Comparative Transcriptome Analysis Reveals Potential Molecular Regulation of Organic Acid Metabolism During Fruit Development in Late-Maturing Hybrid Citrus Varieties
Abstract
1. Introduction
2. Results
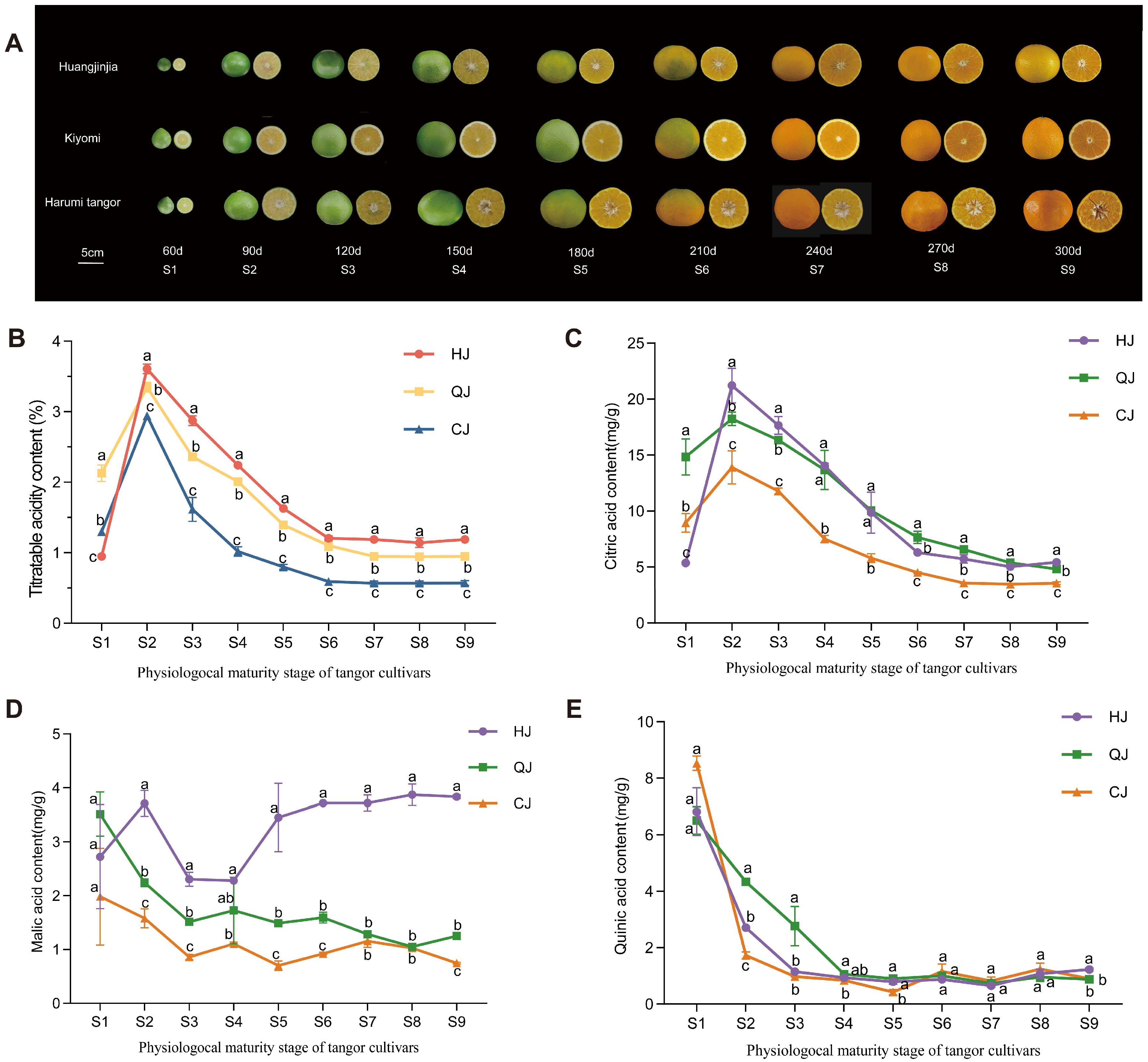
2.1. TA and Organic Acid Content in the Growth of Late-Maturing Hybrid Citrus Varieties with Different Acidities
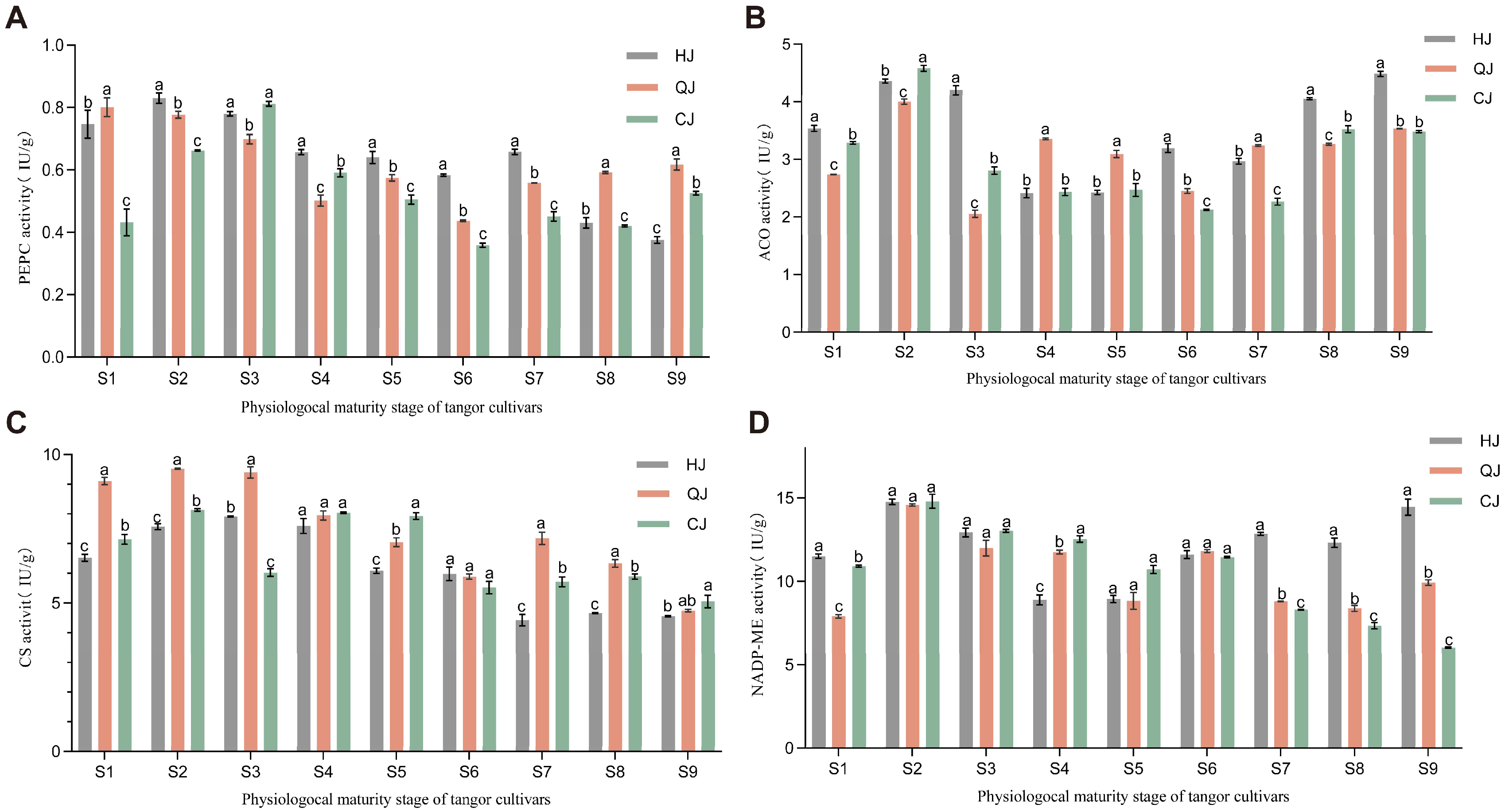
2.2. Determination and Analysis of Enzyme Activities Related to Organic Acid Metabolism in Fruit of Citrus Varieties
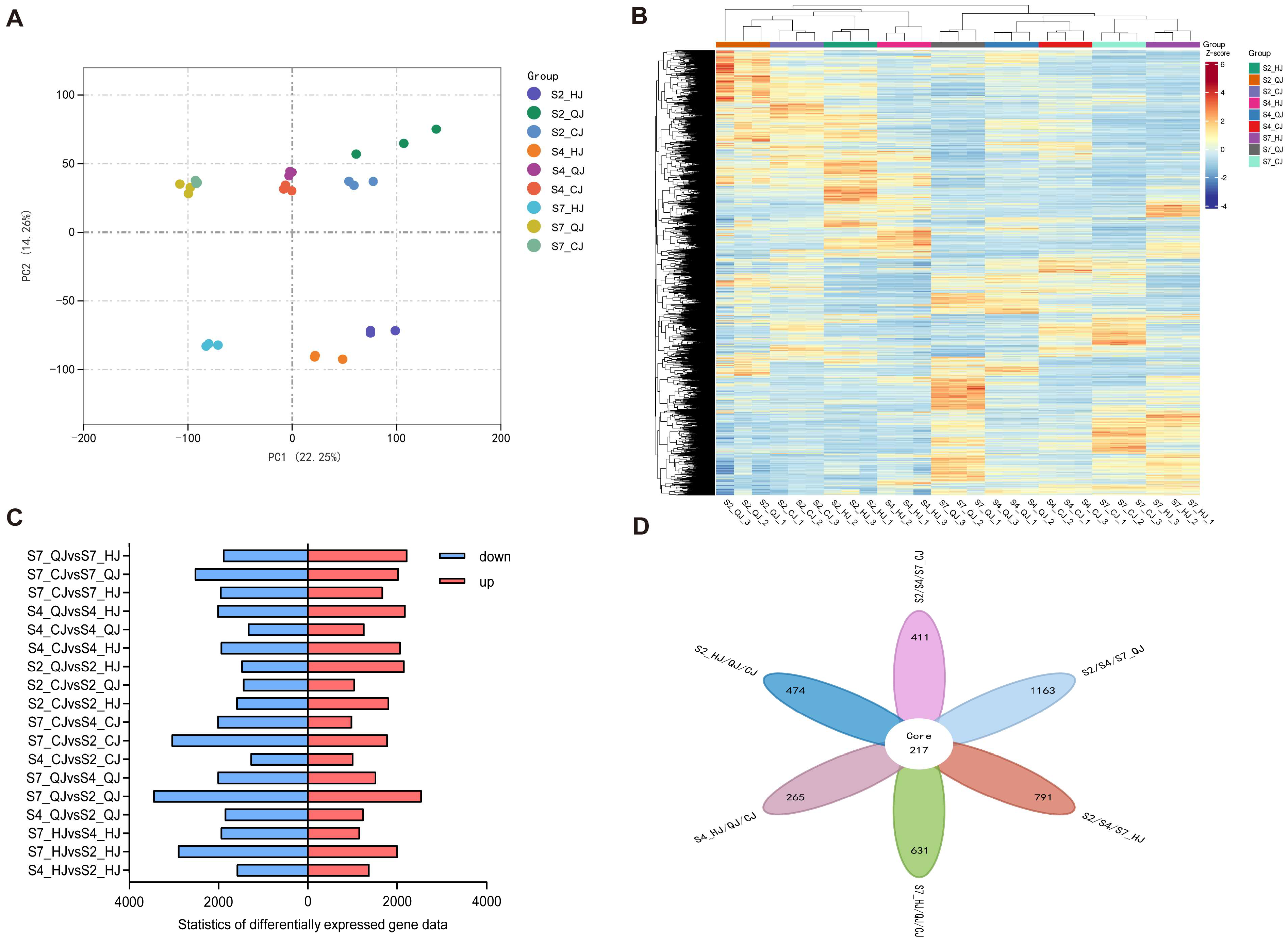
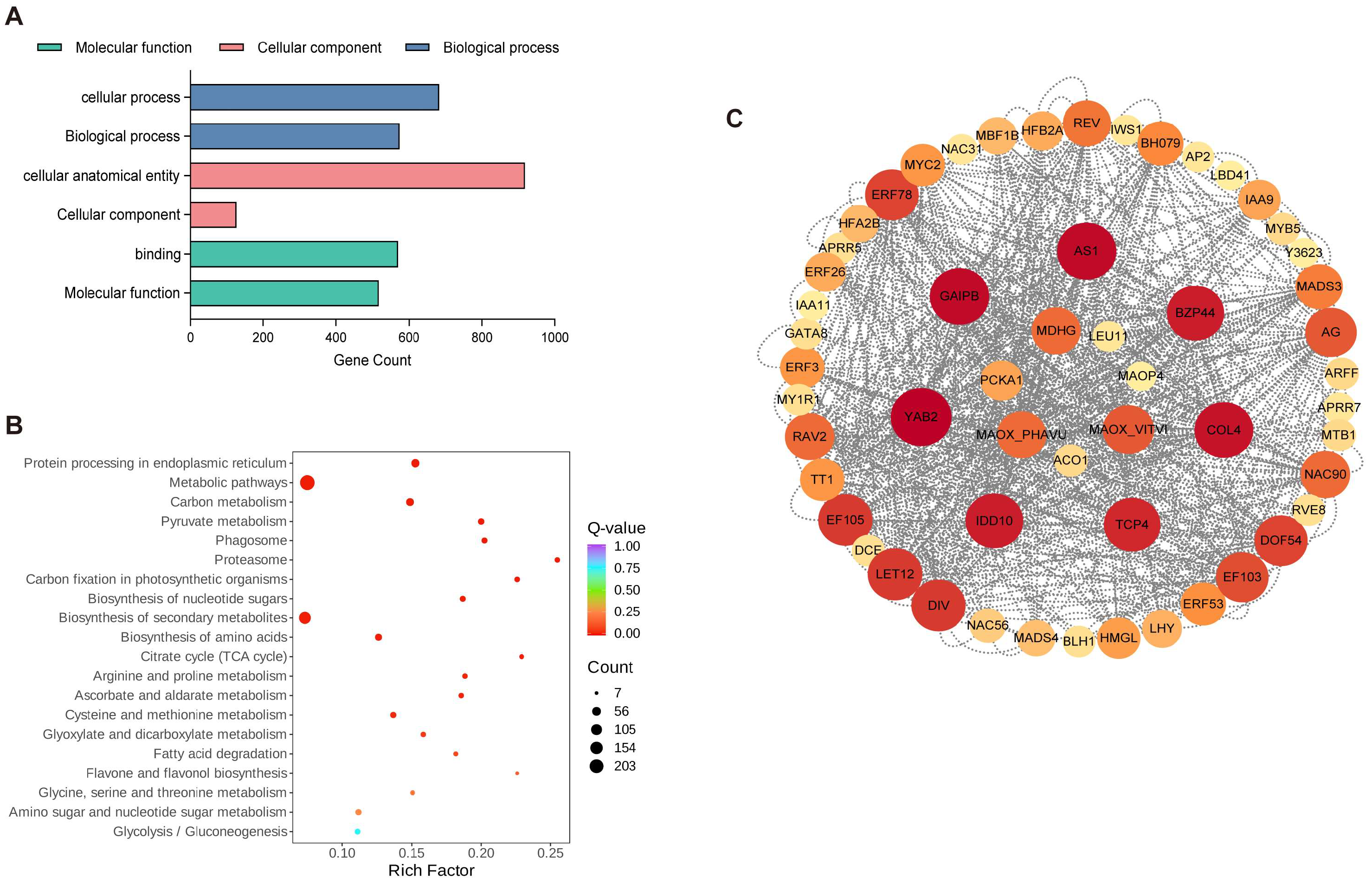
2.3. Identification of DEGs Associated with Organic Acid Degradation Using Transcriptome Analysis
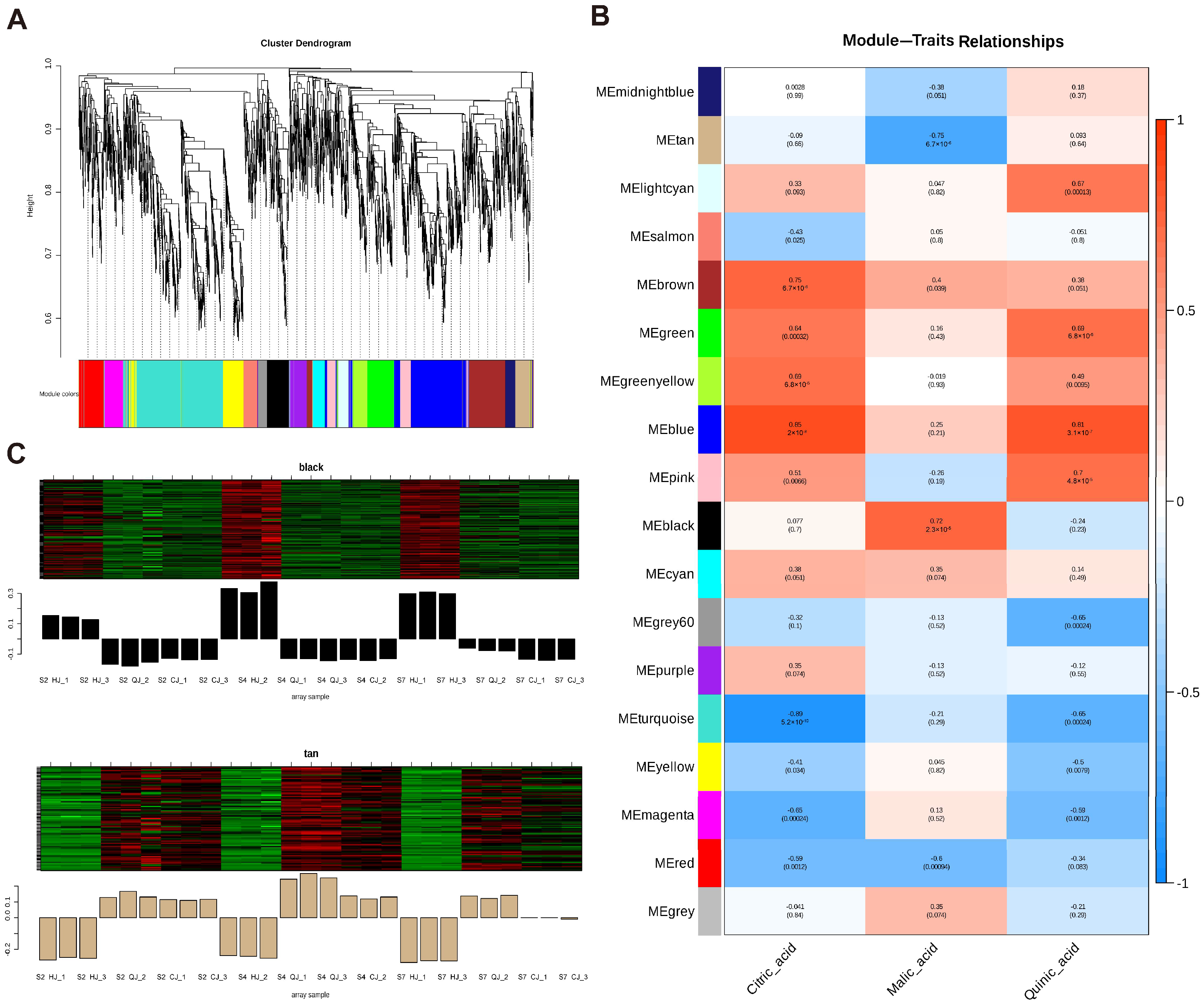
2.4. Weighted Gene Co-Expression Network Analysis of RNA-Seq Data
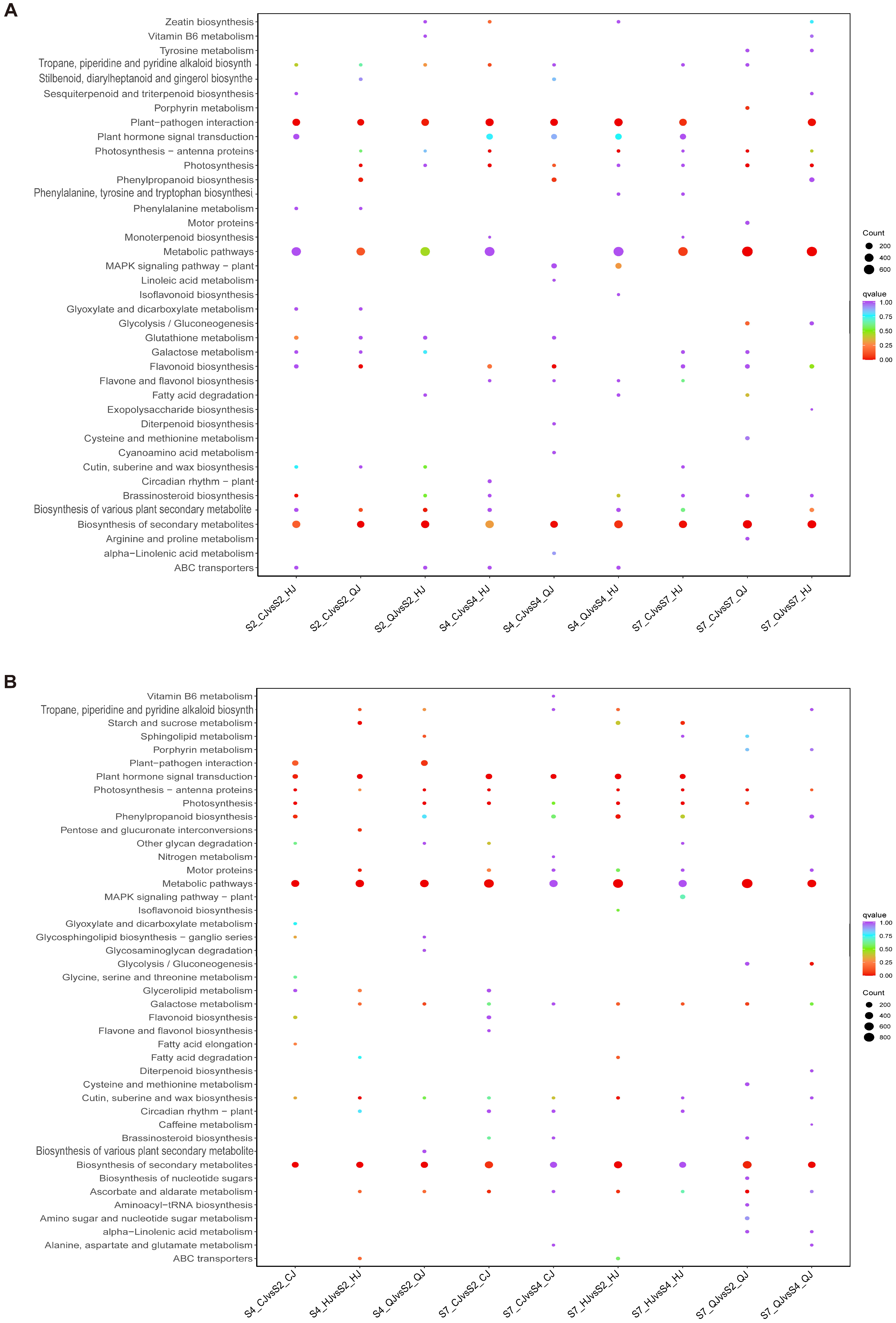
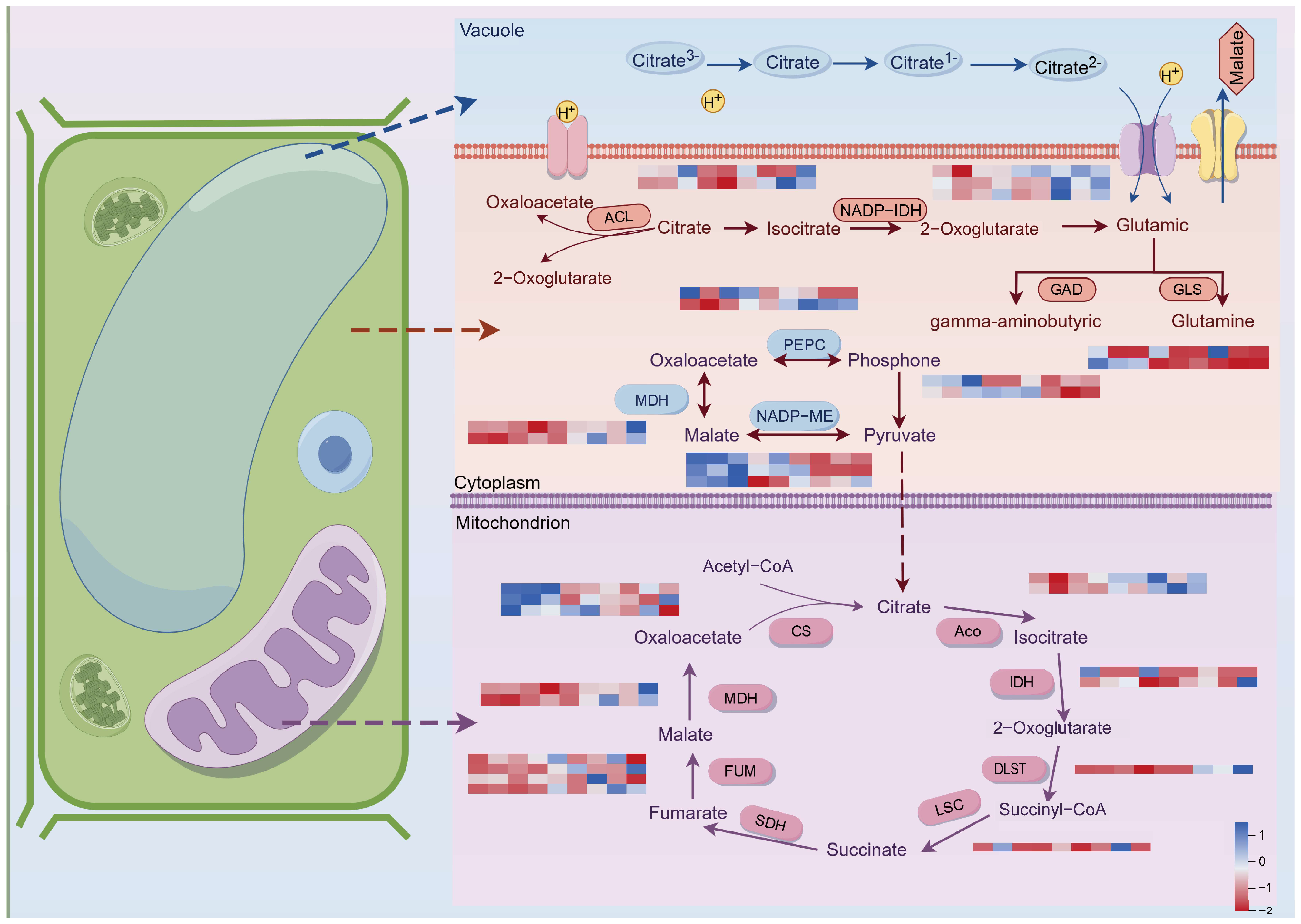
2.5. Organic Acid Metabolic Pathways Involved in the Development of Late-Maturing Hybrid Citrus
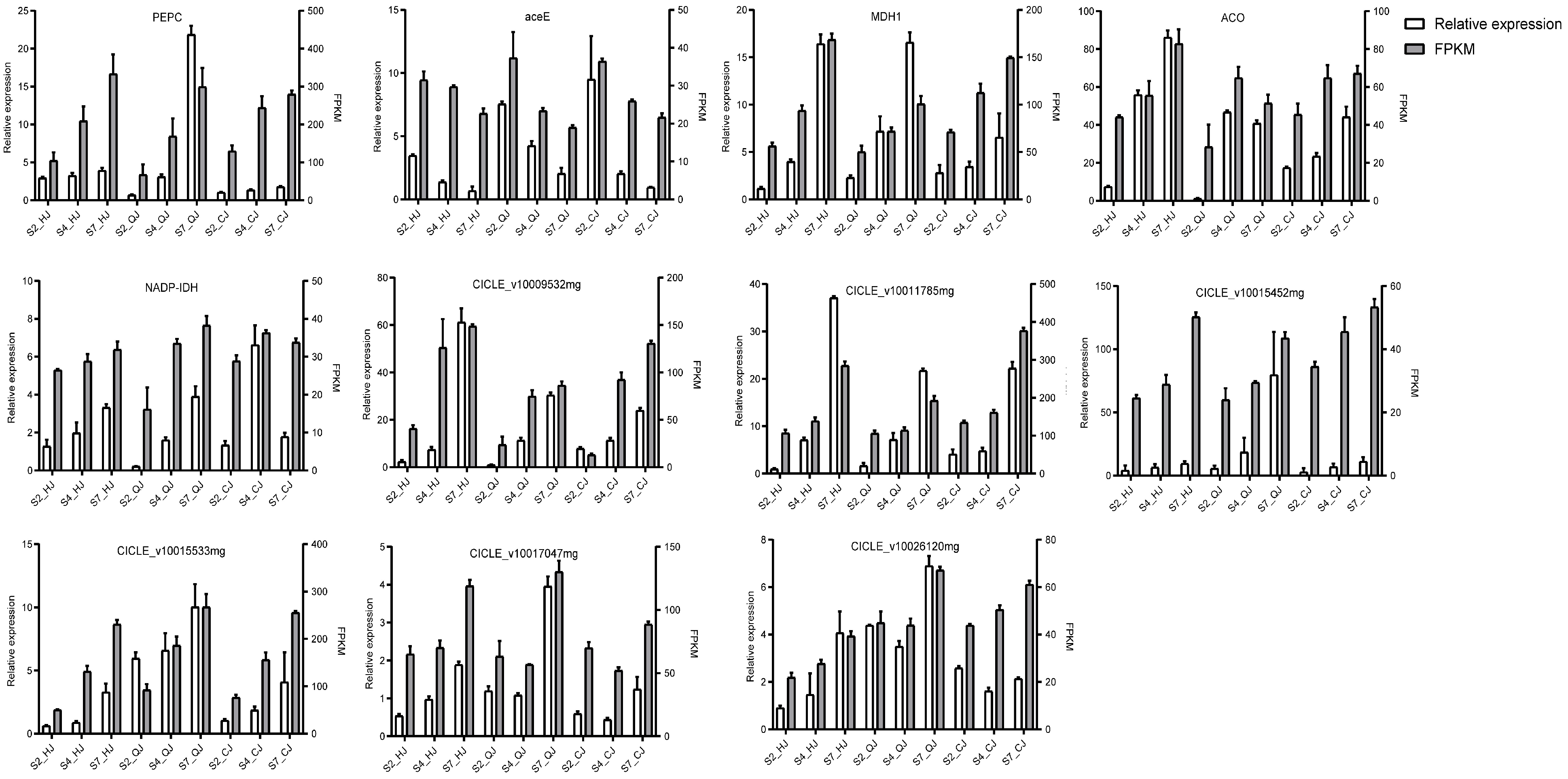
2.6. Validation of RNA-Seq Data Using qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Sampling
4.2. Assessment of Titratable Acidity, Organic Acid Content, and Enzyme Activity
4.3. RNA Library Preparation and Analysis of the Differentially Expressed Genes (DEGs)
4.4. Co-Expression Network Analysis
4.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.6. Statistical Analysis
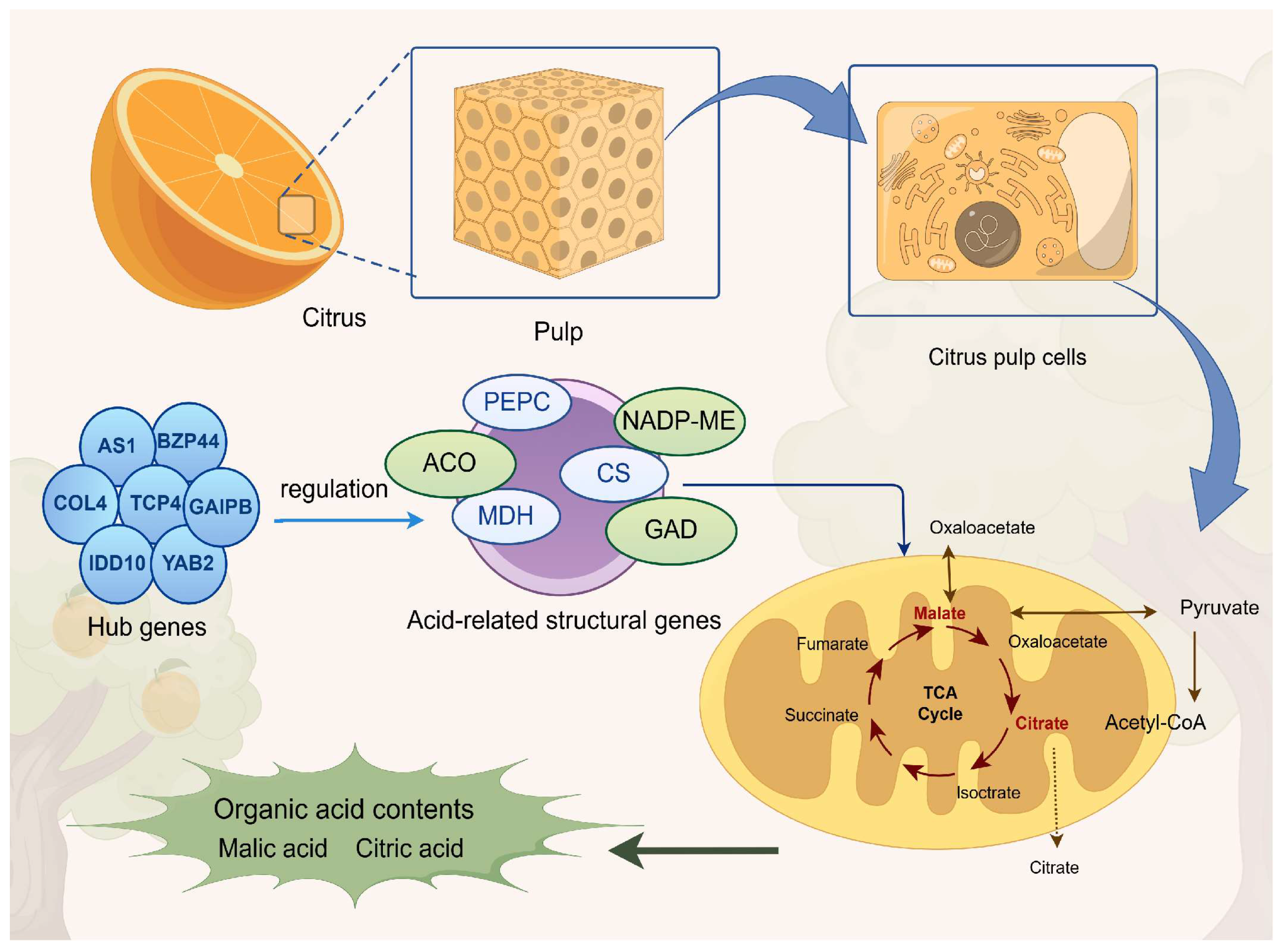
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bi, X.; Liao, L.; Deng, L.; Jin, Z.; Huang, Z.; Sun, G.; Xiong, B.; Wang, Z. Combined transcriptome and metabolome analyses reveal candidate genes involved in tangor (Citrus reticulata × Citrus sinensis) fruit development and quality formation. Int. J. Mol. Sci. 2022, 23, 5457. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Li, J.; Lan, X.; Li, Y.; Li, Y.; Huang, Z.; Jin, Z.; Yang, Y.; Wang, X.; Zhang, M.; et al. Exogenous melatonin and interstock treatments confer chilling tolerance in citrus fruit during cold storage. Sci. Hortic. 2024, 327, 112802. [Google Scholar] [CrossRef]
- Alfaro, J.M.; Bermejo, A.; Navarro, P.; Ones, A.Q.; Salvador, A. Effect of rootstock on citrus fruit quality: A review. Food Rev. Int. 2023, 39, 2835–2853. [Google Scholar] [CrossRef]
- Finkemeier, I.; Konig, A.C.; Heard, W.; Nunes-Nesi, A.; Pham, P.A.; Leister, D.; Fernie, A.R.; Sweetlove, L.J. Transcriptomic analysis of the role of carboxylic acids in metabolite signaling in arabidopsis leaves. Plant Physiol. 2013, 162, 239–253. [Google Scholar] [CrossRef]
- Jiang, Y.; Qi, Y.; Chen, X.; Yan, Q.; Chen, J.; Liu, H.; Shi, F.; Wen, Y.; Cai, C.; Ou, L. Combined metabolome and transcriptome analyses unveil the molecular mechanisms of fruit acidity variation in litchi (Litchi chinensis Sonn.). Int. J. Mol. Sci. 2023, 24, 1871. [Google Scholar] [CrossRef]
- Sheng, L.; Shen, D.; Luo, Y.; Sun, X.; Wang, J.; Luo, T.; Zeng, Y.; Xu, J.; Deng, X.; Cheng, Y. Exogenous gamma-aminobutyric acid treatment affects citrate and amino acid accumulation to improve fruit quality and storage performance of postharvest citrus fruit. Food Chem. 2017, 216, 138–145. [Google Scholar] [CrossRef]
- Yao, S.; Cao, Q.; Xie, J.; Deng, L.; Zeng, K. Alteration of sugar and organic acid metabolism in postharvest granulation of ponkan fruit revealed by transcriptome profiling. Postharvest Biol. Technol. 2018, 139, 2–11. [Google Scholar] [CrossRef]
- Liao, H.-Z.; Lin, X.-K.; Du, J.-J.; Peng, J.-J.; Zhou, K.-B. Transcriptomic analysis reveals key genes regulating organic acid synthesis and accumulation in the pulp of Litchi chinensis Sonn. cv. Feizixiao. Sci. Hortic. 2022, 303, 111220. [Google Scholar]
- Muhammad, A. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem. Biophys. 2013, 68, 475–478. [Google Scholar]
- Li, X.; Li, C.; Sun, J.; Jackson, A. Dynamic changes of enzymes involved in sugar and organic acid level modification during blueberry fruit maturation. Food Chem. 2020, 309, 125617. [Google Scholar] [CrossRef]
- Gao, M.; Yang, N.; Shao, Y.; Shen, T.; Li, W.; Ma, B.; Wei, X.; Ruan, Y.L.; Ma, F.; Li, M. An insertion in the promoter of a malate dehydrogenase gene regulates malic acid content in apple fruit. Plant Physiol. 2024, 196, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.M.; Shi, C.Y.; Liu, X.; Jin, L.F.; Liu, Y.Z.; Peng, S.A. Genome-wide identification of citrus ATP-citrate lyase genes and their transcript analysis in fruits reveals their possible role in citrate utilization. Mol. Genet. Genom. 2015, 290, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, H.; Zeng, K.; Yao, S. New insights into vesicle granulation in citrus grandis revealed by systematic analysis of sugar- and acid-related genes and metabolites. Postharvest Biol. Technol. 2022, 194, 112063. [Google Scholar] [CrossRef]
- Gao, M.; Zhao, H.; Zheng, L.; Zhang, L.; Peng, Y.; Ma, W.; Tian, R.; Yuan, Y.; Ma, F.; Li, M.; et al. Overexpression of apple Ma12, a mitochondrial pyrophosphatase pump gene, leads to malic acid accumulation and the upregulation of malate dehydrogenase in tomato and apple calli. Hortic. Res. 2022, 9, uhab053. [Google Scholar] [CrossRef]
- Sweetman, C.; Deluc, L.G.; Cramer, G.R.; Ford, C.M.; Soole, K.L. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 2009, 70, 1329–1344. [Google Scholar] [CrossRef]
- Lin, Q.; Li, S.; Dong, W.; Feng, C.; Yin, X.; Xu, C.; Sun, C.; Chen, K. Involvement of citchx and citdic in developmental-related and postharvest-hot-air driven citrate degradation in citrus fruits. PLoS ONE 2015, 10, e119410. [Google Scholar] [CrossRef]
- Fu, B.L.; Wang, W.Q.; Li, X.; Qi, T.H.; Shen, Q.F.; Li, K.F.; Liu, X.F.; Li, S.J.; Allan, A.C.; Yin, X.R. A dramatic decline in fruit citrate induced by mutagenesis of a NAC transcription factor, AcNAC1. Plant Biotechnol. J. 2023, 21, 1695–1706. [Google Scholar] [CrossRef]
- Zheng, L.; Ma, W.; Liu, P.; Song, S.; Wang, L.; Yang, W.; Ren, H.; Wei, X.; Zhu, L.; Peng, J.; et al. Transcriptional factor MdESE3 controls fruit acidity by activating genes regulating malic acid content in apple. Plant Physiol. 2024, 196, 261–272. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, L.; Yang, M.; Xie, X.; Sun, P.; Fang, C.; Zhao, J. R2R3-MYB transcription factor FaMYB5 is involved in citric acid metabolism in strawberry fruits. J. Plant Physiol. 2022, 277, 153789. [Google Scholar] [CrossRef]
- Yu, J.Q.; Gu, K.D.; Sun, C.H.; Zhang, Q.Y.; Wang, J.H.; Ma, F.F.; You, C.X.; Hu, D.G.; Hao, Y.J. The apple bHLH transcription factor MdbHLH3 functions in determining the fruit carbohydrates and malate. Plant Biotechnol. J. 2021, 19, 285–299. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, H.; Ma, Y.; Jiang, G.; Mei, X.; Li, X.; Yang, Q.; Kan, J.; Xu, Y.; Yang, T.; et al. WGCNA analysis revealing molecular mechanism that bio-organic fertilizer improves pear fruit quality by increasing sucrose accumulation and reducing citric acid metabolism. Front. Plant Sci. 2022, 13, 1039671. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.P.; Paoletti, A.; Leegood, R.C.; Famiani, F. Phosphorylation of phosphoenolpyruvate carboxykinase (PEPCK) and phosphoenolpyruvate carboxylase (PEPC) in the flesh of fruits. Plant Physiol. Biochem. 2016, 108, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Scheibe, R. Malate valves to balance cellular energy supply. Physiol. Plant. 2004, 120, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Beruter, J. Carbohydrate metabolism in two apple genotypes that differ in malate accumulation. J. Plant Physiol. 2004, 161, 1011–1029. [Google Scholar] [CrossRef]
- Zhou, Y.; He, W.; Zheng, W.; Tan, Q.; Xie, Z.; Zheng, C.; Hu, C. Fruit sugar and organic acid were significantly related to fruit mg of six citrus cultivars. Food Chem. 2018, 259, 278–285. [Google Scholar] [CrossRef]
- Yang, L.; Chen, Y.; Wang, M.; Hou, H.; Li, S.; Guan, L.; Yang, H.; Wang, W.; Hong, L. Metabolomic and transcriptomic analyses reveal the effects of grafting on blood orange quality. Front. Plant Sci. 2023, 14, 1169220. [Google Scholar] [CrossRef]
- Lu, X.; Cao, X.; Li, F.; Li, J.; Xiong, J.; Long, G.; Cao, S.; Xie, S. Comparative transcriptome analysis reveals a global insight into molecular processes regulating citrate accumulation in sweet orange (Citrus sinensis). Physiol. Plant. 2016, 158, 463–482. [Google Scholar] [CrossRef]
- Sun, X.; Zhu, A.; Liu, S.; Sheng, L.; Ma, Q.; Zhang, L.; Nishawy, E.M.; Zeng, Y.; Xu, J.; Ma, Z.; et al. Integration of metabolomics and subcellular organelle expression microarray to increase understanding the organic acid changes in post-harvest citrus fruit. J. Integr. Plant Biol. 2013, 55, 1038–1053. [Google Scholar] [CrossRef]
- Gou, N.; Chen, C.; Huang, M.; Zhang, Y.; Bai, H.; Li, H.; Wang, L.; Wuyun, T. Transcriptome and metabolome analyses reveal sugar and acid accumulation during apricot fruit development. Int. J. Mol. Sci. 2023, 24, 16992. [Google Scholar] [CrossRef]
- Sadka, A.; Dahan, E.; Cohen, L.; Marsh, K.B. Aconitase activity and expression during the development of lemon fruit. Physiol. Plant. 2000, 108, 255–262. [Google Scholar] [CrossRef]
- Gao, Y.; Yao, Y.; Chen, X.; Wu, J.; Wu, Q.; Liu, S.; Guo, A.; Zhang, X. Metabolomic and transcriptomic analyses reveal the mechanism of sweet-acidic taste formation during pineapple fruit development. Front. Plant Sci. 2022, 13, 971506. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, R.; Li, X.; Yue, Y.; Geng, Z. Effect of bamboo (Phyllostachys pubescens) extract on broiler chickens under cold stress. S. Afr. J. Anim. Sci. 2011, 41, 57–62. [Google Scholar] [CrossRef]
- Shi, C.Y.; Song, R.Q.; Hu, X.M.; Liu, X.; Jin, L.F.; Liu, Y.Z. Citrus PH5-like H+-ATPase genes: Identification and transcript analysis to investigate their possible relationship with citrate accumulation in fruits. Front. Plant Sci. 2015, 6, 135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lu, W.; Yang, X.; Li, Q.; Lin, X.; Liu, K.; Yin, C.; Xiong, B.; Liao, L.; Sun, G.; et al. Comprehensive analyses of the citrus wrky gene family involved in the metabolism of fruit sugars and organic acids. Front. Plant Sci. 2023, 14, 1264283. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Liu, S.C.; Lin, X.H.; Grierson, D.; Yin, X.R.; Chen, K.S. Citrus heat shock transcription factor CitHsfA7-mediated citric acid degradation in response to heat stress. Plant Cell Environ. 2022, 45, 95–104. [Google Scholar] [CrossRef]
- Alabd, A.; Cheng, H.; Ahmad, M.; Wu, X.; Peng, L.; Wang, L.; Yang, S.; Bai, S.; Ni, J.; Teng, Y. Abre-binding factor3-wrky dna-binding protein44 module promotes salinity-induced malate accumulation in pear. Plant Physiol. 2023, 192, 1982–1996. [Google Scholar] [CrossRef]
- Muller, M.; Irkens-Kiesecker, U.; Rubinstein, B.; Taiz, L. On the mechanism of hyperacidification in lemon. Comparison of the vacuolar H+-ATPase activities of fruits and epicotyls. J. Biol. Chem. 1996, 271, 1916–1924. [Google Scholar]
- Wu, Z.; Tu, M.; Yang, X.; Xu, J.; Yu, Z. Effect of cutting and storage temperature on sucrose and organic acids metabolism in postharvest melon fruit. Postharvest Biol. Technol. 2020, 161, 111081. [Google Scholar] [CrossRef]
- Terol, J.; Soler, G.; Talon, M.; Cercos, M. The aconitate hydratase family from citrus. BMC Plant Biol. 2010, 10, 222. [Google Scholar] [CrossRef]
- Degu, A.; Hatew, B.; Nunes-Nesi, A.; Shlizerman, L.; Zur, N.; Katz, E.; Fernie, A.R.; Blumwald, E.; Sadka, A. Inhibition of aconitase in citrus fruit callus results in a metabolic shift towards amino acid biosynthesis. Planta 2011, 234, 501–513. [Google Scholar] [CrossRef]
- Xu, J.; Yan, J.; Li, W.; Wang, Q.; Wang, C.; Guo, J.; Geng, D.; Guan, Q.; Ma, F. Integrative analyses of widely targeted metabolic profiling and transcriptome data reveals molecular insight into metabolomic variations during apple (Malus domestica) fruit development and ripening. Int. J. Mol. Sci. 2020, 21, 4797. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yin, X.; Chen, K. The citrus transcription factor, CitERF13, regulates citric acid accumulation via a protein-protein interaction with the vacuolar proton pump, CitVHA-c4. Sci. Rep. 2016, 6, 20151. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, Y.; Zhang, X.; Hu, W.; Lin, L.; Deng, Q.; Xia, H.; Liang, D.; Lv, X. Effects of potassium-containing fertilizers on sugar and organic acid metabolism in grape fruits. Int. J. Mol. Sci. 2024, 25, 2828. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Gueguen, L.; Coppee, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e157022. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. Kegg for linking genomes to life and the environment. Nucleic. Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, X.; Huang, M.; Deng, L.; Li, Y.; Jin, X.; Xu, J.; Xiong, B.; Liao, L.; Zhang, M.; He, J.; et al. Comparative Transcriptome Analysis Reveals Potential Molecular Regulation of Organic Acid Metabolism During Fruit Development in Late-Maturing Hybrid Citrus Varieties. Int. J. Mol. Sci. 2025, 26, 803. https://doi.org/10.3390/ijms26020803
Tang X, Huang M, Deng L, Li Y, Jin X, Xu J, Xiong B, Liao L, Zhang M, He J, et al. Comparative Transcriptome Analysis Reveals Potential Molecular Regulation of Organic Acid Metabolism During Fruit Development in Late-Maturing Hybrid Citrus Varieties. International Journal of Molecular Sciences. 2025; 26(2):803. https://doi.org/10.3390/ijms26020803
Chicago/Turabian StyleTang, Xiaoyu, Mengqi Huang, Lijun Deng, Yixuan Li, Xiaojun Jin, Jiaqi Xu, Bo Xiong, Ling Liao, Mingfei Zhang, Jiaxian He, and et al. 2025. "Comparative Transcriptome Analysis Reveals Potential Molecular Regulation of Organic Acid Metabolism During Fruit Development in Late-Maturing Hybrid Citrus Varieties" International Journal of Molecular Sciences 26, no. 2: 803. https://doi.org/10.3390/ijms26020803
APA StyleTang, X., Huang, M., Deng, L., Li, Y., Jin, X., Xu, J., Xiong, B., Liao, L., Zhang, M., He, J., Sun, G., He, S., & Wang, Z. (2025). Comparative Transcriptome Analysis Reveals Potential Molecular Regulation of Organic Acid Metabolism During Fruit Development in Late-Maturing Hybrid Citrus Varieties. International Journal of Molecular Sciences, 26(2), 803. https://doi.org/10.3390/ijms26020803





