Glycosylation Pathways Targeted by Deregulated miRNAs in Autism Spectrum Disorder
Abstract
:1. Introduction
2. Results
2.1. Recognition of Differentially Expressed ASD-miRNAs
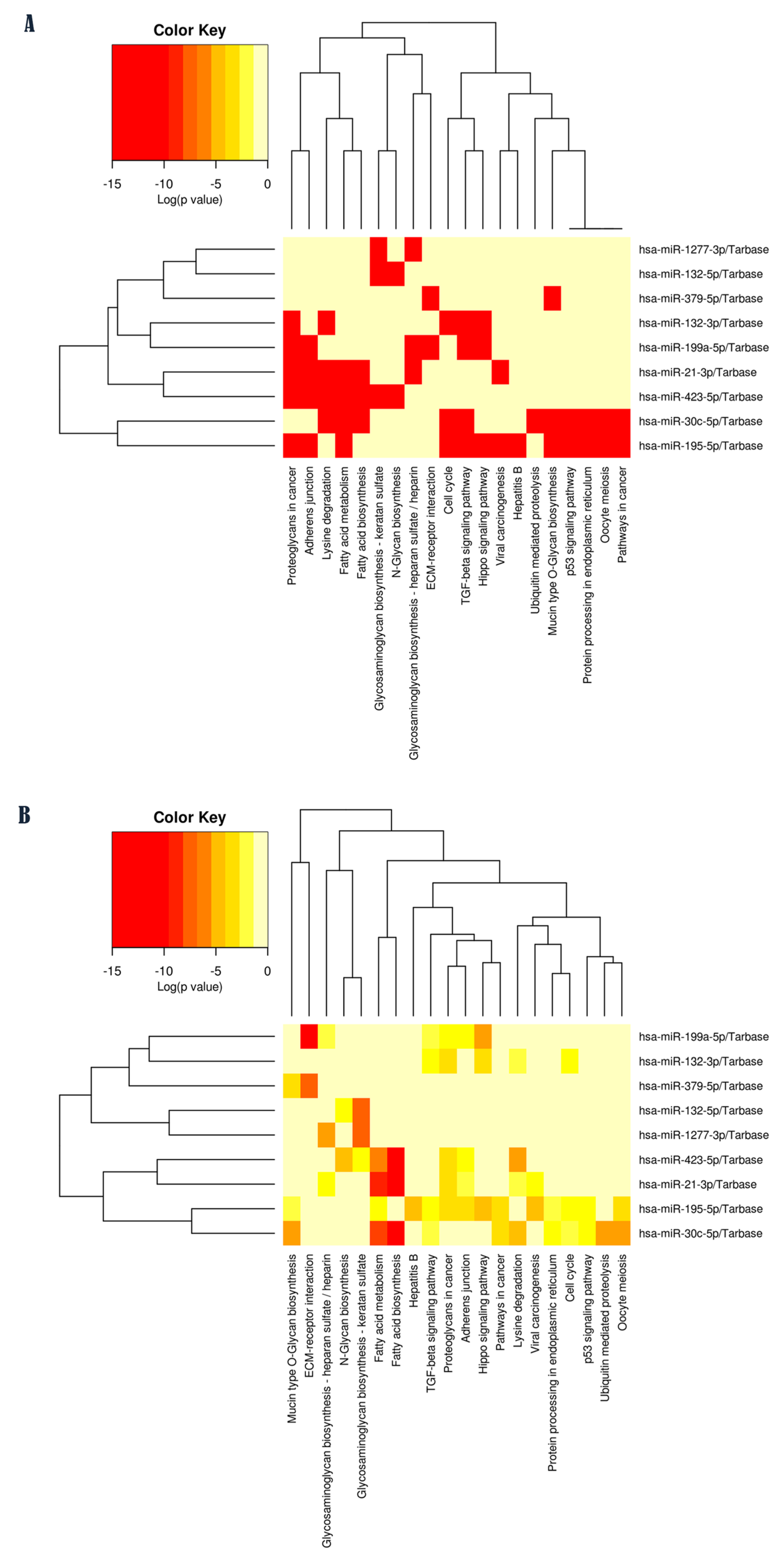
2.2. Pathway Enrichment Analysis of Validated Targets for Each ASD-miRNA
2.3. Identification of Experimentally Validated Target Genes of Each miRNA
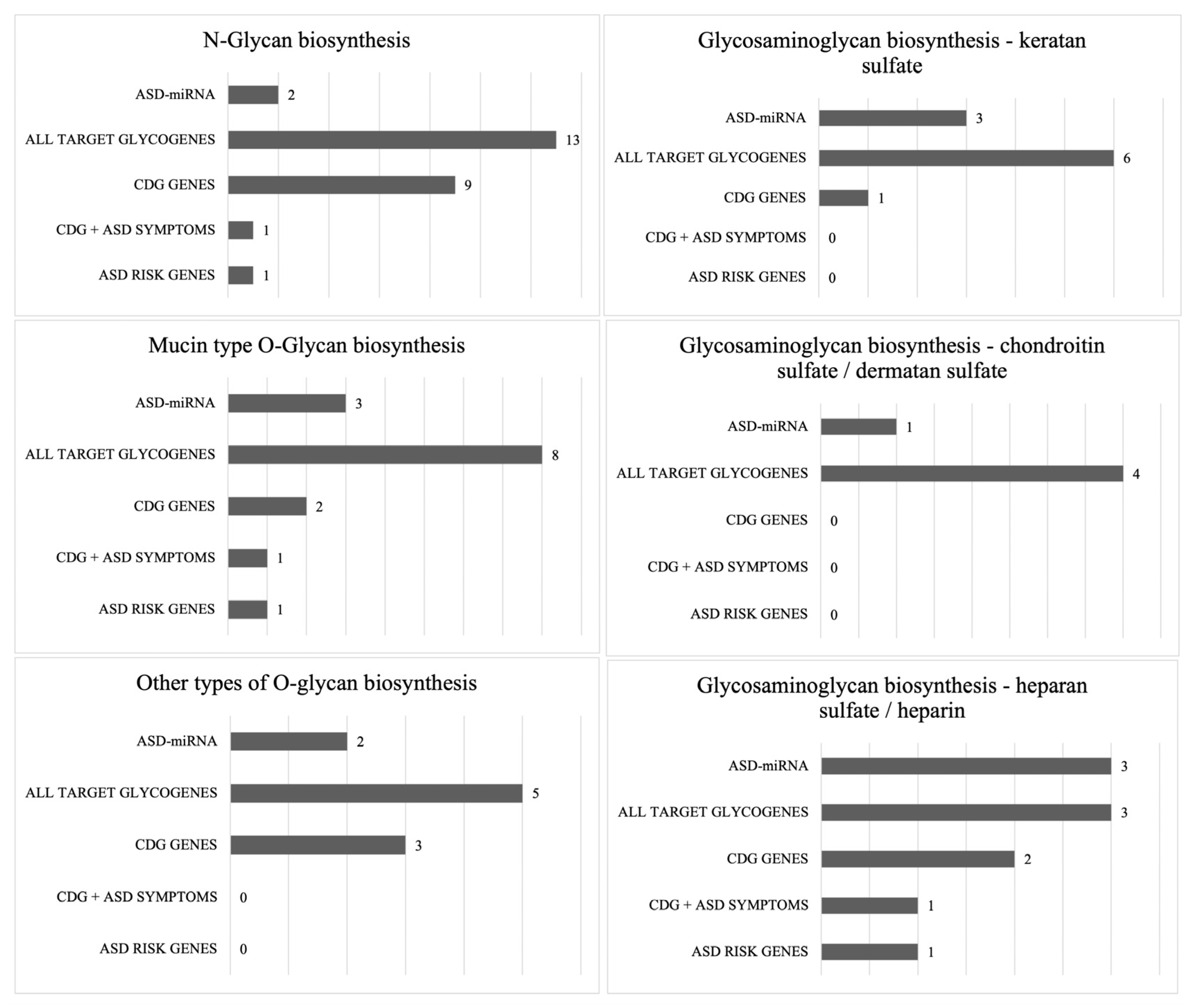
2.4. Enrichment for Glycogenes and ASD Risk Genes
2.5. Reconstruction of the Whole miRNA-Mediated Regulatory Network
2.6. Protein Function Analysis for Each Target Gene
3. Discussion
4. Materials and Methods
4.1. Criteria for Considering Deregulated miRNAs in ASD for This Analysis
4.2. Selection of Studies
4.3. Computational Analysis
4.3.1. Computational Pathway Enrichment Analysis of Validated Targets for Each ASD-miRNAs
4.3.2. Exploring the Experimentally Validated Target Genes of Each miRNA
4.3.3. Enrichment for Glycogenes and ASD Risk Genes Among Validated Targets
4.3.4. Reconstruction of miRNA-Mediated Regulatory Networks
4.3.5. Exploring KEGG Pathways That Are Commonly Targeted by Multiple miRNAs and Their Regulatory Networks
4.3.6. Exploring the Protein Function for Each Target Gene
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2022. [Google Scholar]
- Hodges, H.; Fealko, C.; Soares, N. Autism spectrum disorder: Definition, epidemiology, causes, and clinical evaluation. Transl. Pediatr. 2020, 9 (Suppl. S1), S55–S65. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.T.; Chen, L.; Du, Y.; Jiang, Z.Y.; Cheng, Y. MicroRNAs as Regulators, Biomarkers, and Therapeutic Targets in Autism Spectrum Disorder. Mol. Neurobiol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.X.; Chen, Y.; Guo, H.R.; Chen, G.F. Systematic review and bioinformatic analysis of microRNA expression in autism spectrum disorder identifies pathways associated with cancer, metabolism, cell signaling, and cell adhesion. Front. Psychiatry 2021, 12, 630876. [Google Scholar] [CrossRef] [PubMed]
- Senarathne, U.D.; Indika, N.R.; Jezela-Stanek, A.; Ciara, E.; Frye, R.E.; Chen, C.; Stepien, K.M. Biochemical, genetic and clinical diagnostic approaches to autism-associated inherited metabolic disorders. Genes 2023, 14, 803. [Google Scholar] [CrossRef] [PubMed]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Polenghi, M.; Taverna, E. Intracellular traffic and polarity in brain development. Front. Neurosci. 2023, 17, 1172016. [Google Scholar] [CrossRef]
- Pradeep, P.; Kang, H.; Lee, B. Glycosylation and behavioral symptoms in neurological disorders. Transl. Psychiatry 2023, 13, 154. [Google Scholar] [CrossRef]
- Dwyer, C.A.; Esko, J.D. Glycan susceptibility factors in autism spectrum disorders. Mol. Asp. Med. 2016, 51, 104–114. [Google Scholar] [CrossRef]
- Liu, Y.; Di, Y.; Zheng, Q.; Qian, Z.; Fan, J.; Ren, W.; Wei, Z.; Tian, Y. Altered expression of glycan patterns and glycan-related genes in the medial prefrontal cortex of the valproic acid rat model of autism. Front. Cell. Neurosci. 2022, 16, 1057857. [Google Scholar] [CrossRef]
- Pivac, N.; Knezević, A.; Gornik, O.; Pucić, M.; Igl, W.; Peeters, H.; Crepel, A.; Steyaert, J.; Novokmet, M.; Redzić, I.; et al. Human plasma glycome in attention-deficit hyperactivity disorder and autism spectrum disorders. Mol. Cell. Proteom. 2011, 10, M110.004200. [Google Scholar] [CrossRef]
- Kianičková, K.; Pažitná, L.; Kundalia, P.H.; Pakanová, Z.; Nemčovič, M.; Baráth, P.; Katrlíková, E.; Šuba, J.; Trebatická, J.; Katrlík, J. Alterations in the Glycan Composition of Serum Glycoproteins in Attention-Deficit Hyperactivity Disorder. Int. J. Mol. Sci. 2023, 24, 8745. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Chen, Y.; Li, X.; Huang, Q.; Lu, R.; Ye, J.; Meng, W.; Fan, C.; Mo, X. Predicting psychiatric risk: IgG N-glycosylation traits as biomarkers for mental health. Front. Psychiatry 2024, 15, 1431942. [Google Scholar] [CrossRef]
- Cast, T.P.; Boesch, D.J.; Smyth, K.; Shaw, A.E.; Ghebrial, M.; Chanda, S. An autism-associated mutation impairs neuroligin-4 glycosylation and enhances excitatory synaptic transmission in human neurons. J. Neurosci. 2021, 41, 392–407. [Google Scholar] [CrossRef] [PubMed]
- Barone, R.; Sturiale, L.; Fiumara, A.; Palmigiano, A.; Bua, R.O.; Rizzo, R.; Zappia, M.; Garozzo, D. CSF N-glycan profile reveals sialylation deficiency in a patient with GM2 gangliosidosis presenting as childhood disintegrative disorder. Autism Res. 2016, 9, 423–428. [Google Scholar] [CrossRef]
- Qin, Y.; Cao, L.; Zhang, J.; Zhang, H.; Cai, S.; Guo, B.; Wu, F.; Zhao, L.; Li, W.; Ni, L.; et al. Whole-transcriptome analysis of serum L1CAM-captured extracellular vesicles reveals neural and glycosylation changes in autism spectrum disorder. J. Mol. Neurosci. 2022, 72, 1274–1292. [Google Scholar] [CrossRef] [PubMed]
- Quelhas, D.; Jaeken, J. Treatment of congenital disorders of glycosylation: An overview. Mol. Genet. Metab. 2024, 143, 108567. [Google Scholar] [CrossRef] [PubMed]
- Freeze, H.H.; Eklund, E.A.; Ng, B.G.; Patterson, M.C. Neurology of inherited glycosylation disorders. Lancet Neurol. 2012, 11, 453–466. [Google Scholar] [CrossRef]
- Uzunyayla-Inci, G.; Kiykim, E.; Zubarioglu, T.; Yesil, G.; Aktuglu Zeybek, C. Autism spectrum disorder in two unrelated patients with homozygous variants in either ALG8 or ALG11. Mol. Syndromol. 2023, 14, 428–432. [Google Scholar] [CrossRef]
- Ng, B.G.; Freeze, H.H.; Himmelreich, N.; Blau, N.; Ferreira, C.R. Clinical and biochemical footprints of congenital disorders of glycosylation: Proposed nosology. Mol. Genet. Metab. 2024, 142, 108476. [Google Scholar] [CrossRef]
- Godfrey, C.; Foley, A.R.; Clement, E.; Muntoni, F. Dystroglycanopathies: Coming into focus. Curr. Opin. Genet. Dev. 2011, 21, 278–285. [Google Scholar] [CrossRef]
- Indellicato, R.; Trinchera, M. Epigenetic regulation of glycosylation. Adv. Exp. Med. Biol. 2021, 1325, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Cirnigliaro, M.; Barbagallo, C.; Gulisano, M.; Domini, C.N.; Barone, R.; Barbagallo, D.; Ragusa, M.; di Pietro, C.; Rizzo, R.; Purrello, M. Expression and Regulatory Network Analysis of miR-140-3p, a New Potential Serum Biomarker for Autism Spectrum Disorder. Front. Mol. Neurosci. 2017, 10, 250. [Google Scholar] [CrossRef] [PubMed]
- Bleazard, T. Investigating the Role of microRNAs in Autism. Ph.D. Thesis, University of Manchester, Manchester, UK, 2018. Corpus ID: 149752101. [Google Scholar]
- Kim, J.Y.; Kim, W.; Lee, K.H. The role of microRNAs in the molecular link between circadian rhythm and autism spectrum disorder. Anim. Cells Syst. 2023, 27, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Sarachana, T.; Zhou, R.; Chen, G.; Manji, H.K.; Hu, V.W. Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med. 2010, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Hicks, S.D.; Middleton, F.A. A comparative review of microRNA expression patterns in autism spectrum disorder. Front. Psychiatry 2016, 7, 176. [Google Scholar] [CrossRef]
- Huang, F.; Long, Z.; Chen, Z.; Li, J.; Hu, Z.; Qiu, R.; Huang, W.; Tang, B.; Xia, K.; Jiang, H. Investigation of gene regulatory networks associated with autism spectrum disorder based on miRNA expression in China. PLoS ONE 2015, 10, e0129052. [Google Scholar] [CrossRef]
- Vasu, M.M.; Anitha, A.; Thanseem, I.; Suzuki, K.; Yamada, K.; Takahashi, T.; Wakuda, T.; Iwata, K.; Tsujii, M.; Sugiyama, T.; et al. Serum microRNA profiles in children with autism. Mol. Autism 2014, 5, 40. [Google Scholar] [CrossRef]
- Seno, M.M.G.; Hu, P.; Gwadry, F.G.; Pinto, D.; Marshall, C.R.; Casallo, G.; Scherer, S.W. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res. 2011, 1380, 85–97. [Google Scholar] [CrossRef]
- Vaccaro, T.D.S.; Sorrentino, J.M.; Salvador, S.; Veit, T.; Souza, D.O.; de Almeida, R.F. Alterations in the MicroRNA of the blood of autism spectrum disorder patients: Effects on epigenetic regulation and potential biomarkers. Behav. Sci. 2018, 8, 75. [Google Scholar] [CrossRef]
- Talebizadeh, Z.; Butler, M.G.; Theodoro, M.F. Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism Res. 2008, 1, 240–250. [Google Scholar] [CrossRef]
- Abu-Elneel, K.; Liu, T.; Gazzaniga, F.S.; Nishimura, Y.; Wall, D.P.; Geschwind, D.H.; Lao, K.; Kosik, K.S. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics 2008, 9, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Mor, M.; Nardone, S.; Sams, D.S.; Elliott, E. Hypomethylation of miR-142 promoter and upregulation of microRNAs that target the oxytocin receptor gene in the autism prefrontal cortex. Mol. Autism 2015, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Hicks, S.D.; Carpenter, R.L.; Wagner, K.E.; Pauley, R.; Barros, M.; Tierney-Aves, C.; Barns, S.; Greene, C.D.; Middleton, F.A. Saliva microRNA differentiates children with autism from peers with typical and atypical development. J. Am. Acad. Child Adolesc. Psychiatry 2020, 59, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Parikshak, N.; Belgard, T.; Geschwind, D.H. Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat. Neurosci. 2016, 19, 1463–1476. [Google Scholar] [CrossRef]
- Wu, X.; Li, W.; Zheng, Y. Recent Progress on Relevant microRNAs in Autism Spectrum Disorders. Int. J. Mol. Sci. 2020, 21, 5904. [Google Scholar] [CrossRef]
- Fregeac, J.; Colleaux, L.; Nguyen, L.S. The emerging roles of MicroRNAs in autism spectrum disorders. Neurosci. Biobehav. Rev. 2016, 71, 729–738. [Google Scholar] [CrossRef]
- Hu, Y.; Ehli, E.A.; Boomsma, D.I. MicroRNAs as biomarkers for psychiatric disorders with a focus on autism spectrum disorder: Current progress in genetic association studies, expression profiling, and translational research. Autism Res. 2017, 10, 1184–1203. [Google Scholar] [CrossRef]
- Thu, C.T.; Mahal, L.K. Sweet control: MicroRNA regulation of the glycome. Biochemistry 2020, 59, 3098–3110. [Google Scholar] [CrossRef]
- Helander, A.; Stödberg, T.; Jaeken, J.; Matthijs, G.; Eriksson, M.; Eggertsen, G. Dolichol kinase deficiency (DOLK-CDG) with a purely neurological presentation caused by a novel mutation. Mol. Genet. Metab. 2013, 110, 342–344. [Google Scholar] [CrossRef]
- Xiong, J.; Chen, S.; Pang, N.; Deng, X.; Yang, L.; He, F. Neurological Diseases With Autism Spectrum Disorder: Role of ASD Risk Genes. Front. Neurosci. 2019, 13, 349. [Google Scholar] [CrossRef]
- Zilmer, M.; Edmondson, A.C.; Khetarpal, S.A.; Alesi, V.; Zaki, M.S.; Rostasy, K.; Madsen, C.G.; Lepri, F.R.; Sinibaldi, L.; Cusmai, R.; et al. Novel congenital disorder of O-linked glycosylation caused by GALNT2 loss of function. Brain 2020, 143, 1114–1126. [Google Scholar] [CrossRef] [PubMed]
- Pearson, B.L.; Corley, M.J.; Vasconcellos, A.; Blanchard, D.C.; Blanchard, R.J. Heparan sulfate deficiency in autistic postmortem brain tissue from the subventricular zone of the lateral ventricles. Behav. Brain Res. 2013, 243, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Mercier, F.; Kwon, Y.C.; Douet, V. Hippocampus/amygdala alterations, loss of heparan sulfates, fractones and ventricle wall reduction in adult BTBR T+ tf/J mice, animal model for autism. Neurosci. Lett. 2012, 506, 208–213. [Google Scholar] [CrossRef]
- Irie, F.; Badie-Mahdavi, H.; Yamaguchi, Y. Autism-like socio-communicative deficits and stereotypies in mice lacking heparan sulfate. Proc. Natl. Acad. Sci. USA 2012, 109, 5052–5056. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yamagata, T.; Mori, M.; Momoi, M.Y. Association of autism in two patients with hereditary multiple exostoses caused by novel deletion mutations of EXT1. J. Hum. Genet. 2002, 47, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 2017, 8, 21. [Google Scholar] [CrossRef]
- Nickolls, A.R.; Bönneman, C.G. The roles of dystroglycan in the nervous system: Insights from animal models of muscular dystrophy. Dis. Models Mech. 2018, 11, dmm035931. [Google Scholar] [CrossRef]
- Egan, S.; Herbrick, J.A.; Tsui, L.C.; Cohen, B.; Flock, G.; Beatty, B.; Scherer, S.W. Mapping of the human Lunatic Fringe (LFNG) gene to 7p22 and Manic Fringe (MFNG) to 22q12. Genomics 1998, 54, 576–577. [Google Scholar] [CrossRef]
- Nikolaou, N.; Watanabe-Asaka, T.; Gerety, S.; Distel, M.; Köster, R.W.; Wilkinson, D.G. Lunatic fringe promotes the lateral inhibition of neurogenesis. Development 2009, 136, 2523–2533. [Google Scholar] [CrossRef]
- Kato, T.M.; Kawaguchi, A.; Kosodo, Y.; Niwa, H.; Matsuzaki, F. Lunatic fringe potentiates Notch signaling in the developing brain. Mol. Cell. Neurosci. 2010, 45, 12–25. [Google Scholar] [CrossRef]
- Cirnigliaro, L.; Clericò, L.; Russo, L.C.; Prato, A.; Caruso, M.; Rizzo, R.; Barone, R. Head circumference growth in children with autism spectrum disorder: Trend and clinical correlates in the first five years of life. Front. Psychiatry 2024, 15, 1431693. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Nayak, S.; Raidas, S.; Guo, L.; Gatta, G.D.; Koppolu, S.; Halasz, G.; Montasser, M.E.; Shuldiner, A.R.; Mao, Y.; et al. In-Depth Mass Spectrometry Analysis Reveals the Plasma Proteomic and N-Glycoproteomic Impact of an Amish-Enriched Cardioprotective Variant in B4GALT1. Mol. Cell. Proteom. 2023, 22, 100595. [Google Scholar] [CrossRef] [PubMed]
- Guerra, D.J. The molecular genetics of autism spectrum disorders: Genomic mechanisms, neuroimmunopathology, and clinical implications. Autism Res. Treat. 2011, 2011, 398636. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef]
- Abrahams, B.S.; Arking, D.E.; Campbell, D.B.; Mefford, H.C.; Morrow, E.M.; Weiss, L.; Menashe, I.; Wadkins, T.; Banerjee-Basu, S.; Packer, A. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 2013, 4, 36. [Google Scholar] [CrossRef]
- Chang, L.; Xia, J. MicroRNA regulatory network analysis using miRNet 2.0. Methods Mol. Biol. 2023, 2594, 185–204. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Chen, C.Y.; Seward, C.H.; Song, Y.; Inamdar, M.; Leddy, A.M.; Zhang, H.; Yoo, J.; Kao, W.-C.; Pawlowski, H.; Stubbs, L.J. Galnt17 loss-of-function leads to developmental delay and abnormal coordination, activity, and social interactions with cerebellar vermis pathology. Dev. Biol. 2022, 490, 155–171. [Google Scholar] [CrossRef]
- Howerton, C.L.; Morgan, C.P.; Fischer, D.B.; Bale, T.L. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc. Natl. Acad. Sci. USA 2013, 110, 5169–5174. [Google Scholar] [CrossRef]
- Van der Zwaag, B.; Franke, L.; Poot, M.; Hochstenbach, R.; Spierenburg, H.A.; Vorstman, J.A.; van Daalen, E.; de Jonge, M.V.; Verbeek, N.E.; Brilstra, E.H.; et al. Gene-network analysis identifies susceptibility genes related to glycobiology in autism. PLoS ONE 2009, 4, e5324. [Google Scholar] [CrossRef] [PubMed]
- Volpi, S.; Yamazaki, Y.; Brauer, P.M.; van Rooijen, E.; Hayashida, A.; Slavotinek, A.; Kuehn, H.S.; Di Rocco, M.; Rivolta, C.; Bortolomai, I.; et al. EXTL3 mutations cause skeletal dysplasia, immune deficiency, and developmental delay. J. Exp. Med. 2017, 214, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Germain, N.D.; Chung, W.K.; Sarmiere, P.D. RNA interference (RNAi)-based therapeutics for treatment of rare neurologic diseases. Mol. Asp. Med. 2023, 91, 101148. [Google Scholar] [CrossRef] [PubMed]
- Goel, H.; Goel, A. MicroRNA and Rare Human Diseases. Genes 2024, 15, 1243. [Google Scholar] [CrossRef]
- Almehmadi, K.A.; Tsilioni, I.; Theoharides, T.C. Increased expression of miR-155p5 in amygdala of children with autism spectrum disorder. Autism Res. 2020, 13, 18–23. [Google Scholar] [CrossRef]
- Jyonouchi, H.; Geng, L. Associations between monocyte and T cell cytokine profiles in autism spectrum disorders: Effects of dysregulated innate immune responses on adaptive responses to recall antigens in a subset of ASD children. Int. J. Mol. Sci. 2019, 20, 4731. [Google Scholar] [CrossRef]
- Jyonouchi, H.; Geng, L.; Streck, D.L.; Dermody, J.J.; Toruner, G.A. MicroRNA expression changes in association with changes in interleukin-1ß/interleukin10 ratios produced by monocytes in autism spectrum disorders: Their association with neuropsychiatric symptoms and comorbid conditions (observational study). J. Neuroinflamm. 2017, 14, 229. [Google Scholar] [CrossRef]
- Kichukova, T.M.; Popov, N.T.; Ivanov, I.S.; Vachev, T.I. Profiling of circulating serum microRNAs in children with autism spectrum disorder using stem-loop qRT-PCR assay. Folia Med. 2017, 59, 43–52. [Google Scholar] [CrossRef]
- Mor, E.; Shomron, N. Species-specific microRNA regulation influences phenotypic variability: Perspectives on species-specific microRNA regulation. BioEssays 2013, 35, 881–888. [Google Scholar] [CrossRef]
- Nakata, M.; Kimura, R.; Funabiki, Y.; Awaya, T.; Murai, T.; Hagiwara, M. MicroRNA profiling in adults with high-functioning autism spectrum disorder. Mol. Brain 2019, 12, 82. [Google Scholar] [CrossRef]
- Safdar, A.; Khurshid, S.; Farwa, U.; Bakhtiar, S.M. The analyzing miR-106b-5p and miR-93-5p as promising diagnostic markers for autism spectrum disorder. CTO 2021, 1, 36–44. [Google Scholar] [CrossRef]
- Salloum-Asfar, S.; Elsayed, A.K.; Elhag, S.F.; Abdulla, S.A. Circulating non-coding RNAs as a signature of autism spectrum disorder symptomatology. Int. J. Mol. Sci. 2021, 22, 6549. [Google Scholar] [CrossRef] [PubMed]
- Schumann, C.M.; Sharp, F.R.; Ander, B.P.; Stamova, B. Possible sexually dimorphic role of miRNA and other sncRNA in ASD brain. Mol. Autism 2017, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Winden, K.D.; Ebrahimi-Fakhari, D.; Sahin, M. Abnormal mTOR Activation in Autism. Annu. Rev. Neurosci. 2018, 41, 1–23. [Google Scholar] [CrossRef]
- Abdelkarem, O.A.; Zaki, M.A.; Elwafa, R.A.; Elmaksoud, M.A.; El Banna, A. Evaluation of the diagnostic performance of circulating microRNAs for the diagnosis of autism spectrum disorders. Alex. J. Pediatr. 2024, 37, 130–136. [Google Scholar] [CrossRef]
| ASD-miRNA | Glycosylation Pathways | FDR Corrected p-Value | Targeted Genes of Glycosylation Pathways | Prediction Score of Interaction | No. of Glycogenes | CDG Genes | ASD-Genes |
|---|---|---|---|---|---|---|---|
| hsa-miR-423-5p | N-glycan biosynthesis (hsa00510) | 4.45 × 10−5 | RPN1, DPM2, B4GALT1, ALG10B, ALG14, MAN1B1, DDOST, DOLK, B4GALT3, MGAT1, ALG3, MGAT4B | 0.474 (DPM2) 0.661 (ALG10B) | 9 | RPN1, DPM2, B4GALT1, ALG14, MAN1B1, DDOST, DOLK, MGAT1, ALG3 | DOLK |
| Glycosaminoglycan biosynthesis–keratan sulfate (hsa00533) | 5.82 × 10−3 | ST3GAL1, B4GALT1, B4GALT3 | N/A | 1 | B4GALT1 | / | |
| hsa-miR-30c-5p | Mucin-type O-glycan biosynthesis (hsa00512) | 1.64 × 10−6 | GALNT7, B4GALT5, ST3GAL1, GCNT3, GALNT1, GALNT3, GALNT2 | 1 (GALNT7) 0.559 (B4GALT5) 0.507 (GCNT3) 0.999 (GALNT1) 0.992 (GALNT3) 0.996 (GALNT2) | 2 | GALNT3, GALNT2 | GALNT2 |
| hsa-miR-195-5p | Glycosaminoglycan biosynthesis–chondroitin sulfate/dermatan sulfate (hsa00532) | 1.62 × 10−2 | UST, DSE, CHPF, CHPF2 | 0.457 (UST) 0.564 (CHPF) | 0 | / | / |
| Mucin-type O-glycan biosynthesis (hsa00512) | 3.87 × 10−2 | GALNT7, GALNT1, GALNT3, GALNT2 | 0.784 (GALNT7) 0.659 (GALNT1) | 2 | GALNT3, GALNT2 | GALNT2 | |
| hsa-miR-132-3p | Other types of O-glycan biosynthesis (hsa00514) | 2.69 × 10−2 | LFNG, B3GAT1, POMT1, EOGT | N/A | 2 | LFNG, POMT1 | / |
| hsa-miR-21-3p | Glycosaminoglycan biosynthesis–heparan sulfate/heparin (hsa00534) | 5.14 × 10−3 | EXT1, EXTL3, NDST2 | 0.503 (EXT1) | 2 | EXT1, EXTL3 | EXT1 |
| hsa-miR-132-5p | Glycosaminoglycan biosynthesis–keratan sulfate (hsa00533) | 1.85 × 10−8 | B4GALT1, B3GNT1 | 0.537 (B4GALT1) | 1 | B4GALT1 | / |
| N-glycan biosynthesis (hsa00510) | 6.53 × 10−3 | GANAB, B4GALT1 | 0.537 (B4GALT1) | 1 | B4GALT1 | / | |
| Other types of O-glycan biosynthesis (hsa00514) | 9.66 × 10−3 | B4GALT1 | 0.537 (B4GALT1) | 1 | B4GALT1 | / | |
| hsa-miR-199a-5p | Glycosaminoglycan biosynthesis–heparan sulfate/heparin (hsa00534) | 4.90 × 10−2 | EXT1 | 0.67 (EXT1) | 1 | EXT1 | EXT1 |
| hsa-miR-1277-3p | Glycosaminoglycan biosynthesis–keratan sulfate (hsa00533) | 1.06 × 10−8 | CHST1, B3GNT2 | N/A | 0 | / | / |
| Glycosaminoglycan biosynthesis–heparan sulfate/heparin (hsa00534) | 1.66 × 10−6 | EXT1 | N/A | 1 | EXT1 | EXT1 | |
| hsa-miR-379-5p | Mucin-type O-glycan biosynthesis (hsa00512) | 6.53 × 10−4 | GALNT11 | N/A | 0 | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirabella, F.; Randazzo, M.; Rinaldi, A.; Pettinato, F.; Rizzo, R.; Sturiale, L.; Barone, R. Glycosylation Pathways Targeted by Deregulated miRNAs in Autism Spectrum Disorder. Int. J. Mol. Sci. 2025, 26, 783. https://doi.org/10.3390/ijms26020783
Mirabella F, Randazzo M, Rinaldi A, Pettinato F, Rizzo R, Sturiale L, Barone R. Glycosylation Pathways Targeted by Deregulated miRNAs in Autism Spectrum Disorder. International Journal of Molecular Sciences. 2025; 26(2):783. https://doi.org/10.3390/ijms26020783
Chicago/Turabian StyleMirabella, Federica, Martina Randazzo, Alessandro Rinaldi, Fabio Pettinato, Renata Rizzo, Luisa Sturiale, and Rita Barone. 2025. "Glycosylation Pathways Targeted by Deregulated miRNAs in Autism Spectrum Disorder" International Journal of Molecular Sciences 26, no. 2: 783. https://doi.org/10.3390/ijms26020783
APA StyleMirabella, F., Randazzo, M., Rinaldi, A., Pettinato, F., Rizzo, R., Sturiale, L., & Barone, R. (2025). Glycosylation Pathways Targeted by Deregulated miRNAs in Autism Spectrum Disorder. International Journal of Molecular Sciences, 26(2), 783. https://doi.org/10.3390/ijms26020783





