Comprehensive Analysis of the GiTCP Gene Family and Its Expression Under UV-B Radiation in Glycyrrhiza inflata Bat
Abstract
:1. Introduction
2. Results
2.1. Identification and Physicochemical Properties of Members of the GiTCP Gene Family of G. inflata
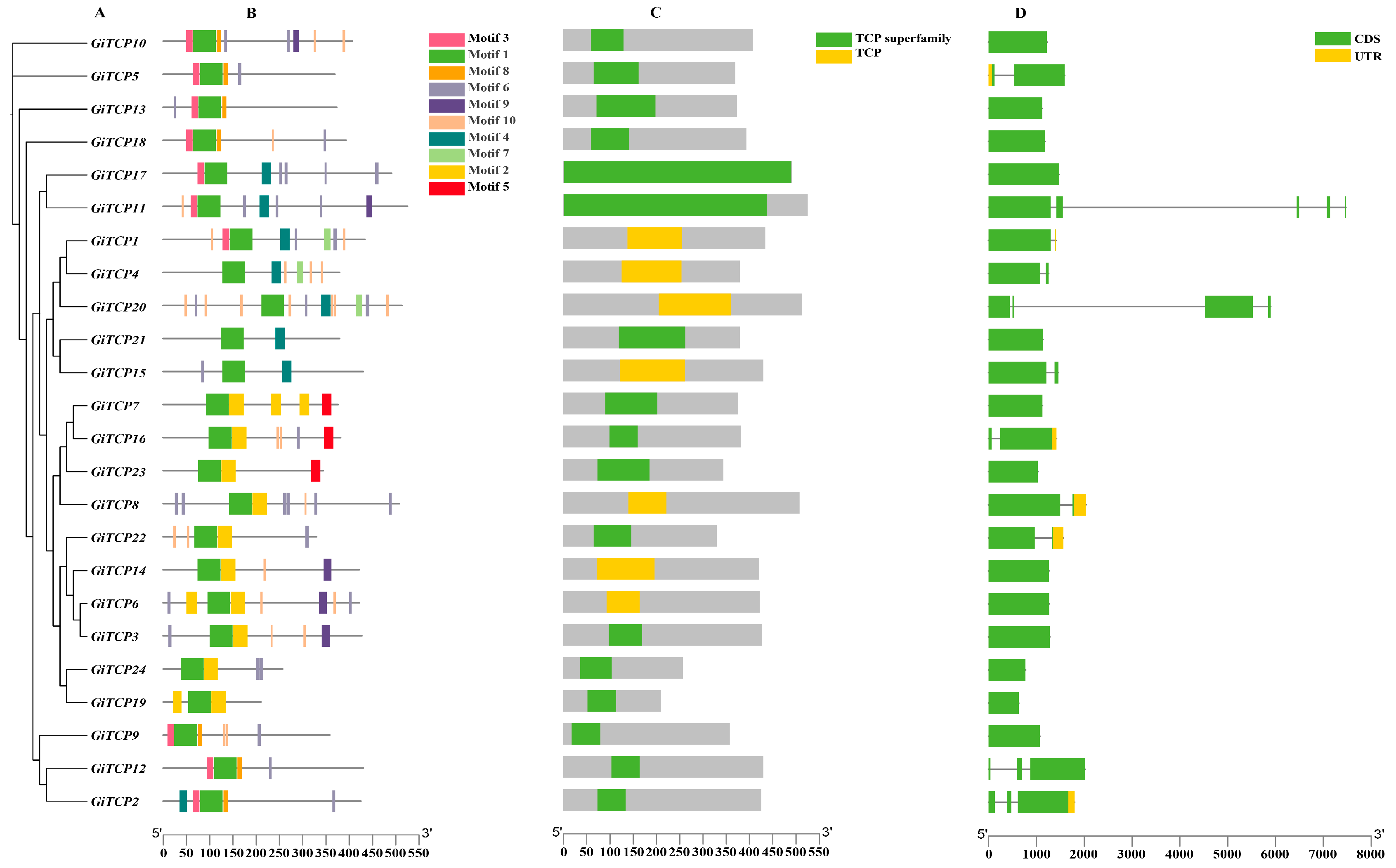
2.2. Phylogenetic Construction, Motif, Domain, and Gene Structure Analysis of the GiTCP Gene Family in G. inflata
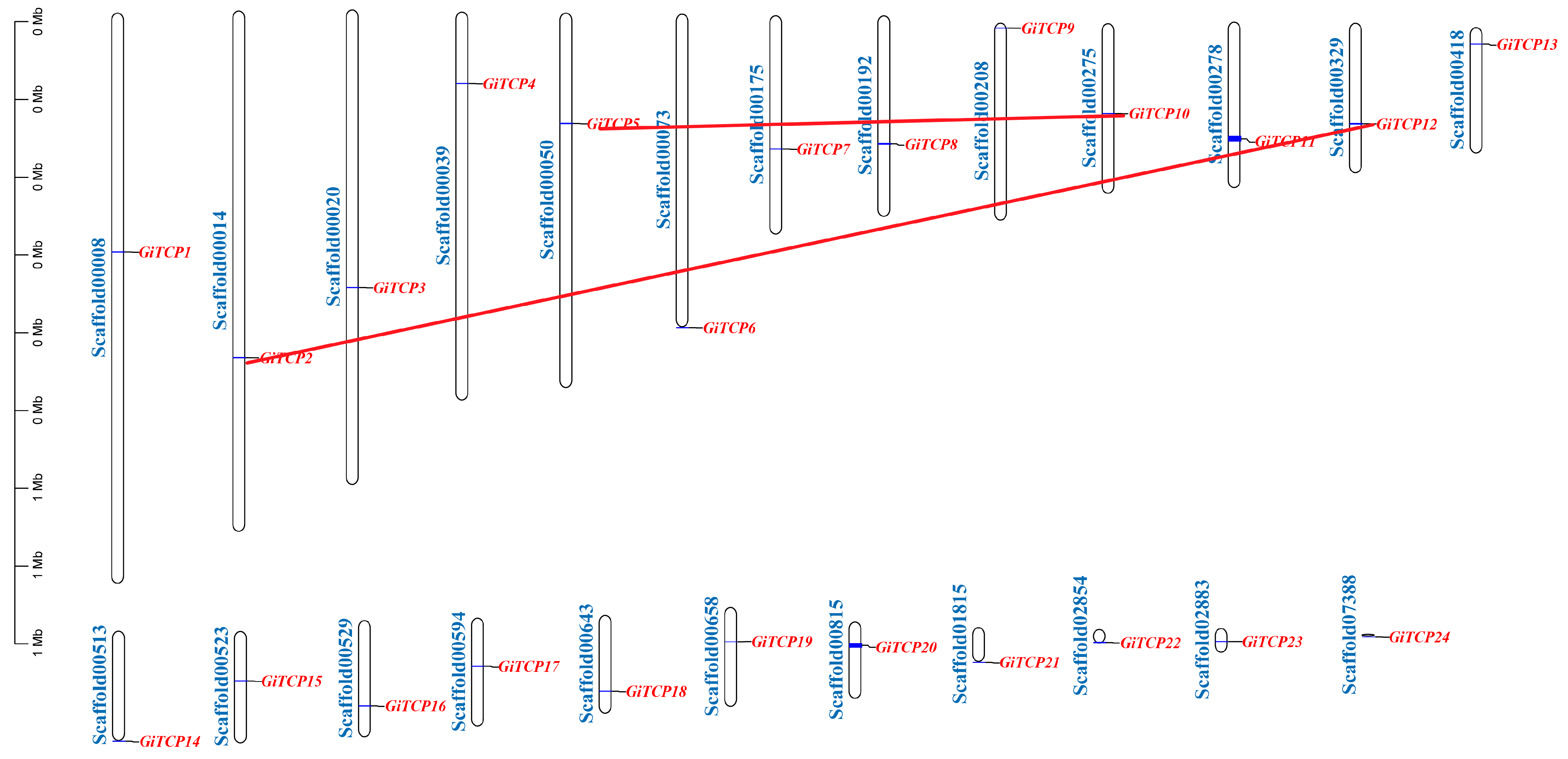
2.3. Distribution of GiTCP Genes on Chromosomes and Predictive Analysis of Secondary Structure in G. inflata
2.4. Phylogenetic Tree Analysis of TCP Genes
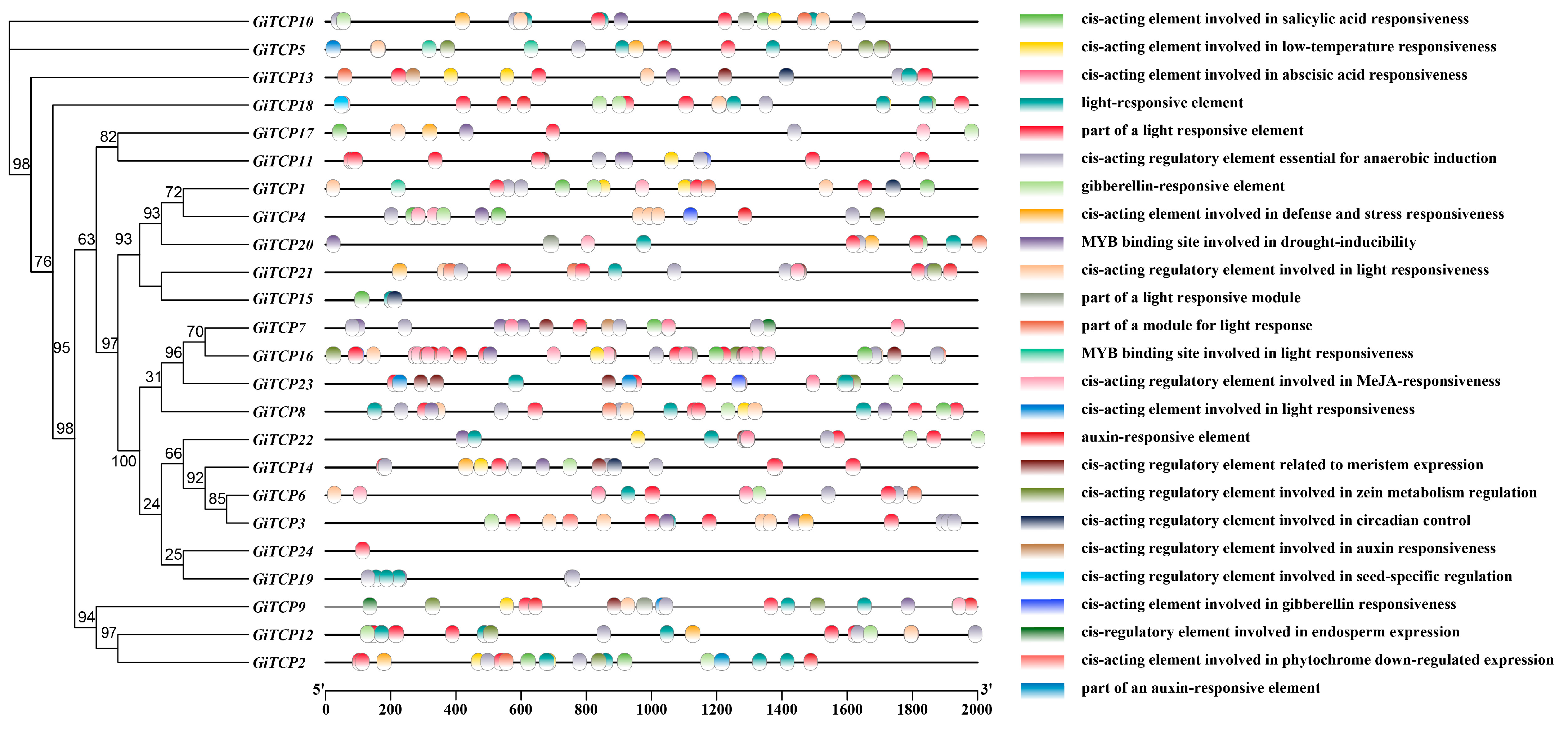
2.5. Cis-Acting Element Analysis of GiTCP Genes in G. inflata
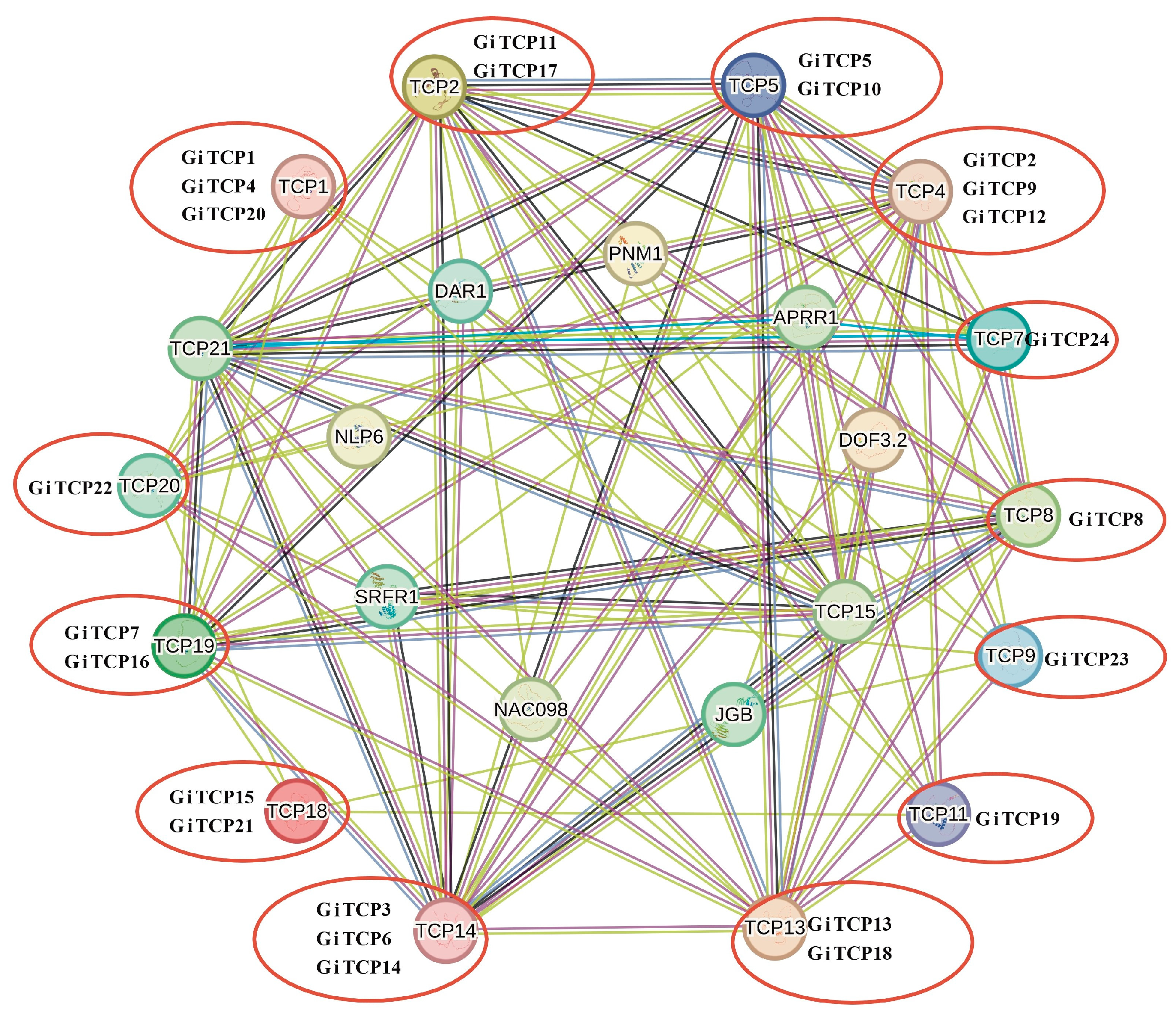
2.6. Protein Interaction Network Analysis of GiTCPs
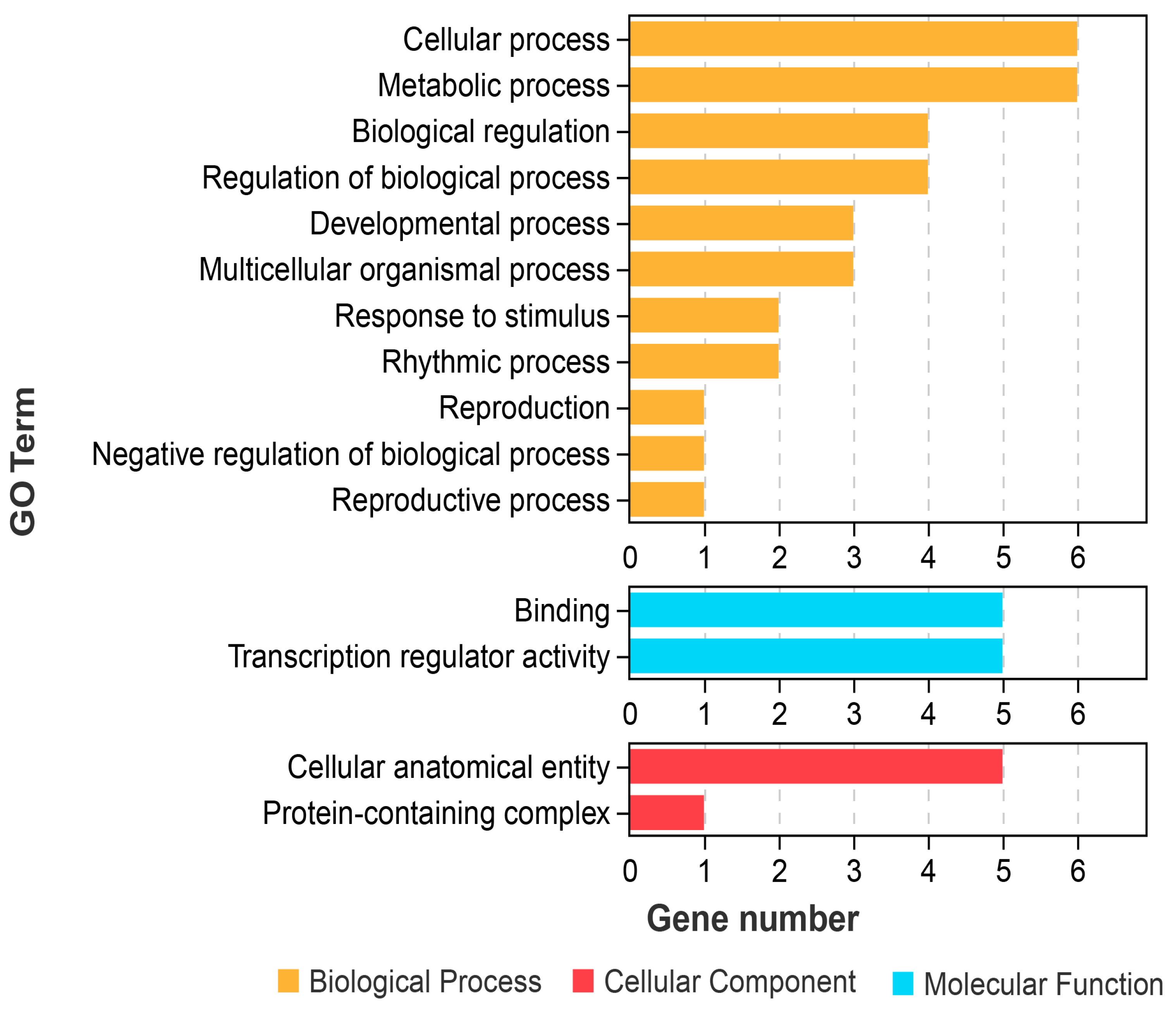
2.7. GO Functional Annotation of GiTCP Genes in G. inflata
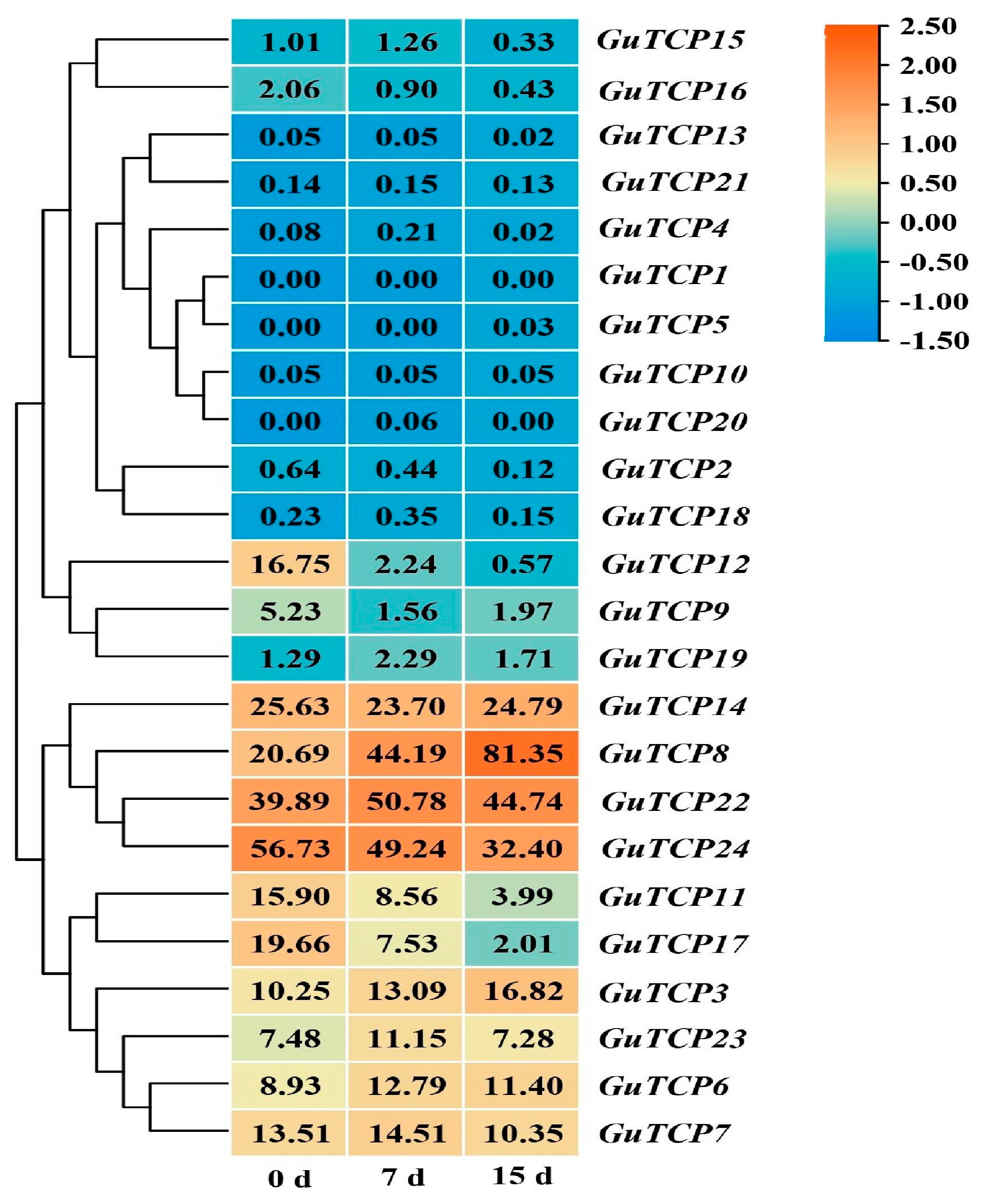
2.8. Effect of UV-B Radiation on G. inflata
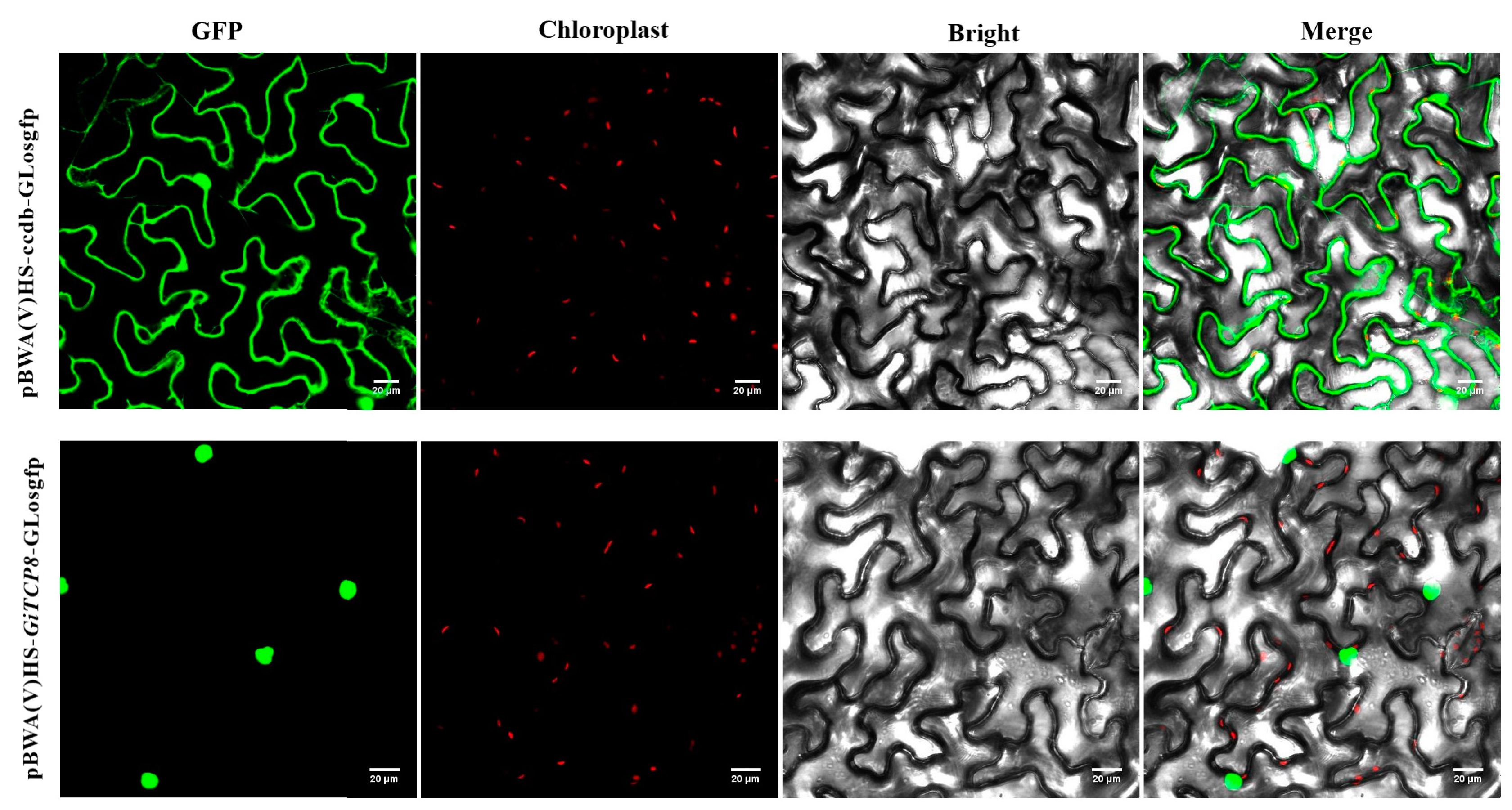
2.9. Subcellular Localization Analysis
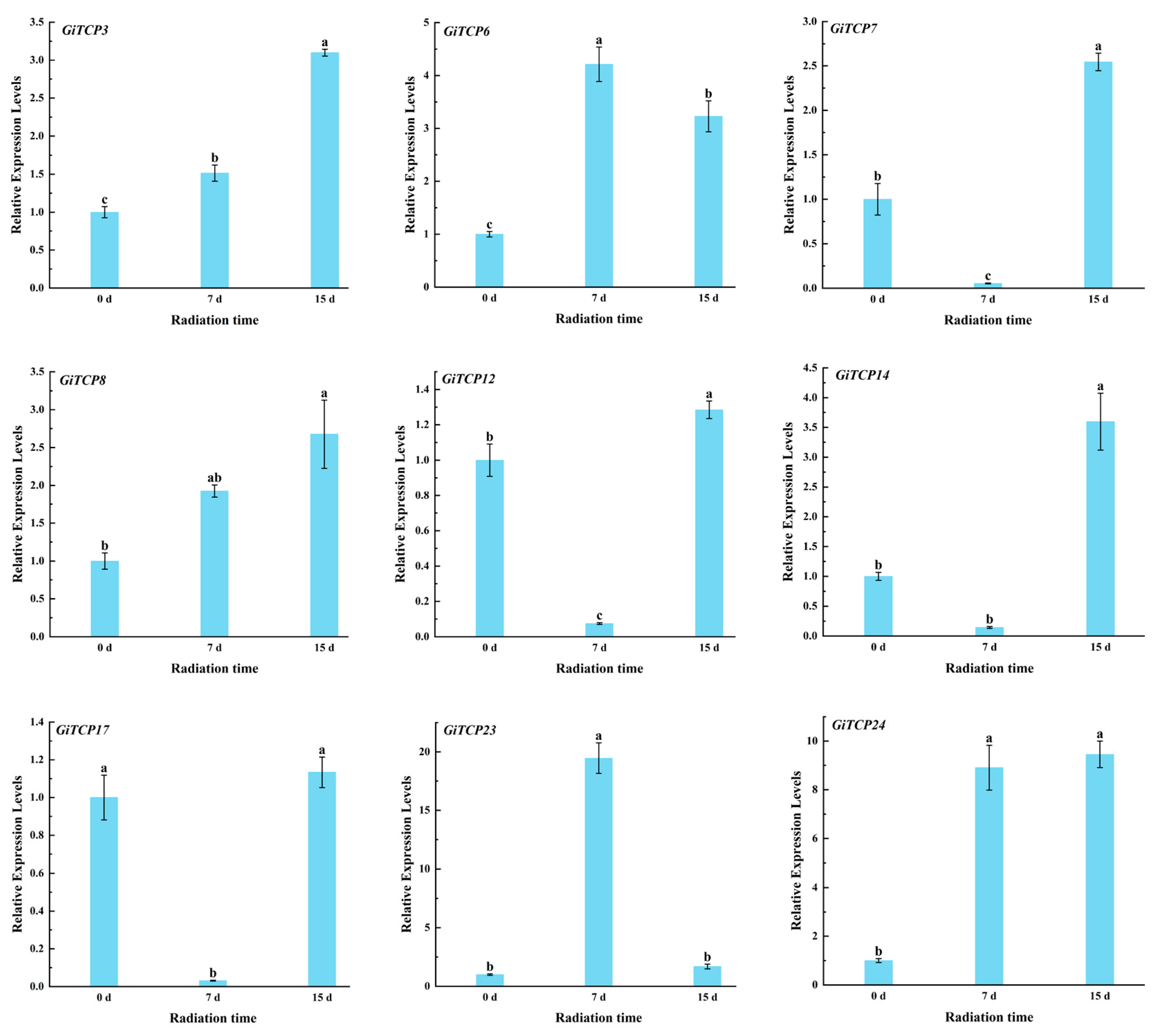
2.10. Expression Pattern Analysis of GiTCP Genes in Licorice Under UV-B Radiation
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatments
4.2. Identification of TCP Gene Family Members in Licorice
4.3. Domain, Gene Structure, and Conserved Motifs of TCP Proteins in Licorice
4.4. Chromosomal Mapping and Secondary Structure Analysis of TCP Genes in Licorice
4.5. Phylogenetic Tree Analysis of TCP Genes in Licorice
4.6. Analysis of Cis-Acting Elements of TCP Gene Family in G. inflata
4.7. Construction of TCP Protein Interaction Network in G. inflata
4.8. Subcellular Localization Assays
4.9. Transcriptome Sequencing Analysis
4.10. GO Analysis of TCP Genes in Licorice
4.11. RNA Isolation and qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TCP | teosinte branched 1/cincinnata/proliferating cell factor |
| TB1 | teosinte branched 1 |
| PCF | proliferating cell factor |
| CYC | cycloidea |
| Ms | Medicago sativa L |
| Ma | Musa acuminata |
| Pg | Panax ginseng |
| Ap | Andrographis paniculata |
| Dch | Dendrobium |
| Gi | G. inflata Bat |
| GA3 | gibberellic acid 3 |
| MeJA | methyl jasmonate |
| bZIP | basic region/leucine zipper |
| AA | amino acid sequence length |
| MW | molecular weight |
| pI | isoelectric point |
| GRAVY | grand average of hydropathicity |
| II | instability index |
| AI | aliphatic index |
| SL | subcellular localization |
| N | nucleus |
| C | cytoplasm |
| CP | chloroplast |
| E | extracellular matrix |
| M | mitochondria |
| Pm | plasma membrane |
| At | Arabidopsis thaliana |
| Eu | Eucommia ulmoides |
| GO | gene ontology |
| qRT-PCR | quantitative real-time PCR |
| UV-B | ultraviolet B |
| Cr | Catharanthus roseus |
References
- Almeida, D.M.; Gregorio, G.B.; Oliveira, M.M.; Saibo, N.J. Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol. Biol. 2017, 93, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Cubas, P.; Coen, E.; Zapater, J.M. Ancient asymmetries in the evolution of flowers. Curr. Biol. 2001, 11, 1050–1052. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997, 9, 1607–1619. [Google Scholar] [PubMed]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef]
- An, J.X.; Guo, Z.X.; Gou, X.P.; Li, J. TCP1 positively regulates the expression of DWF4 in Arabidopsis thaliana. Plant Signal. Behav. 2011, 6, 1117–1118. [Google Scholar] [CrossRef]
- Luo, M.; Zhang, Z.M.; Gao, J.; Zeng, X.; Pan, G.T. The role of miR319 in plant development regulation. Hereditas 2011, 33, 1203–1211. [Google Scholar] [CrossRef]
- Kim, Y.; Wang, J.; Ma, C.; Jong, C.; Jin, M.; Cha, J.; Wang, J.; Peng, Y.; Ni, H.; Li, H.; et al. GmTCP and GmNLP Underlying Nodulation Character in Soybean Depending on Nitrogen. Int. J. Mol. Sci. 2023, 24, 7750. [Google Scholar] [CrossRef]
- Wei, N.; Li, Y. Genome-wide identification of the alfalfa TCP gene family and analysis of gene transcription patterns in alfalfa (Medicago sativa) under drought stress. Acta Prataculturae Sin. 2021, 31, 118. [Google Scholar]
- Wang, J.Y.; Wang, Z.; Jia, C.H.; Miao, H.X.; Zhang, J.B.; Liu, J.H.; Xu, B.Y.; Jin, Z.Q. Genome-wide identification and transcript analysis of TCP gene family in Banana (Musa acuminata L.). Biochem. Genet. 2022, 60, 204–222. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Jiao, H.H.; Yu, Q.R.; Hua, Z.Y.; Zhao, Y.Y.; Zhou, J.H.; Yuan, Y.; Huang, L.Q. Bioinformatics analysis and function prediction of ginseng TCP transcription factor family. China J. Chin. Mater. Medica. 2021, 46, 3838–3845. [Google Scholar]
- Zhang, L.; Xu, S.Q.; Li, J.Y.; Hu, X.B.; Gu, Y.; Wang, J.H. Genome-wide identification and expression analysis of the TCP gene family of Andrographis paniculata under abiotic stress. China J. Chin. Mater. Medica. 2024, 49, 379–388. [Google Scholar]
- Huang, Y.; Zhao, X.; Zheng, Q.; He, X.; Zhang, M.M.; Ke, S.; Li, Y.; Zhang, C.; Ahmad, S.; Lan, S.; et al. Genome-wide identification of TCP gene family in dendrobium and their expression patterns in Dendrobium chrysotoxum. Int. J. Mol. Sci. 2023, 24, 14320. [Google Scholar] [CrossRef]
- Li, J.; Li, X.; Cao, M.M.; Dong, W.R.; Jiang, J.Q. Research progress in pharmacological actions of liquorice and proportion of couplet medicines in combination. Shanghai J. Tradit. Chin. Med. 2019, 53, 83–87. [Google Scholar]
- Zhang, D.; Liu, Y.; Yang, Z.R.; Song, X.Q.; Ma, Y.L.; Zhao, J.Y.; Wang, X.Y.; Liu, H.; Fan, L. Widely target metabolomics analysis of the differences in metabolites of licorice under drought stress. Ind. Crops Prod. 2023, 202, 117071. [Google Scholar] [CrossRef]
- Wang, C.C.; Chen, L.H.; Cai, Z.C.; Chen, C.H.; Liu, Z.X.; Liu, S.J.; Zhou, L.S.; Tan, M.X.; Chen, J.L.; Liu, X.H.; et al. Metabolite profiling and transcriptome analysis explains difference in accumulation of bioactive constituents in licorice (Glycyrrhiza uralensis) under salt stress. Front. Plant Sci. 2021, 12, 727882. [Google Scholar] [CrossRef]
- Yao, H.; Wang, F.; Bi, Q.; Liu, H.L.; Liu, L.; Xiao, G.H.; Zhu, J.B.; Shen, H.T.; Li, H.B. Combined analysis of pharmaceutical active ingredients and transcriptomes of Glycyrrhiza uralensis under PEG6000-induced drought stress revealed glycyrrhizic acid and flavonoids accumulation via ja-mediated signaling. Front. Plant Sci. 2022, 13, 920172. [Google Scholar] [CrossRef]
- Liang, X.W.; Yang, Q.; Li, D.; Chen, X.X.; Tang, X.M.; Pan, L.M.; Zhang, C.R. Regulation of methyl jasmonate on secondary metabolism of Glycyrrhiza uralensis. Guangdong Agric. Sci. 2017, 44, 57–62. [Google Scholar]
- Mochida, K.; Sakurai, T.; Seki, H.; Yoshida, T.; Takahagi, K.; Sawai, S.; Uchiyama, H.; Muranaka, T.; Saito, K. Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J. 2017, 89, 181–194. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, L.Q.; Fan, F.H.; Xu, Z.C.; Luo, H.M.; Sun, W.; Wang, Y.; Zheng, S.H.; Song, J.Y.; Yao, H. Systematic screening and analysis of bZIP transcription factors in Glycyrrhiza uralensis and their response to ABA stress. Acta Pharm. Sin. 2023, 25, 1207–1217. [Google Scholar]
- Goyal, P.; Manzoor, M.M.; Vishwakarma, R.A.; Sharma, D.; Dhar, M.K.; Gupta, K. A comprehensive transcriptome-wide identification and screening of WRKY gene family engaged in abiotic stress in Glycyrrhiza glabra. Sci. Rep. 2020, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Zhang, Z.; Wang, Y.; Li, S.H.; Liang, Z.C. Genome-wide identification and characterization of the NF-Y gene family in grape (Vitis vinifera L.). BMC Genom. 2016, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, L.; Wu, Y.S.; Liu, Y.; Ren, S.S.; Chen, Y.L. Identification and bioinformatics analysis of TCP family genes in Eucommia ulmoides. Chin. Tradit. Herb. Drugs. 2022, 53, 5813–5824. [Google Scholar]
- Abel, S.; Savchenko, T.; Levy, M. Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa. Bmc Evol. Biol. 2005, 5, 1–25. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yama Gichi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 39–47. [Google Scholar] [CrossRef]
- Taoka, K.; Yanagimoto, Y.; Daimon, Y.; Hibara, K.; Aida, M.; Tasaka, M. The NAC domain mediates functional specificity of CUP-SHAPED COTYLEDON proteins. Plant J. 2004, 40, 462–473. [Google Scholar] [CrossRef]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef]
- Pruneda-Paz, J.L.; Breton, G.; Para, A.; Kay, S.A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 2009, 323, 1481–1485. [Google Scholar] [CrossRef]
- Rueda-Romero, P.; Barrero-Sicilia, C.; Gomez-Cadenas, A.; Carbonero, P.; Oñate-Sánchez, L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 2012, 63, 1937–1949. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.C.; Chen, L.L.; Lu, Y.; Wu, Y.B.; Dumenil, J.; Zhu, Z.G.; Bevan, M.W.; Li, Y.H. The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell. 2015, 27, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Hammani, K.; Gobert, A.; Hleibieh, K.; Choulier, L.; Small, I.; Giegé, P. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell. 2011, 23, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Halane, M.K.; Kim, S.H.; Gassmann, W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 2011, 334, 1405–1408. [Google Scholar] [CrossRef]
- Zheng, L.; Bai, X.T.; Li, H.Y. Genome-wide identification and expression analysis of Sorghum bicolor TCP gene family. Henan Agric. Sci. 2019, 537, 36–42. [Google Scholar]
- Liu, Y.; Zhang, H.; Xin, D.W.; Wang, L.L.; Zhang, L.W.; Liu, C.Y.; Chen, Q.S.; Hu, G.H. Analysis and function prediction of Glycine max TCP transcription factor domains. Soybean Sci. 2012, 31, 707–713. [Google Scholar]
- Li, K.J.; Tan, S.S.; Sun, B.; Rao, D.; He, Q.; Min, A.L.; Li, M.Y. Genome-wide identification and expression analysis of Brassica juncea TCP transcription factor. J. Sichuan Agric. Univ. 2019, 37, 459–468. [Google Scholar]
- Lei, D.; Wu, Y.; Su, Z. Research progress on the interaction between TCP transcription factors and hormone signals. Mol. Plant Breed. 2019, 17, 2868–2875. [Google Scholar]
- Zhang, M.; Agassin, R.H.; Huang, Z.; Wang, D.; Yao, S.; Ji, K. Transcriptome-wide identification of TCP transcription factor family members in Pinus massoniana and their expression in regulation of development and in response to stress. Int. J. Mol. Sci. 2023, 24, 15938. [Google Scholar] [CrossRef]
- Lei, Q.D.; Sun, X.D.; Xu, H.N. Research progress in transcription factor TCP4 participating in plant growth, development and stress resistance regulation. Acta Agric. Boreali-Sin. 2021, 36, 210–214. [Google Scholar]
- Wang, J.L.; Wang, H.W.; Cao, Y.N.; Kan, S.L.; Liu, Y.Y. Comprehensive evolutionary analysis of the TCP gene family: Further insights for its origin, expansion, and diversification. Front. Plant Sci. 2021, 13, 994567. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.M.; Wang, M.M.; Yang, J.; Wen, J.; Guo, P.C.; Wu, Y.W.; Ke, Y.Z.; Li, P.F.; Li, J.N.; Du, H. Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. Int. J. Mol. Sci. 2019, 20, 3591. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.; Gierringue, Y.; Ranty, B.; Pouzet, C.; Jauneau, A.; Robe, E.; Mazars, C.; Galaud, J.P.; Aldon, D. Specific TCP transcription factors interact with and stabilize PRR2 within different nuclear sub-domains. Plant Sci. 2019, 287, 110197. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; Wang, H.W.; Liu, R.; Xu, Y.T.; Lu, Z.C.; Zhou, C.E. Genome-wide identification of TCP family transcription factors in Medicago truncatula reveals significant roles of miR319-Targeted TCPs in nodule development. Front. Plant Sci. 2018, 9, 774. [Google Scholar] [CrossRef]
- He, Z.M.; Zhao, X.Y.; Kong, F.N.; Zuo, Z.C.; Liu, X.M. TCP2 positively regulates HY5/HYH and photomorphogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 775–785. [Google Scholar] [CrossRef]
- Viola, I.L.; Uberti Manassero, N.G.; Ripoll, R.; Gonzalez, D.H. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 2011, 435, 143–155. [Google Scholar] [CrossRef]
- Li, Z.; Li, B.; Dong, A. The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant. 2012, 5, 270–280. [Google Scholar] [CrossRef]
- Si, C.; Zhan, D.; Wang, L.; Sun, X.; Zhong, Q.; Yang, S. Systematic investigation of TCP gene family: Genome-wide identification and light-regulated gene expression analysis in pepino (Solanum Muricatum). Cells. 2023, 12, 1015. [Google Scholar] [CrossRef]
- Zhou, L.X.; Zhao, Z.H.; Yang, M.D.; Cao, H.X.; Yang, Y.D. Bioinformatics identification and gene expression analysis of TCP transcription factor family in oil palm. Mol. Plant Breed. 2022, 20, 5637–5648. [Google Scholar]
- Ling, L.; Zhang, W.R.; An, Y.M.; Du, B.H.; Wang, D.; Guo, C.H. Genome-wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Funct. Integr. Genom. 2020, 20, 537–550. [Google Scholar] [CrossRef]
- Ren, L.; Wu, H.X.; Zhang, T.T.; Ge, X.Y.; Wang, T.L.; Zhou, W.Y.; Zhang, L.; Ma, D.F.; Wang, A.M. Genome-wide identification of TCP transcription factors family in sweet potato reveals significant roles of miR319-targeted TCPs in leaf anatomical morphology. Front. Plant Sci. 2021, 12, 686698. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.D.; Zhao, Y.R.; Gao, H.C.; Li, Y.H.; Tang, W.; Ma, L.C.; Yang, G.F.; Sun, J. Genomic characterization and expression analysis of TCP transcription factors in Setaria italica and Setaria viridis. Plant Signal. Behav. 2022, 17, 2075158. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.X.; Hou, Z.N.; He, Q.L.; Zhang, X.M.; Yan, K.J.; Han, R.L.; Liang, Z.S. Genome-wide identification and expression analysis of TCP family genes in Catharanthus roseus. Front. Plant Sci. 2023, 14, 1161534. [Google Scholar]
- Li, Y.; Zhuang, F.; Zeng, J.; Yang, Z.; Li, Y.Q.; Luo, M.; Wang, Y. Identification of the histone demethylases gene family in Glycyrrhiza inflata reveals genes responding to abiotic stresses. J. Cell. Biochem. 2022, 123, 1780–1792. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.; Rouzé, P.; Rombauts, S. Plant CARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.Y.; Minguez, P.; Bork, P.; Mering, C.V.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, G.Y.; Ding, Q.; Wei, B.Q. Identification of the PIP5K gene family in watermelon (Citrullus lanatus) and its expression analysis in male sterile floral buds. Acta Agric. Boreali-Occident. Sin. 2021, 30, 883–893. [Google Scholar]


| Gene Name | Sequence ID | AA | MW | pI | II | AI | GRAVY | SL |
|---|---|---|---|---|---|---|---|---|
| GiTCP1 | Glyur000008s00001296.1 | 433 | 48,419.6 | 6.35 | 56.22 | 58 | −0.688 | N,C |
| GiTCP2 | Glyur000014s00002559.1 | 424 | 46,637.73 | 6.56 | 55.14 | 60 | −0.806 | N,C,CP |
| GiTCP3 | Glyur000020s00001802.1 | 426 | 46,357.61 | 6.39 | 57.12 | 53 | −0.877 | N,C |
| GiTCP4 | Glyur000039s00004191.1 | 378 | 42,805.57 | 6.46 | 55.23 | 46 | −1.098 | N,C |
| GiTCP5 | Glyur000050s00005694.1 | 369 | 40,912.51 | 8.97 | 59.19 | 62 | −0.729 | N,C,E |
| GiTCP6 | Glyur000073s00007830.1 | 421 | 45,416.08 | 7.41 | 70.02 | 58 | −0.769 | N,C |
| GiTCP7 | Glyur000175s00012061.1 | 375 | 39,457.06 | 6.26 | 47.75 | 62 | −0.461 | N |
| GiTCP8 | Glyur000192s00009341.1 | 507 | 54,268.62 | 7.26 | 60.82 | 58 | −0.736 | N |
| GiTCP9 | Glyur000208s00014261.1 | 357 | 39,483.73 | 6.17 | 66.29 | 59 | −0.815 | N |
| GiTCP10 | Glyur000275s00017056.1 | 406 | 44,812.15 | 9.32 | 54.38 | 67 | −0.623 | N,C,E |
| GiTCP11 | Glyur000278s00017293.1 | 524 | 57,436.66 | 7.42 | 52.22 | 61 | −0.736 | N,CP |
| GiTCP12 | Glyur000329s00019843.1 | 429 | 46,829.39 | 6.33 | 41.64 | 64 | −0.559 | N,CP,M |
| GiTCP13 | Glyur000418s00021337.1 | 372 | 41,089.66 | 7.13 | 53.33 | 69 | −0.617 | N,CP |
| GiTCP14 | Glyur000513s00025388.1 | 420 | 43,972.18 | 6.42 | 55.38 | 57 | −0.606 | N |
| GiTCP15 | Glyur000523s00021974.1 | 429 | 48,570.05 | 7.31 | 51.61 | 57 | −0.943 | N,Pm |
| GiTCP16 | Glyur000529s00019352.1 | 380 | 40,865.72 | 6.15 | 53.77 | 61 | −0.611 | N,CP,M |
| GiTCP17 | Glyur000594s00025967.1 | 490 | 54,254.93 | 6.68 | 58.91 | 51 | −1.027 | N,CP |
| GiTCP18 | Glyur000643s00026942.1 | 392 | 43,118.88 | 7.91 | 52.49 | 67 | −0.754 | N,C,CP |
| GiTCP19 | Glyur000658s00024843.1 | 209 | 22,115.54 | 8.46 | 63.38 | 70 | −0.266 | N,C,M |
| GiTCP20 | Glyur000815s00035011.1 | 512 | 57,147.71 | 9.42 | 55.92 | 57 | −0.972 | N |
| GiTCP21 | Glyur001815s00041763.1 | 378 | 42,273.69 | 9.2 | 56.38 | 72 | −0.598 | N,CP |
| GiTCP22 | Glyur002854s00044943.1 | 329 | 34,959.7 | 9.01 | 51.47 | 66 | −0.751 | N,CP,E |
| GiTCP23 | Glyur002883s00039141.1 | 343 | 36,224.26 | 9.72 | 62.61 | 73 | −0.334 | N,Pm |
| GiTCP24 | Glyur007388s00045515.1 | 256 | 27,486.88 | 9.38 | 54.13 | 68 | −0.627 | N,CP,M |
| Gene Name | Sequence ID | Alpha Helix | Extended Strand | Beta Turn | Random Coil |
|---|---|---|---|---|---|
| GiTCP1 | Glyur000008s00001296.1 | 27.25% | 12.24% | 1.85% | 58.66% |
| GiTCP2 | Glyur000014s00002559.1 | 19.81% | 11.56% | 3.77% | 64.86% |
| GiTCP3 | Glyur000020s00001802.1 | 15.49% | 12.44% | 3.99% | 68.08% |
| GiTCP4 | Glyur000039s00004191.1 | 33.07% | 8.73% | 2.38% | 55.82% |
| GiTCP5 | Glyur000050s00005694.1 | 15.18% | 13.28% | 3.25% | 68.29% |
| GiTCP6 | Glyur000073s00007830.1 | 20.67% | 11.88% | 4.75% | 62.71% |
| GiTCP7 | Glyur000175s00012061.1 | 12.53% | 12.80% | 3.47% | 71.02% |
| GiTCP8 | Glyur000192s00009341.1 | 19.13% | 10.65% | 3.35% | 66.86% |
| GiTCP9 | Glyur000208s00014261.1 | 14.85% | 12.32% | 3.08% | 69.75% |
| GiTCP10 | Glyur000275s00017056.1 | 18.72% | 13.05% | 5.42% | 62.81% |
| GiTCP11 | Glyur000278s00017293.1 | 28.05% | 11.64% | 3.63% | 56.68% |
| GiTCP12 | Glyur000329s00019843.1 | 22.38% | 14.22% | 3.03% | 60.37% |
| GiTCP13 | Glyur000418s00021337.1 | 18.82% | 7.80% | 3.49% | 69.89% |
| GiTCP14 | Glyur000513s00025388.1 | 22.14% | 11.90% | 3.57% | 62.71% |
| GiTCP15 | Glyur000523s00021974.1 | 36.60% | 6.76% | 2.33% | 54.31% |
| GiTCP16 | Glyur000529s00019352.1 | 14.47% | 11.84% | 3.42% | 70.28% |
| GiTCP17 | Glyur000594s00025967.1 | 25.21% | 14.17% | 7.5% | 53.12% |
| GiTCP18 | Glyur000643s00026942.1 | 13.27% | 13.52% | 4.59% | 68.62% |
| GiTCP19 | Glyur000658s00024843.1 | 19.62% | 20.57% | 2.87% | 56.94% |
| GiTCP20 | Glyur000815s00035011.1 | 20.70% | 15.23% | 4.10% | 59.96% |
| GiTCP21 | Glyur001815s00041763.1 | 38.89% | 12.70% | 5.29% | 43.12% |
| GiTCP22 | Glyur002854s00044943.1 | 23.40% | 8.81% | 6.08% | 61.70% |
| GiTCP23 | Glyur002883s00039141.1 | 20.41% | 16.62% | 4.66% | 58.31% |
| GiTCP24 | Glyur007388s00045515.1 | 25.0% | 16.80% | 3.12% | 55.08% |
| Class | GO Term | Annotation | GiTCP Genes |
|---|---|---|---|
| MF | GO:0005488 | Binding | GiTCP1, GiTCP14, GiTCP16, GiTCP19, GiTCP23 |
| GO:0140110 | Transcription regulator activity | GiTCP14, GiTCP16, GiTCP19, GiTCP23, GiTCP24 | |
| CC | GO:0032991 | Protein-containing complex | GiTCP24 |
| GO:0110165 | Cellular anatomical entity | GiTCP14, GiTCP16, GiTCP19, GiTCP23, GiTCP24 | |
| BP | GO:0000003 | Reproduction | GiTCP14 |
| GO:0008152 | Metabolic process | GiTCP1, GiTCP14, GiTCP16, GiTCP19, GiTCP23, GiTCP24 | |
| GO:0009987 | Cellular process | GiTCP1, GiTCP14, GiTCP16, GiTCP19, GiTCP23, GiTCP24 | |
| GO:0022414 | Reproductive process | GiTCP14 | |
| GO:0032501 | Multicellular organismal process | GiTCP14, GiTCP19, GiTCP23 | |
| GO:0032502 | Developmental process | GiTCP14, GiTCP19, GiTCP23 | |
| GO:0048511 | Rhythmic process | GiTCP14, GiTCP19 | |
| GO:0048519 | Negative regulation of biological process | GiTCP23 | |
| GO:0050789 | Regulation of biological process | GiTCP14, GiTCP16, GiTCP19, GiTCP23 | |
| GO:0050896 | Response to stimulus | GiTCP14, GiTCP16 | |
| GO:0065007 | Biological regulation | GiTCP14, GiTCP16, GiTCP19, GiTCP23 |
| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
| GiTCP3 | CAAACTCCTCGTCAACCCGA | CTTTGTGTGCCGGTCCTTTG |
| GiTCP6 | CCGAATTAGCCGCGAACAAG | GAGCTCACGTGTCAGTTGGA |
| GiTCP7 | GCAGCGACGAGTATTCCAGA | CGCTGGAACTAACACTGGGT |
| GiTCP8 | TCTACCAGCCCTCTCAGCAT | GATCGGAATTGTCGGTGGGT |
| GiTCP11 | CGTGTCTGGCCTTTTCCGAT | CCCAAATGGTCTTTCCTTGCG |
| GiTCP12 | GCTCCTGTTGGGTTTGATGC | AGGGTCCCCCTATGGGAAAA |
| GiTCP14 | GCGAGACGATAGAGTGGCTC | CGGAGAAGGTGCTAGGGTTG |
| GiTCP17 | GGCACCACCACTCATCAAGA | GCTGTTGTCACTGAGAGCCT |
| GiTCP22 | CCTGGTTTGGAACTGGGGTT | GGCCATGCAATTCCACCTTG |
| GiTCP23 | TGTTCAGCTCCAACCAGCAA | TGGAAGAAGAGCGCGACAAT |
| GiTCP24 | TACCCTCGCGGTGAAGAAAC | GCGGCGATAATTGACGGTTC |
| β-actin | CCTCATGCCATCCTTCGTC | TCTTTGCAGTCTCGAGTTCTTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Zhao, J.; Xiao, Y.; Li, C.; Miao, R.; Chen, S.; Zhang, D.; Zhou, X.; Li, M. Comprehensive Analysis of the GiTCP Gene Family and Its Expression Under UV-B Radiation in Glycyrrhiza inflata Bat. Int. J. Mol. Sci. 2025, 26, 159. https://doi.org/10.3390/ijms26010159
Liu Z, Zhao J, Xiao Y, Li C, Miao R, Chen S, Zhang D, Zhou X, Li M. Comprehensive Analysis of the GiTCP Gene Family and Its Expression Under UV-B Radiation in Glycyrrhiza inflata Bat. International Journal of Molecular Sciences. 2025; 26(1):159. https://doi.org/10.3390/ijms26010159
Chicago/Turabian StyleLiu, Ziliang, Jiaang Zhao, Ying Xiao, Caijuan Li, Rong Miao, Sijin Chen, Dan Zhang, Xiangyan Zhou, and Mengfei Li. 2025. "Comprehensive Analysis of the GiTCP Gene Family and Its Expression Under UV-B Radiation in Glycyrrhiza inflata Bat" International Journal of Molecular Sciences 26, no. 1: 159. https://doi.org/10.3390/ijms26010159
APA StyleLiu, Z., Zhao, J., Xiao, Y., Li, C., Miao, R., Chen, S., Zhang, D., Zhou, X., & Li, M. (2025). Comprehensive Analysis of the GiTCP Gene Family and Its Expression Under UV-B Radiation in Glycyrrhiza inflata Bat. International Journal of Molecular Sciences, 26(1), 159. https://doi.org/10.3390/ijms26010159







