Identification and Analysis of Cuticular Wax Biosynthesis Related Genes in Salicornia europaea Under NaCl Treatment
Abstract
1. Introduction
2. Results
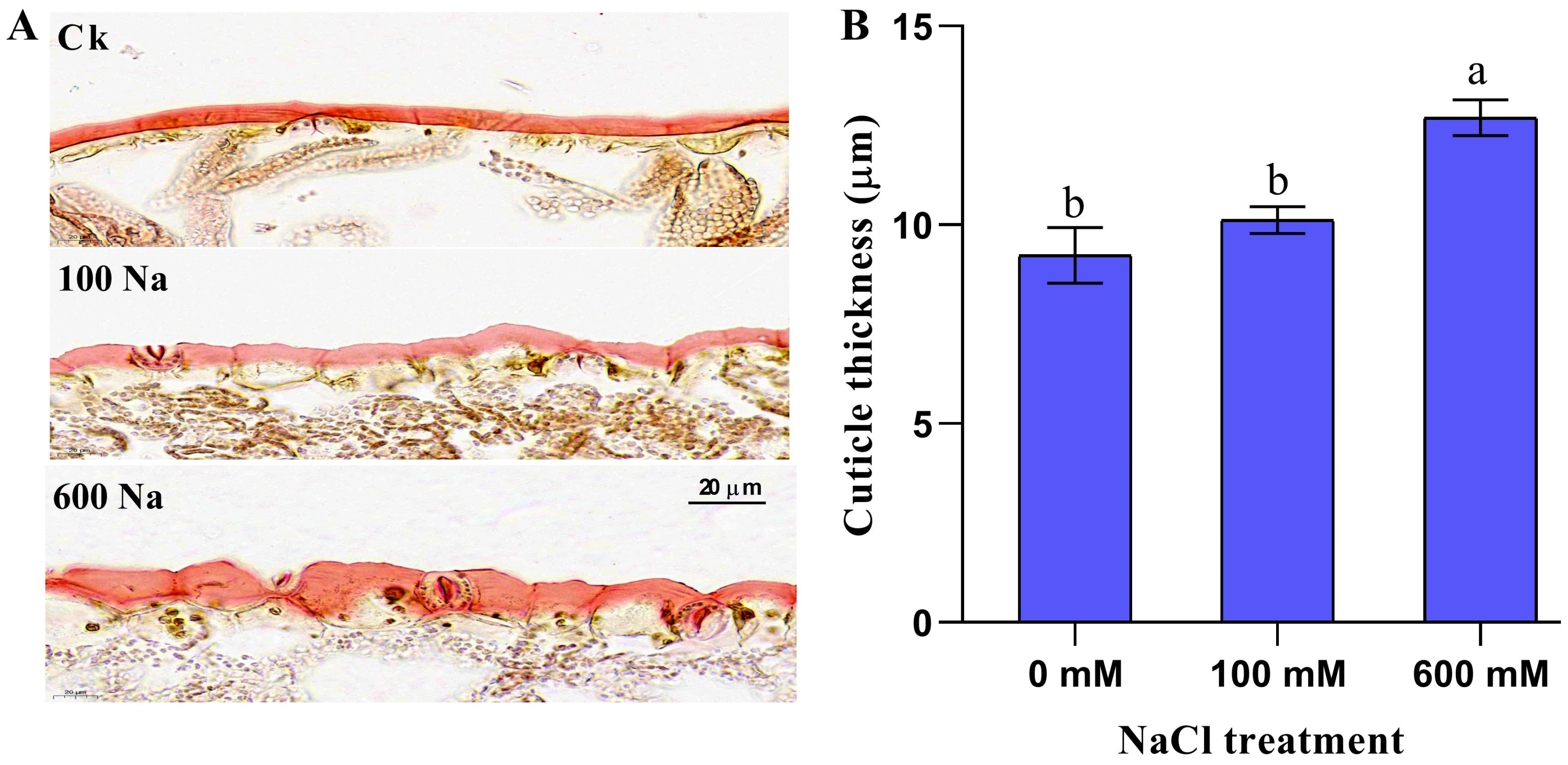
2.1. Shoot Cuticle Thickness Response to NaCl Treatment
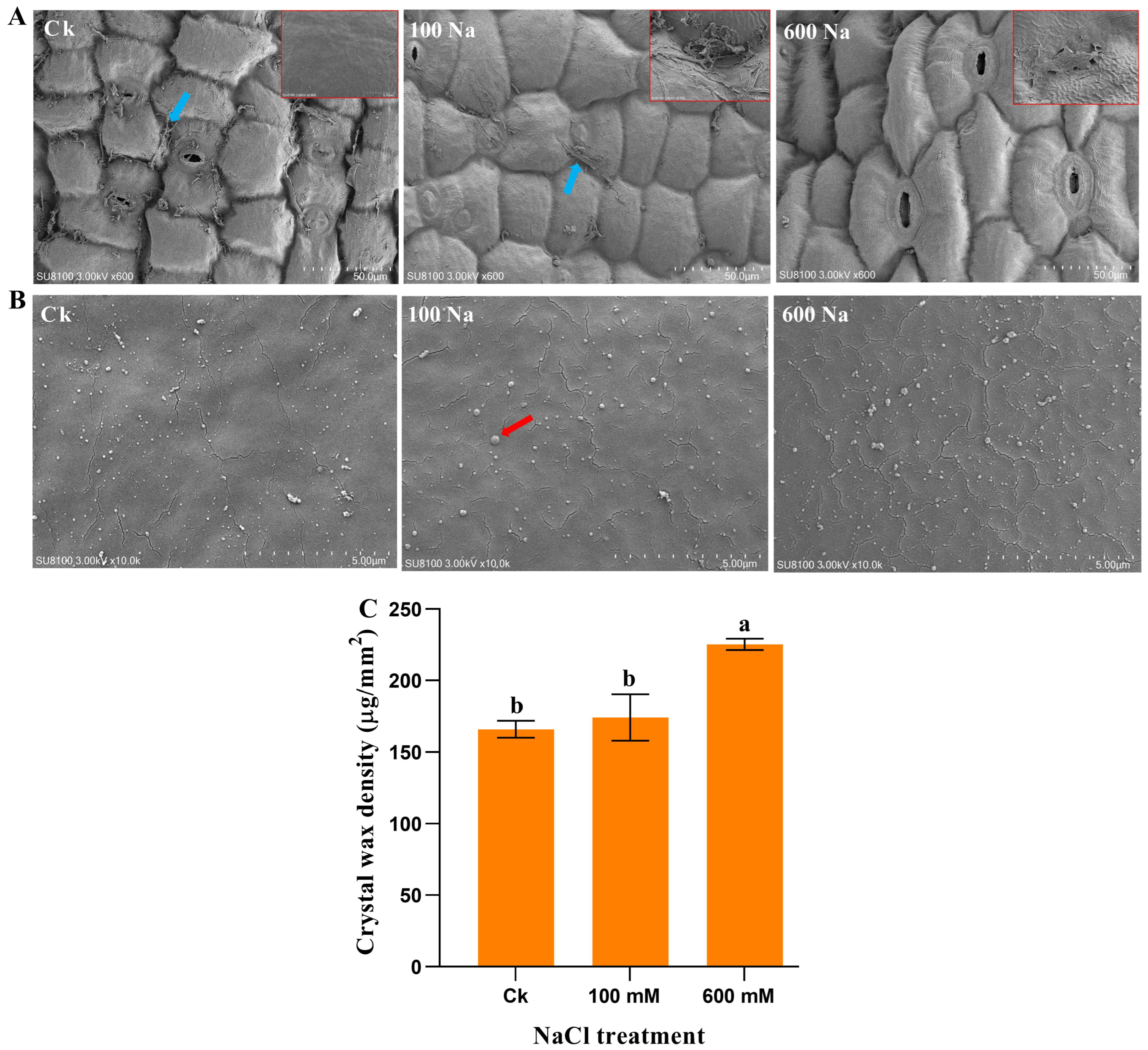
2.2. Cuticular Wax Morphology Under NaCl Treatment
2.3. Wax Content and GC-MS Analysis of the Wax Composition in S. europaea
2.4. Identification of Cuticular Wax Biosynthesis Proteins in S. europaea
2.5. Multiple Sequence Alignment of Cuticular Wax Biosynthesis Proteins
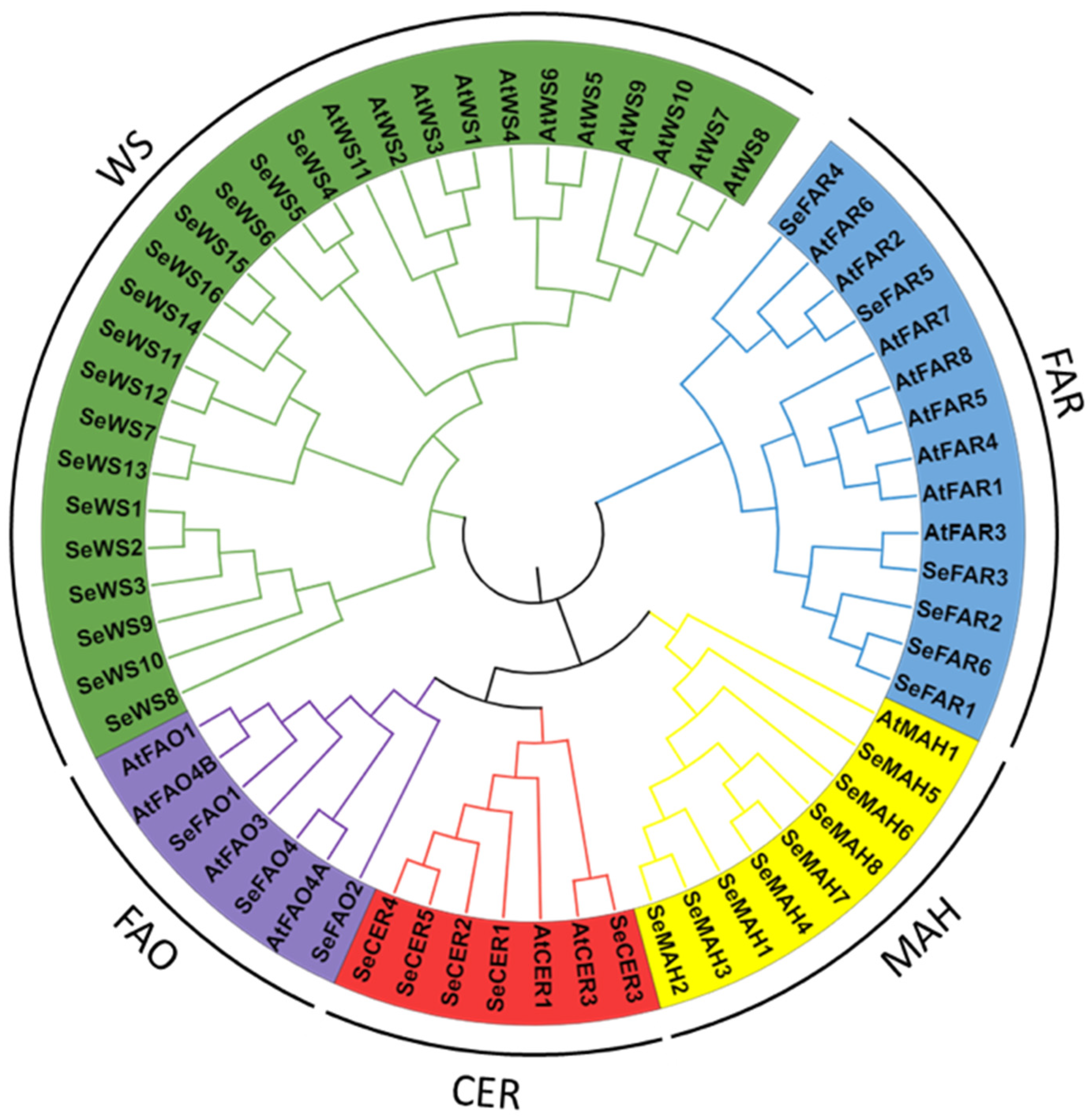
2.6. Phylogenetic Relationship
2.7. Conserved Motifs of Wax Biosynthesis Proteins in S. europaea
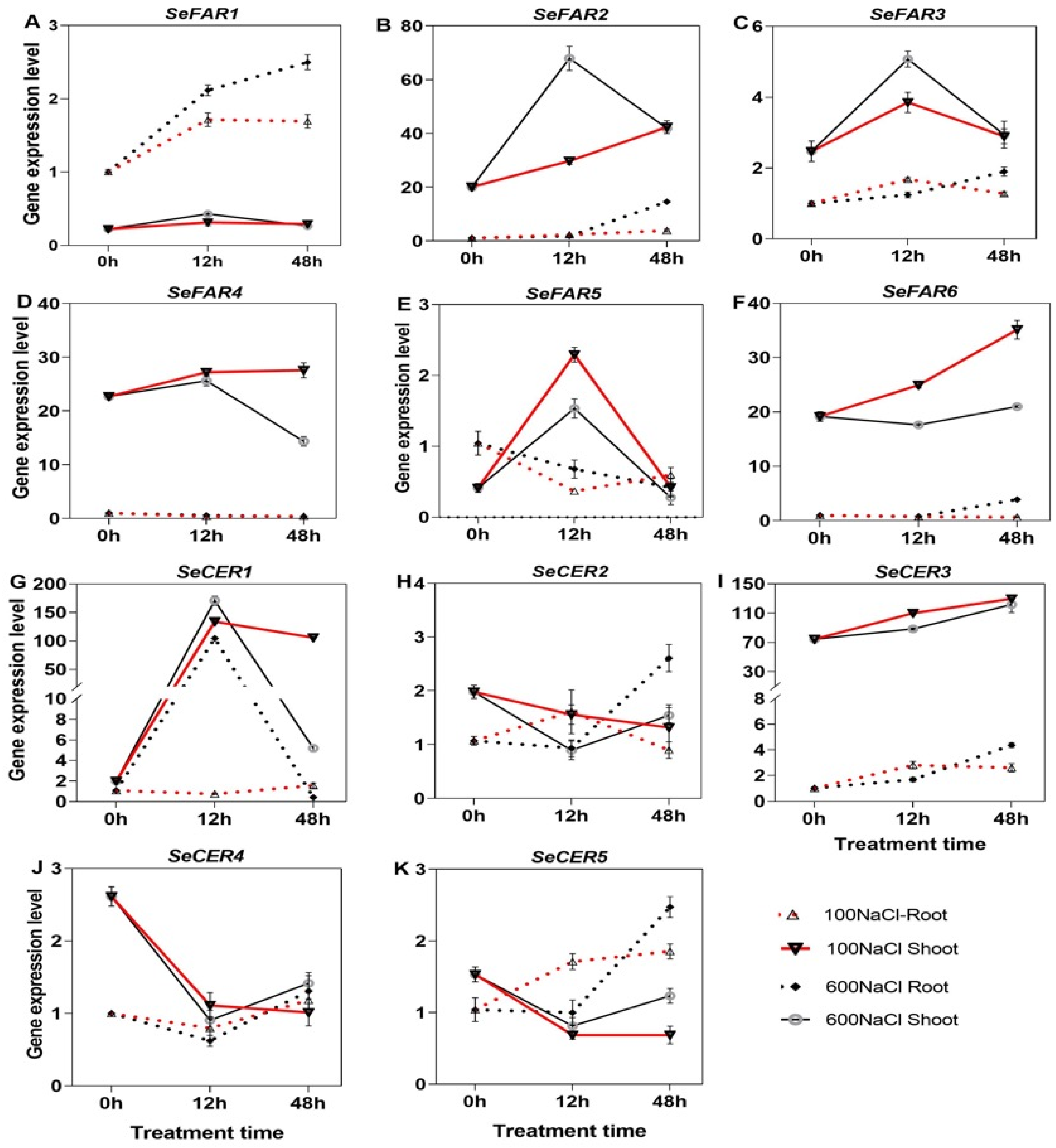
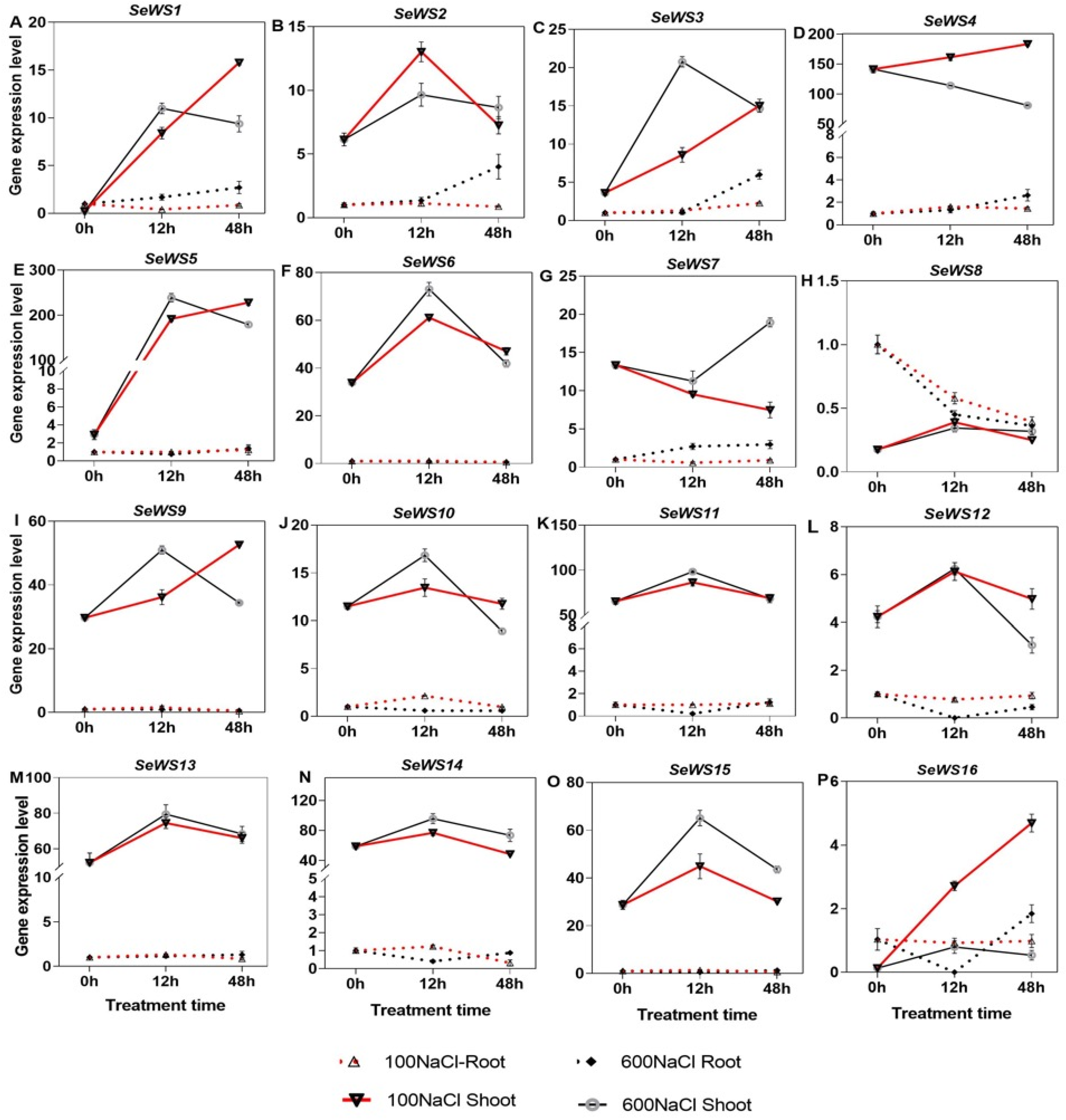
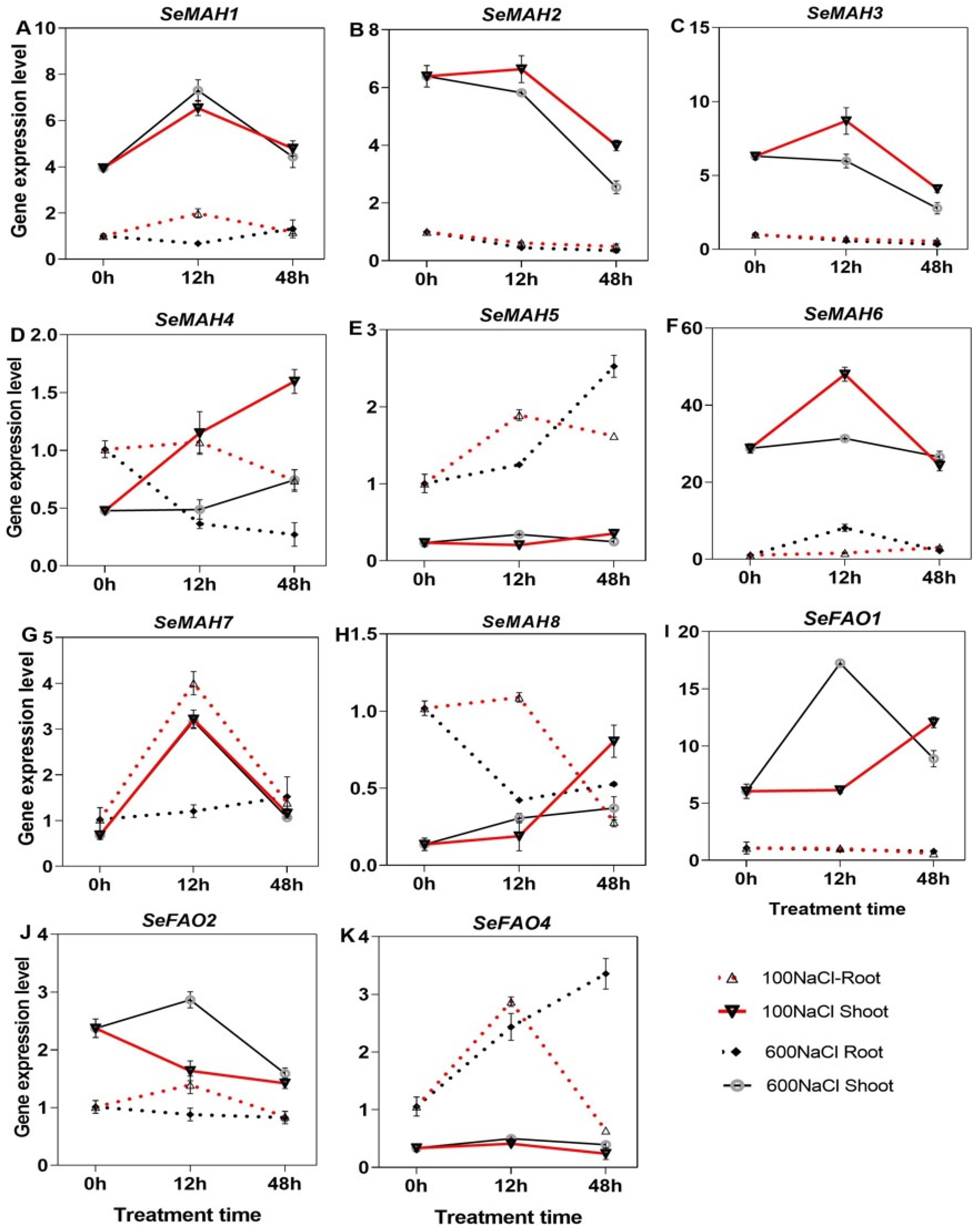
2.8. Expression of Wax Biosynthesis Genes Under NaCl Treatment
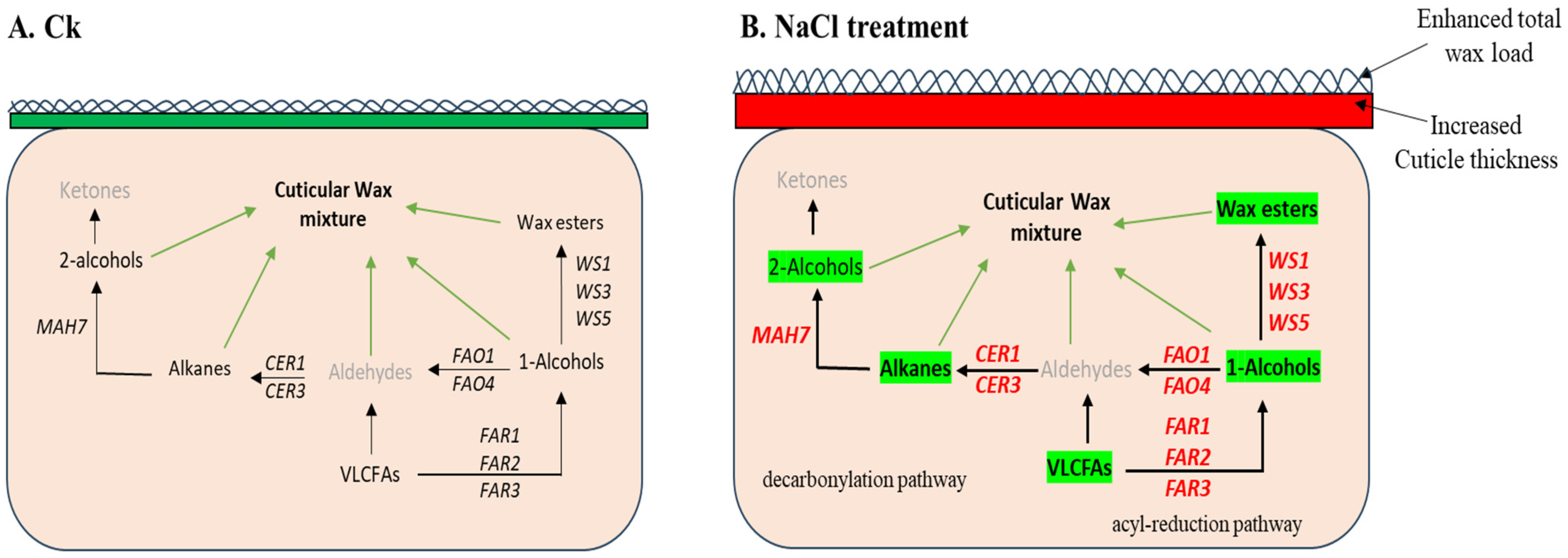
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions and Stress Treatments
4.2. Scanning Electron Microscopy (SEM) Analysis
4.3. Oil Red Staining Procedure for Cuticle Analysis
4.4. Determination of Cuticular Wax Composition
4.5. Cuticular Wax Biosynthesis Protein Sequence Search
4.6. Analysis of Physicochemical Properties and Subcellular Localization of Proteins
4.7. Sequence Alignment, Phylogenetic, and Conservative Motif Analysis
4.8. RNA Extraction and RT-qPCR Analysis
4.9. Data Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Z.; Li, X.; Wang, Y. Evaluation of salt tolerance of 59 alfalfa germplasm materials at seedling stage and screening of salt tolerance indexes. Acta Grassl. 2020, 28, 112–121. [Google Scholar]
- Zörb, C.; Geilfus, C.M.; Dietz, K.-J. Salinity and Crop Yield. Plant Biol. 2018, 21, 31–38. [Google Scholar] [CrossRef]
- Shaheenuzzamn, M.; Shi, S.; Sohail, K.; Wu, H.; Liu, T.; An, P.; Wang, Z.; Hasanuzzaman, M. Regulation of cuticular wax biosynthesis in plants under abiotic stress. Plant Biotechnol. Rep. 2021, 15, 1–12. [Google Scholar] [CrossRef]
- Zhang, P.; Zou, Y.; Song, B.; Zhou, M.; He, J.; Chen, Y.; Zhou, Y.; Xu, X. Cuticular lipids and associated gene expression analysis under NaCl stress in Thellungiella salsuginea. Physiol. Plant. 2022, 174, e13625. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Suh, M.C. Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 2015, 34, 557–572. [Google Scholar] [CrossRef]
- Bernard, A.; Joubès, J. Arabidopsis cuticular waxes: Advances in synthesis, export and regulation. Prog. Lipid Res. 2013, 52, 110–129. [Google Scholar] [CrossRef] [PubMed]
- Nawrath, C.; Schreiber, L.; Franke, R.B.; Geldner, N.; Reina-Pinto, J.J.; Kunst, L. Apoplastic diffusion barriers in Arabidopsis. Arab. Book/Am. Soc. Plant Biol. 2013, 11, e0167. [Google Scholar] [CrossRef]
- Xue, D.; Zhang, X.; Lu, X.; Chen, G.; Chen, Z.H. Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front. Plant Sci. 2017, 8, 621. [Google Scholar] [CrossRef]
- Vogg, G.; Fischer, S.; Leide, J.; Emmanuel, E.; Jetter, R.; Levy, A.A.; Riederer, M. Tomato fruit cuticular waxes and their effects on transpiration barrier properties: Functional characterization of a mutant deficient in a very-long-chain fatty acid β-ketoacyl-CoA synthase. J. Exp. Bot. 2004, 55, 1401–1410. [Google Scholar] [CrossRef]
- Xu, X.; Xue, K.; Tang, S.; He, J.; Song, B.; Zhou, M.; Zou, Y.; Zhou, Y.; Jenks, M.A. The relationship between cuticular lipids and associated gene expression in above ground organs of Thellungiella salsugineum (Pall.) Al-Shehbaz & Warwick. Plant Sci. 2019, 287, 110200. [Google Scholar] [CrossRef]
- Lv, S.; Tai, F.; Guo, J.; Jiang, P.; Lin, K.; Wang, D.; Zhang, X.; Li, Y. Phosphatidylserine Synthase from Salicornia europaea Is Involved in Plant Salt Tolerance by Regulating Plasma Membrane Stability. Plant Cell Physiol. 2021, 62, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Feng, J.; Fan, P.; Chen, X.; Guo, J.; Lv, S.; Bao, H.; Jia, W.; Tai, F.; Jiang, P.; et al. Comparative proteomics of root plasma membrane proteins reveals the involvement of calcium signalling in NaCl-facilitated nitrate uptake in Salicornia europaea. J. Exp. Bot. 2015, 66, 4497–4510. [Google Scholar] [CrossRef]
- Singh, D.; Buhmann, A.K.; Flowers, T.J.; Seal, C.E.; Papenbrock, J. Salicornia as a crop plant in temperate regions: Selection of genetically characterized ecotypes and optimization of their cultivation conditions. AoB Plants 2014, 6, plu071. [Google Scholar] [CrossRef] [PubMed]
- Shpigel, M.; Ben-Ezra, D.; Shauli, L.; Sagi, M.; Ventura, Y.; Samocha, T.; Lee, J.J. Constructed wetland with Salicornia as a biofilter for mariculture effluents. Aquaculture 2013, 412, 52–63. [Google Scholar] [CrossRef]
- Lv, S.; Jiang, P.; Nie, L.; Chen, X.; Tai, F.; Wang, D.; Fan, P.; Feng, J.; Bao, H.; Wang, J. H+-pyrophosphatase from S alicornia europaea confers tolerance to simultaneously occurring salt stress and nitrogen deficiency in Arabidopsis and wheat. Plant Cell Environ. 2015, 38, 2433–2449. [Google Scholar] [CrossRef]
- Duan, H.; Tiika, R.J.; Tian, F.; Lu, Y.; Zhang, Q.; Hu, Y.; Cui, G.; Yang, H. Metabolomics analysis unveils important changes involved in the salt tolerance of Salicornia europaea. Front. Plant Sci. 2023, 13, 1097076. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, J.; Fan, P.; Jia, W.; Nie, L.; Jiang, P.; Chen, X.; Lv, S.; Wan, L.; Chang, S.; et al. High-throughput deep sequencing reveals that microRNAs play important roles in salt tolerance of euhalophyte Salicornia europaea. BMC Plant Biol. 2015, 15, 63. [Google Scholar] [CrossRef]
- Singh, D.; Jha, B. The isolation and identification of salt-responsive novel microRNAs from Salicornia brachiata, an extreme halophyte. Plant Biotechnol. Rep. 2014, 8, 325–336. [Google Scholar] [CrossRef]
- Zhou, D.; Ding, M.; Wen, S.; Tian, Q.; Zhang, X.; Fang, Y.; Xue, D. Characterization of the Fatty Acyl-CoA Reductase (FAR) Gene Family and Its Response to Abiotic Stress in Rice (Oryza sativa L.). Plants 2024, 13, 1010. [Google Scholar] [CrossRef]
- Domergue, F.; Vishwanath, S.J.; Joubès, J.; Ono, J.; Lee, J.A.; Bourdon, M.; Alhattab, R.; Lowe, C.; Pascal, S.; Lessire, R.; et al. Three Arabidopsis Fatty Acyl-Coenzyme A Reductases, FAR1, FAR4, and FAR5, Generate Primary Fatty Alcohols Associated with Suberin Deposition. Plant Physiol. 2010, 153, 1539–1554. [Google Scholar] [CrossRef]
- Rosli, R.; Chan, P.-L.; Chan, K.-L.; Amiruddin, N.; Low, E.-T.L.; Singh, R.; Harwood, J.L.; Murphy, D.J. In silico characterization and expression profiling of the diacylglycerol acyltransferase gene family (DGAT1, DGAT2, DGAT3 and WS/DGAT) from oil palm, Elaeis guineensis. Plant Sci. 2018, 275, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wu, X.; Lam, P.; Bird, D.; Zheng, H.; Samuels, L.; Jetter, R.; Kunst, L. Identification of the Wax Ester Synthase/Acyl-Coenzyme A:Diacylglycerol Acyltransferase WSD1 Required for Stem Wax Ester Biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 97–107. [Google Scholar] [CrossRef]
- Pan, F.; Li, X.; Zhong, D.; Lu, X.; Pan, C.; Hu, J.; Su, W.; Zhang, H.; Zhang, C.; Shi, L.; et al. Eceriferum Genes in Tomato (Solanum lycopersicum): Genome-Wide Identification and Expression Analysis Reveal Their Potential Functions during Domestication. Horticulturae 2023, 9, 748. [Google Scholar] [CrossRef]
- Rizwan, H.M.; Waheed, A.; Ma, S.; Li, J.; Arshad, M.B.; Irshad, M.; Li, B.; Yang, X.; Ali, A.; Ahmed, M.A.A. Comprehensive genome-wide identification and expression profiling of eceriferum (CER) gene family in passion fruit (Passiflora edulis) under fusarium kyushuense and drought stress conditions. Front. Plant Sci. 2022, 13, 898307. [Google Scholar] [CrossRef]
- Li, N.; Li, X.Z.; Song, Y.Q.; Yang, S.T.; Li, L.L. Genome-Wide Identification, Characterization, and Expression Profiling of the ECERIFERUM (CER) Gene Family in Ziziphus jujube. Russ. J. Plant Physiol. 2021, 68, 828–837. [Google Scholar] [CrossRef]
- Chen, G.; Komatsuda, T.; Ma, J.F.; Li, C.; Yamaji, N.; Nevo, E. A functional cutin matrix is required for plant protection against water loss. Plant Signal. Behav. 2011, 6, 1297–1299. [Google Scholar] [CrossRef]
- Mills, D.; Zhang, G.; Benzioni, A. Effect of different salts and of ABA on growth and mineral uptake in Jojoba shoots grown in vitro. J. Plant Physiol. 2001, 158, 1031–1039. [Google Scholar] [CrossRef]
- Liu, N.; Chen, J.; Wang, T.; Li, Q.; Cui, P.; Jia, C.; Hong, Y. Overexpression of WAX INDUCER1/SHINE1 Gene Enhances Wax Accumulation under Osmotic Stress and Oil Synthesis in Brassica napus. Int. J. Mol. Sci. 2019, 20, 4435. [Google Scholar] [CrossRef]
- Wen, X.; Geng, F.; Cheng, Y.; Wang, J. Ectopic expression of CsMYB30 from Citrus sinensis enhances salt and drought tolerance by regulating wax synthesis in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 166, 777–788. [Google Scholar] [CrossRef]
- Tomaszewski, D.; Zieliński, J. Epicuticular wax structures on stems and comparison between stems and leaves—A survey. Flora-Morphol. Distrib. Funct. Ecol. Plants 2014, 209, 215–232. [Google Scholar] [CrossRef]
- Chen, X.; Goodwin, S.M.; Boroff, V.L.; Liu, X.; Jenks, M.A. Cloning and Characterization of the WAX2 Gene of Arabidopsis Involved in Cuticle Membrane and Wax Production. Plant Cell 2003, 15, 1170–1185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Broeckling, C.D.; Blancaflor, E.B.; Sledge, M.K.; Sumner, L.W.; Wang, Z.Y. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 2005, 42, 689–707. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Dommisse, A.; Barthlott, W. Chemistry and crystal growth of plant wax tubules of lotus (Nelumbo nucifera) and nasturtium (Tropaeolum majus) leaves on technical substrates. Cryst. Growth Des. 2006, 6, 2571–2578. [Google Scholar] [CrossRef]
- Borisjuk, N.; Peterson, A.A.; Lv, J.; Qu, G.; Luo, Q.; Shi, L.; Chen, G.; Kishchenko, O.; Zhou, Y.; Shi, J. Structural and biochemical properties of duckweed surface cuticle. Front. Chem. 2018, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Ayaz, A.; Zhao, H.; Lü, S. The Plant Fatty Acyl Reductases. Int. J. Mol. Sci. 2022, 23, 16156. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, S.; Niu, C.; Liu, D.; Yan, T.; Tian, Y.; Liu, S.; Xie, K.; Li, Z.; Wang, Y.; et al. ZmMs25 encoding a plastid-localized fatty acyl reductase is critical for anther and pollen development in maize. J. Exp. Bot. 2021, 72, 4298–4318. [Google Scholar] [CrossRef]
- Stöveken, T.; Kalscheuer, R.; Steinbüchel, A. Both histidine residues of the conserved HHXXXDG motif are essential for wax ester synthase/acyl-CoA: Diacylglycerol acyltransferase catalysis. Eur. J. Lipid Sci. Technol. 2009, 111, 112–119. [Google Scholar] [CrossRef]
- Wang, H.-b.; Lv, X.-y.; Li, Y.-y.; Jiang, H. Identification and Expression Analysis of WSD (Wax Ester Synthase/Diacylglycerol Acyltransferase) Gene Family in Apple. Erwerbs-Obstbau 2023, 65, 633–644. [Google Scholar] [CrossRef]
- Zhang, N.; Mao, Z.; Luo, L.; Wan, X.; Huang, F.; Gong, Y. Two bifunctional enzymes from the marine protist Thraustochytrium roseum: Biochemical characterization of wax ester synthase/acyl-CoA:diacylglycerol acyltransferase activity catalyzing wax ester and triacylglycerol synthesis. Biotechnol. Biofuels 2017, 10, 185. [Google Scholar] [CrossRef]
- Vanhanen, S.; West, M.; Kroon, J.T.; Lindner, N.; Casey, J.; Cheng, Q.; Elborough, K.M.; Slabas, A.R. A consensus sequence for long-chain fatty-acid alcohol oxidases from Candida identifies a family of genes involved in lipid omega-oxidation in yeast with homologues in plants and bacteria. J. Biol. Chem. 2000, 275, 4445–4452. [Google Scholar] [CrossRef]
- Cheng, Q.; Liu, H.T.; Bombelli, P.; Smith, A.; Slabas, A.R. Functional identification of AtFao3, a membrane bound long chain alcohol oxidase in Arabidopsis thaliana. FEBS Lett. 2004, 574, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, K.; Geeta, R.; Panjabi, P. Origin and diversification of ECERIFERUM1 (CER1) and ECERIFERUM3 (CER3) genes in land plants and phylogenetic evidence that the ancestral CER1/3 gene resulted from the fusion of pre-existing domains. Mol. Phylogenet. Evol. 2021, 159, 107101. [Google Scholar] [CrossRef] [PubMed]
- Darnet, S.; Bard, M.; Rahier, A. Functional identification of sterol-4α-methyl oxidase cDNAs from Arabidopsis thaliana by complementation of a yeast erg25 mutant lacking sterol-4α-methyl oxidation1. FEBS Lett. 2001, 508, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Rowland, O.; Zheng, H.; Hepworth, S.R.; Lam, P.; Jetter, R.; Kunst, L. CER4 Encodes an Alcohol-Forming Fatty Acyl-Coenzyme A Reductase Involved in Cuticular Wax Production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef]
- Round, J.; Roccor, R.; Li, S.-N.; Eltis, L.D. A Fatty Acyl Coenzyme A Reductase Promotes Wax Ester Accumulation in Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2017, 83, e00902–e00917. [Google Scholar] [CrossRef]
- Abdullah, H.; Rodriguez, J.; Salacup, J.; Castañeda, I.; Schnell, D.; Pareek, A.; Dhankher, O.P. Increased Cuticle Waxes by Overexpression of WSD1 Improves Osmotic Stress Tolerance in Arabidopsis thaliana and Camelina sativa. Int. J. Mol. Sci. 2021, 22, 5173. [Google Scholar] [CrossRef]
- Bernard, A.; Domergue, F.; Pascal, S.; Jetter, R.; Renne, C.; Faure, J.-D.; Haslam, R.P.; Napier, J.A.; Lessire, R.; Joubès, J. Reconstitution of Plant Alkane Biosynthesis in Yeast Demonstrates That Arabidopsis ECERIFERUM1 and ECERIFERUM3 Are Core Components of a Very-Long-Chain Alkane Synthesis Complex. Plant Cell 2012, 24, 3106–3118. [Google Scholar] [CrossRef]
- Goswami, P.; Chinnadayyala, S.S.R.; Chakraborty, M.; Kumar, A.K.; Kakoti, A. An overview on alcohol oxidases and their potential applications. Appl. Microbiol. Biotechnol. 2013, 97, 4259–4275. [Google Scholar] [CrossRef]
- Yang, X.; Cui, L.; Li, S.; Ma, C.; Kosma, D.K.; Zhao, H.; Lü, S. Fatty alcohol oxidase 3 (FAO3) and FAO4b connect the alcohol- and alkane-forming pathways in Arabidopsis stem wax biosynthesis. J. Exp. Bot. 2021, 73, 3018–3029. [Google Scholar] [CrossRef]
- Wei, J.; Tiika, R.J.; Ma, Y.; Yang, H.; Cui, G.; Tian, F.; Zhang, Y.; Duan, H.; Ma, R. Transcriptome-wide identification and analysis of the KT/HAK/KUP family in black goji under NaCl stress. Agron. J. 2022, 114, 2069–2080. [Google Scholar] [CrossRef]
- Tiika, R.J.; Wei, J.; Cui, G.; Ma, Y.; Yang, H.; Duan, H. Transcriptome-wide characterization and functional analysis of Xyloglucan endo-transglycosylase/hydrolase (XTH) gene family of Salicornia europaea L. under salinity and drought stress. BMC Plant Biol. 2021, 21, 491. [Google Scholar] [CrossRef] [PubMed]
- Lü, X.P.; Gao, H.J.; Zhang, L.; Wang, Y.P.; Shao, K.Z.; Zhao, Q.; Zhang, J.L. Dynamic responses of Haloxylon ammodendron to various degrees of simulated drought stress. Plant Physiol. Biochem. PPB 2019, 139, 121–131. [Google Scholar] [CrossRef]
- Salazar, O.R.; Chen, K.; Melino, V.J.; Reddy, M.P.; Hřibová, E.; Čížková, J.; Beránková, D.; Arciniegas Vega, J.P.; Cáceres Leal, L.M.; Aranda, M.; et al. SOS1 tonoplast neo-localization and the RGG protein SALTY are important in the extreme salinity tolerance of Salicornia bigelovii. Nat. Commun. 2024, 15, 4279. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Xiao, X.; Ma, J.; Wang, J.; Wu, X.; Li, P.; Yao, Y. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 2014, 5, 788. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Protein Name | Gene ID | Amino Acids | Molecular Weight/kD | Theoretical pI | Instability Index (II) | Aliphatic Index | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| SeFAR1 | >Seu_jg19520 | 489 | 55.60 | 7.99 | 28.34 | 96.50 | Golg |
| SeFAR2 | >Seu_jg19521 | 488 | 55.23 | 8.76 | 22.97 | 106.82 | Chlo |
| SeFAR3 | >Seu_jg8291 | 492 | 56.02 | 8.66 | 35.31 | 97.05 | Chlo |
| SeFAR4 | >Seu_jg25353 | 606 | 68.79 | 8.35 | 37.58 | 100.02 | Chlo |
| SeFAR5 | >Seu_jg23269 | 600 | 66.79 | 8.85 | 37.48 | 85.07 | Chlo |
| SeFAR6 | >Seu_jg19519 | 488 | 55.27 | 8.85 | 25.69 | 98.81 | Golg |
| SeMAH1 | >Seu_jg13328 | 486 | 56.52 | 8.81 | 29.10 | 93.42 | ER |
| SeMAH2 | >Seu_jg24080 | 433 | 50.16 | 7.68 | 29.48 | 91.36 | ER |
| SeMAH3 | >Seu_jg24081 | 397 | 46.09 | 8.76 | 28.63 | 92.77 | ER |
| SeMAH4 | >Seu_jg24086 | 488 | 56.79 | 8.45 | 31.65 | 94.02 | ER |
| SeMAH5 | >Seu_jg24089 | 513 | 59.09 | 6.56 | 38.82 | 92.32 | ER |
| SeMAH6 | >Seu_jg17936 | 507 | 58.83 | 8.10 | 36.31 | 88.64 | ER |
| SeMAH7 | >Seu_jg15161 | 314 | 36.62 | 5.94 | 33.22 | 94.87 | ER |
| SeMAH8 | >Seu_jg15162 | 121 | 13.70 | 9.33 | 25.94 | 81.49 | ER |
| SeCER1 | >Seu_jg14489 | 622 | 73.02 | 8.93 | 37.99 | 94.77 | Vac |
| SeCER2 | >Seu_jg6667 | 625 | 71.99 | 8.93 | 32.36 | 101.52 | Vac |
| SeCER3 | >Seu_jg19922 | 623 | 71.52 | 9.19 | 35.87 | 103.56 | Vac |
| SeCER4 | >Seu_jg6668 | 620 | 71.37 | 9.00 | 32.14 | 100.13 | Vac |
| SeCER5 | >Seu_jg6669 | 114 | 12.83 | 5.53 | 47.02 | 71.84 | Chlo |
| SeWS1 | >Seu_jg12815 | 460 | 51.99 | 5.84 | 29.75 | 97.87 | Cyto |
| SeWS2 | >Seu_jg12816 | 149 | 16.87 | 5.91 | 28.51 | 110.60 | Cm, Chlo, Nuc |
| SeWS3 | >Seu_jg12818 | 163 | 18.75 | 5.42 | 38.08 | 84.91 | Nuc |
| SeWS4 | >Seu_jg6079 | 523 | 58.98 | 8.06 | 37.29 | 97.67 | Chlo |
| SeWS5 | >Seu_jg6080 | 541 | 60.68 | 8.36 | 39.12 | 96.38 | Chlo |
| SeWS6 | >Seu_jg6081 | 521 | 59.09 | 9.31 | 30.27 | 94.26 | Per |
| SeWS7 | >Seu_jg18079 | 485 | 53.90 | 9.10 | 34.61 | 90.19 | Chlo |
| SeWS8 | >Seu_jg20467 | 461 | 51.37 | 6.47 | 36.86 | 90.15 | Cm, Chlo, Nuc |
| SeWS9 | >Seu_jg20468 | 480 | 54.06 | 9.06 | 31.77 | 100.44 | Cm, Chlo |
| SeWS10 | >Seu_jg20469 | 476 | 53.23 | 8.70 | 37.97 | 94.98 | Chlo |
| SeWS11 | >Seu_jg20583 | 496 | 56.58 | 9.43 | 38.17 | 94.29 | Chlo |
| SeWS12 | >Seu_jg20584 | 489 | 55.68 | 9.28 | 44.96 | 87.48 | Chlo |
| SeWS13 | >Seu_jg21009 | 473 | 52.30 | 6.39 | 45.66 | 91.10 | Chlo |
| SeWS14 | >Seu_jg21132 | 153 | 16.86 | 8.60 | 35.14 | 98.82 | Cm, Chlo |
| SeWS15 | >Seu_jg21133 | 491 | 54.91 | 8.86 | 38.31 | 99.04 | Chlo |
| SeWS16 | >Seu_jg21134 | 399 | 44.85 | 8.72 | 41.61 | 96.74 | Chlo, Cyto |
| SeFAO1 | >Seu_jg28185 | 770 | 84.66 | 8.70 | 35.63 | 82.31 | Chlo, Vac |
| SeFAO2 | >Seu_jg3513 | 729 | 79.36 | 8.85 | 32.83 | 87.34 | Chlo, Mito |
| SeFAO4 | >Seu_jg12627 | 813 | 90.22 | 8.31 | 36.47 | 81.64 | Chlo, Vac |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiika, R.J.; Yang, H.; Cui, G.; Ma, Y.; Boamah, S.; Li, Y.; Duan, H. Identification and Analysis of Cuticular Wax Biosynthesis Related Genes in Salicornia europaea Under NaCl Treatment. Int. J. Mol. Sci. 2025, 26, 2632. https://doi.org/10.3390/ijms26062632
Tiika RJ, Yang H, Cui G, Ma Y, Boamah S, Li Y, Duan H. Identification and Analysis of Cuticular Wax Biosynthesis Related Genes in Salicornia europaea Under NaCl Treatment. International Journal of Molecular Sciences. 2025; 26(6):2632. https://doi.org/10.3390/ijms26062632
Chicago/Turabian StyleTiika, Richard John, Hongshan Yang, Guangxin Cui, Yanjun Ma, Solomon Boamah, Yi Li, and Huirong Duan. 2025. "Identification and Analysis of Cuticular Wax Biosynthesis Related Genes in Salicornia europaea Under NaCl Treatment" International Journal of Molecular Sciences 26, no. 6: 2632. https://doi.org/10.3390/ijms26062632
APA StyleTiika, R. J., Yang, H., Cui, G., Ma, Y., Boamah, S., Li, Y., & Duan, H. (2025). Identification and Analysis of Cuticular Wax Biosynthesis Related Genes in Salicornia europaea Under NaCl Treatment. International Journal of Molecular Sciences, 26(6), 2632. https://doi.org/10.3390/ijms26062632





