS07815 | ATP-binding cassette domain-containing protein | ↑ | ↑ | ↑↑ | ↑ | ↑ | ↑ | ↑ | ↑ | - | ↑↑ | ↑ | |
| ZCP4_RS03940 | MFS transporter | - | ↑ | - | - | - | - | - | ↑↑ | - | ↑↑ | ↑ | |
| ZCP4_RS08380 | MFS transporter | ↑↑ | ↑↑ | ↑↑ | ↑↑ | ↑↑ | ↑↑ | - | ↑↑ | ↑↑ | ↑↑ | ↑↑ | |
| ZCP4_RS08395 | MFS transporter | - | ↑↑ | ↓ | - | - | - | - | ↑↑ | - | ↑↑ | - | |
| Upregulated DEGs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutant and a pair for comparison | 0567R164C | 0028M99I | 1646aaIns | 0588L118F | 0567R164C and 0028M99I | 0567R164C and 1646aaIns | 0567R164C and 0588L118F | 0028M99I and 1646aaIns | 0028M99I and 0588L118F | 1646aaIns and 0588L118F |
| Number of genes | 165 | 48 | 48 | 84 | 37 | 33 | 48 | 16 | 28 | 23 |
| Shared genes * (%) | 79% | 69% | 57% | 33% | 60% | 48% | ||||
| Downregulated DEGs | ||||||||||
| Mutant and a pair for comparison | 0567R164C | 0028M99I | 1646aaIns | 0588L118F | 0567R164C and 0028M99I | 0567R164C and 1646aaIns | 0567R164C and 0588L118F | 0028M99I and 1646aaIns | 0028M99I and 0588L118F | 1646aaIns and 0588L118F |
| Number of genes | 299 | 215 | 87 | 260 | 162 | 69 | 178 | 60 | 155 | 70 |
| Shared genes * (%) | 75% | 79% | 68% | 69% | 72% | 80% | ||||
| Strain | Description | Reference |
|---|---|---|
| Zymomonas mobilis | ||
| CP4 | H. Yanase [14] | |
| Z4-80a | Thermoadapted mutant of CP4 | [14] |
| Z4-80b | Thermoadapted mutant of CP4 | [14] |
| Z4-80c | Thermoadapted mutant of CP4 | [14] |
| Z4-80d | Thermoadapted mutant of CP4 | [14] |
| 0567R164C | CP4 with a missense mutation (R to C at position 164 in the product of ZCP4_RS02830 (old locus tag ZCP4_0567)) from Z4-80a | This study |
| 1739A89V | CP4 with a missense mutation (A to V at position 89 in the product of ZCP4_RS08695 (old locus tag ZCP4_1739)) from Z4-80a | This study |
| 0125T403A | CP4 with a missense mutation (T to A at position 403 in the product of ZCP4_RS00655 (old locus tag ZCP4_0125)) from Z4-80a | This study |
| 0707P438L | CP4 with a missense mutation (P to L at position 438 in the product of ZCP4_RS03540 (old locus tag ZCP4_0707)) from Z4-80a | This study |
| 1702E136K | CP4 with a missense mutation (E to K at position 136 in the product of ZCP4_RS08505 (old locus tag ZCP4_1702)) from Z4-80a | This study |
| 0028M99I | CP4 with a missense mutation (M to I at position 99 in the product of ZCP4_RS00160 (old locus tag ZCP4_0028)) from Z4-80b | This study |
| 1646aaIns | CP4 with an insertion mutation (IYDGSL at position 230 in the product of ZCP4_RS08220 (old locus tag ZCP4_1646)) from Z4-80b | This study |
| 1739V223M | CP4 with a missense mutation (V to M at position 223 in the product of ZCP4_RS08695 (old locus tag ZCP4_1739)) from Z4-80c | This study |
| 0125R597C | CP4 with a missense mutation (R to C at position 597 in the product of ZCP4_RS00655 (old locus tag ZCP4_0125)) from Z4-80c | This study |
| 1702T144M | CP4 with a missense mutation (T to M at position 144 in the product of ZCP4_RS08505 (old locus tag ZCP4_1702)) from Z4-80c | This study |
| 1739L158F | CP4 with a missense mutation (L to F at position 158 in the product of ZCP4_RS08695 (old locus tag ZCP4_1739)) from Z4-80d | This study |
| 0588L118F | CP4 with a missense mutation (L to F at position 118 in the product of ZCP4_RS02940 (old locus tag ZCP4_0588)) from Z4-80d | This study |
| 1703K462E | CP4 with a missense mutation (K to E at position 462 in the product of ZCP4_RS08510 (old locus tag ZCP4_1703)) from Z4-80d | This study |
| Escherichia coli | ||
| DH5λ | F−, ϕ80lacZΔM15,Δ(lacZYA-argF) U169, deoR, recA1, endA1, hsdR17(rK−, mK+), phoA, supE44, λ−, thi-1, gyrA96, relA1 | Takara Bio |
| S17-1 | F−, recA, pro, thi, hsdR, [RP4-2 Tc::Mu Km::Tn7 Tra+ Tpr Smr] | [41] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pattanakittivorakul, S.; Kato, S.; Kuga, T.; Kosaka, T.; Matsutani, M.; Murata, M.; Ishikawa, M.; Charoenpunthuwong, K.; Thanonkeo, P.; Yamada, M. Limited Diversity of Thermal Adaptation to a Critical Temperature in Zymomonas mobilis: Evidence from Multiple-Parallel Laboratory Evolution Experiments. Int. J. Mol. Sci. 2025, 26, 3052. https://doi.org/10.3390/ijms26073052
Pattanakittivorakul S, Kato S, Kuga T, Kosaka T, Matsutani M, Murata M, Ishikawa M, Charoenpunthuwong K, Thanonkeo P, Yamada M. Limited Diversity of Thermal Adaptation to a Critical Temperature in Zymomonas mobilis: Evidence from Multiple-Parallel Laboratory Evolution Experiments. International Journal of Molecular Sciences. 2025; 26(7):3052. https://doi.org/10.3390/ijms26073052
Chicago/Turabian StylePattanakittivorakul, Sornsiri, Shun Kato, Takashi Kuga, Tomoyuki Kosaka, Minenosuke Matsutani, Masayuki Murata, Morio Ishikawa, Kankanok Charoenpunthuwong, Pornthap Thanonkeo, and Mamoru Yamada. 2025. "Limited Diversity of Thermal Adaptation to a Critical Temperature in Zymomonas mobilis: Evidence from Multiple-Parallel Laboratory Evolution Experiments" International Journal of Molecular Sciences 26, no. 7: 3052. https://doi.org/10.3390/ijms26073052
APA StylePattanakittivorakul, S., Kato, S., Kuga, T., Kosaka, T., Matsutani, M., Murata, M., Ishikawa, M., Charoenpunthuwong, K., Thanonkeo, P., & Yamada, M. (2025). Limited Diversity of Thermal Adaptation to a Critical Temperature in Zymomonas mobilis: Evidence from Multiple-Parallel Laboratory Evolution Experiments. International Journal of Molecular Sciences, 26(7), 3052. https://doi.org/10.3390/ijms26073052






