Yeast Glucan Remodeling Protein Bgl2p: Amyloid Properties and the Mode of Attachment in Cell Wall
Abstract
:1. Introduction
2. Results and Discussion
2.1. Prediction and Choosing of Bgl2p Structural Model for Further Analysis
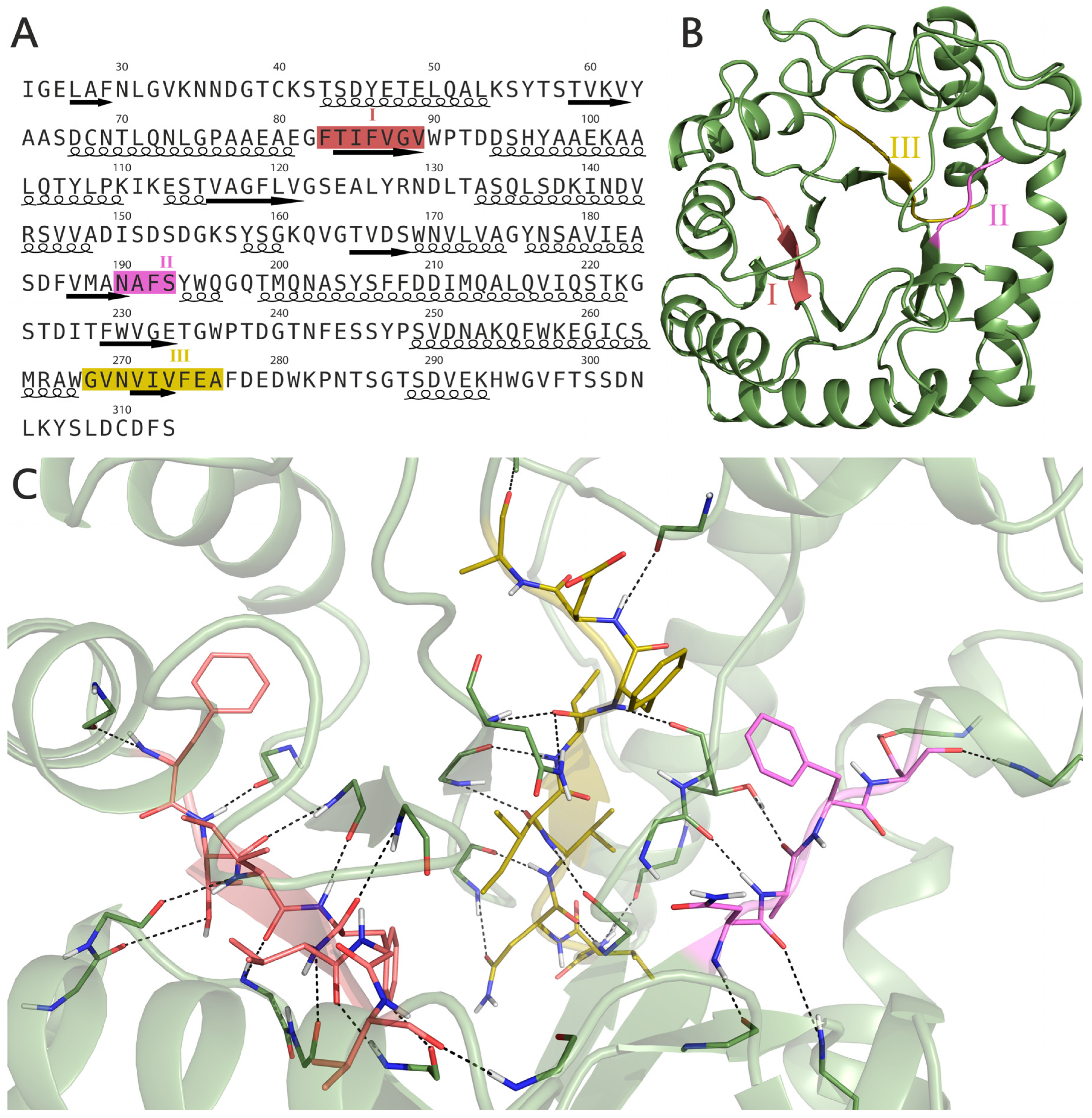
2.2. Structure and Position of Strong Amyloidogenic Sites F83TIFVGV89, N190AFS193 and G268VNVIVFEA276
2.3. Conformation of the C-Terminal Region
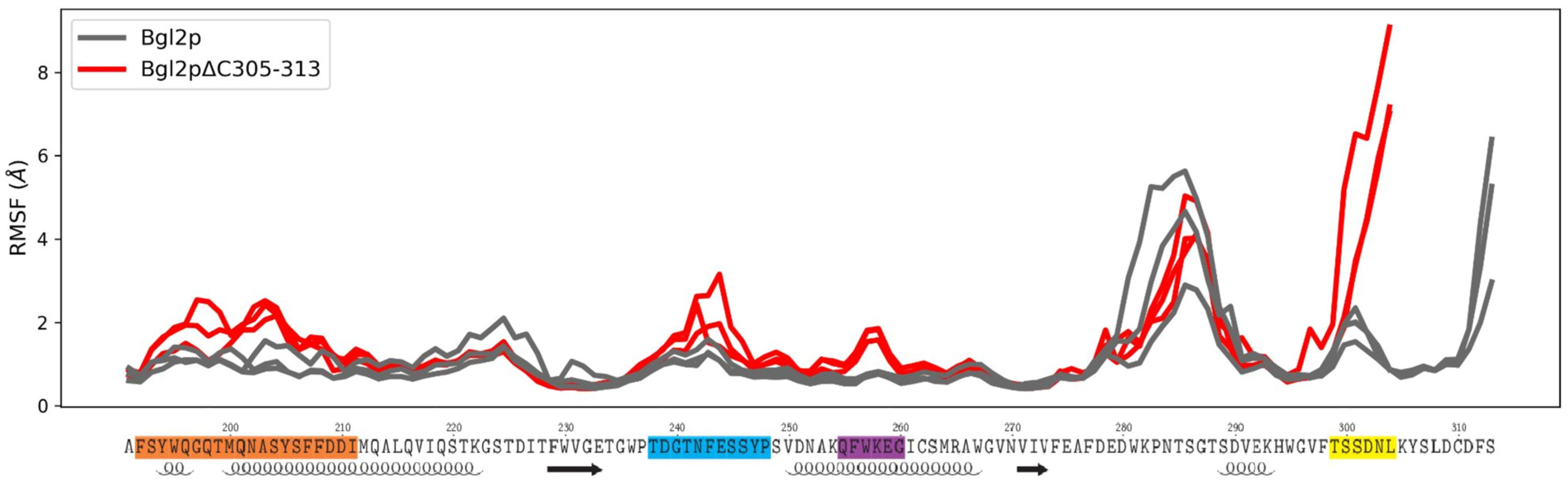
2.4. The Effect of C-Terminal Region Deletion on the Conformational Dynamics of Bgl2p
3. Materials and Methods
3.1. Molecular Dynamics Simulations
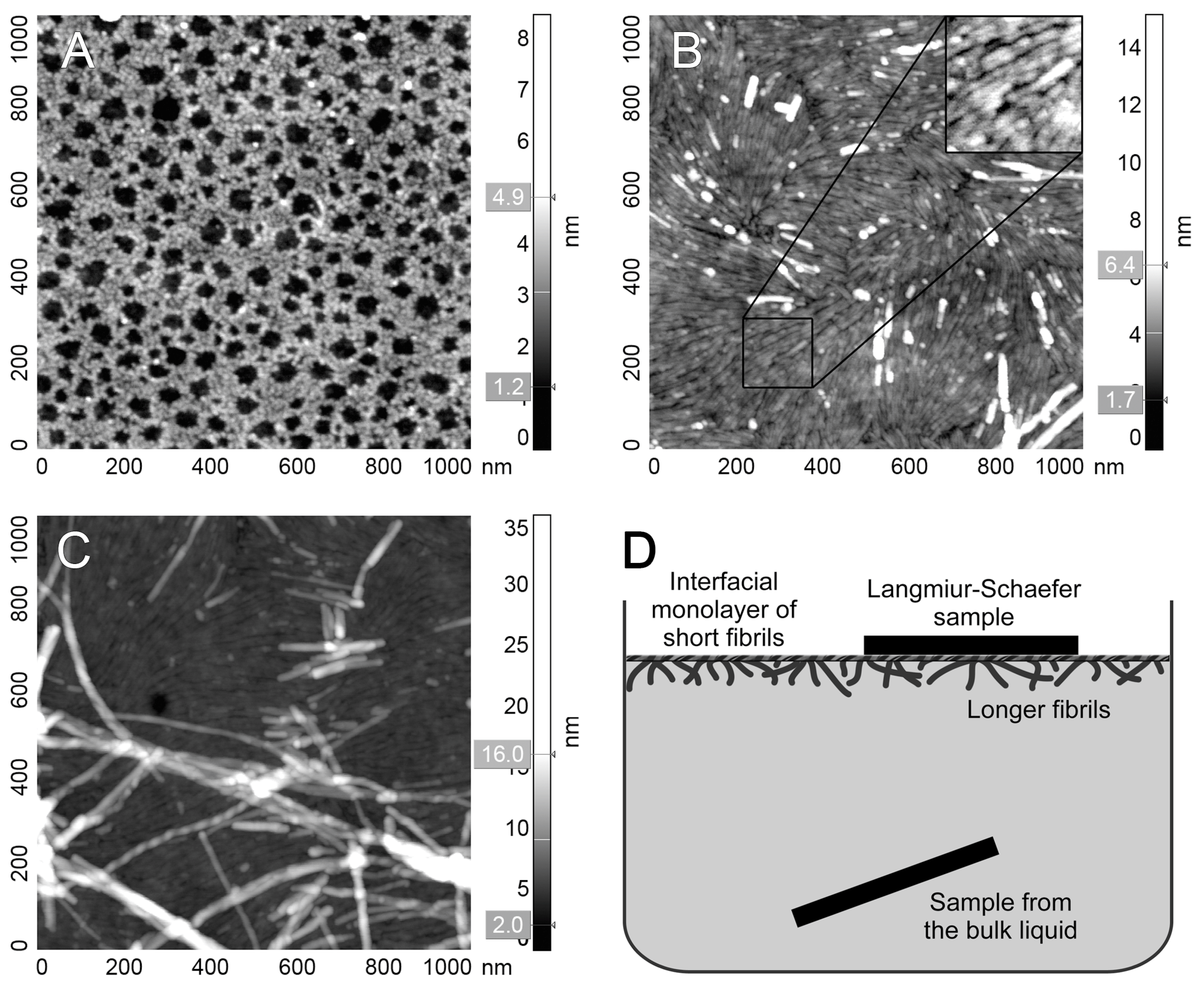
3.2. Atomic Force Microscopy
3.3. Yeast Strains and Cultivation Conditions
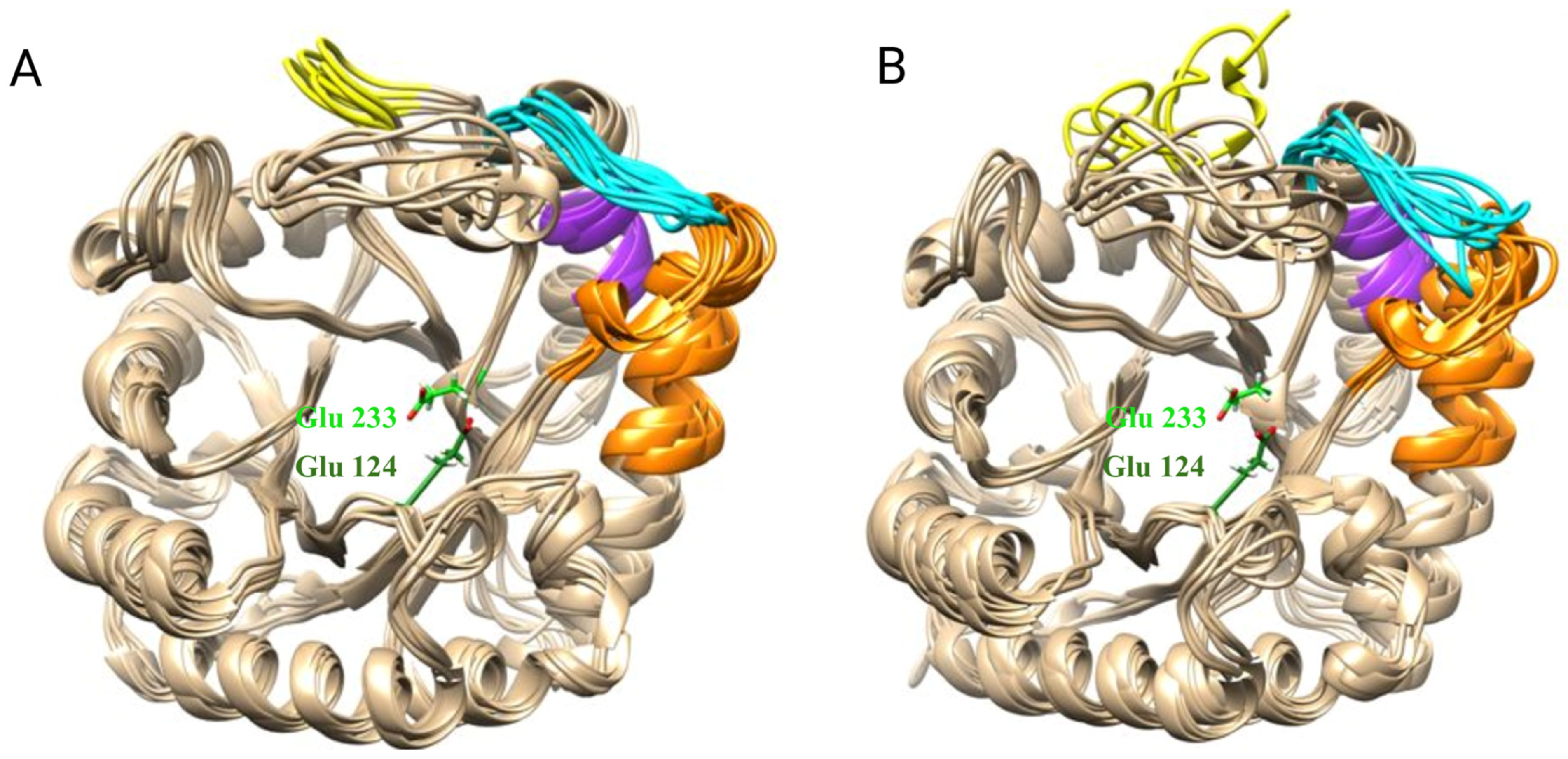
3.4. Construction of E233A Mutant Strain
3.5. Yeast Cell Wall Isolation and Protein Extraction
3.6. Electrophoresis, Western Blot Analysis
3.7. Sample Preparation for LC-MS/MS Analysis
4. Conclusions
- 1
- With the interaction of the active center of its molecule and substrate—glucan;
- 2
- With the interaction of β-turns located in the C-terminal region of the Bgl2p molecule and other cell wall proteins;
- 3
- By amyloid interactions among amino acid sequences localized in the C-terminal region of Bgl2p molecules one with another, possibly enhanced by β-turns and resulting in amyloid structure formation inside the cell wall.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tartaglia, G.G.; Caflisch, A. Computational analysis of the S. cerevisiae proteome reveals the function and cellular localization of the least and most amyloidogenic proteins. Proteins 2007, 68, 273–278. [Google Scholar] [CrossRef]
- Ho, V.; Herman-Bausier, P.; Shaw, C.; Conrad, K.A.; Garcia-Sherman, M.C.; Draghi, J.; Dufrene, Y.F.; Lipke, P.N.; Rauceo, J.M. An amyloid core sequence in the major Candida albicans adhesin Als1p mediates cell-cell adhesion. MBio 2019, 10, e01766-19. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Zhang, R.; Inokawa, N.; Oura, T.; Chen, X.; Iwatani, S.; Niimi, K.; Niimi, M.; Holmes, A.R.; Cannon, R.D.; et al. Candida albicans Bgl2p, Ecm33p, and Als1p proteins are involved in adhesion to saliva-coated hydroxyapatite. J. Oral Microbiol. 2021, 13, 1879497. [Google Scholar] [CrossRef]
- Mourer, T.; Ghalid, M.E.; Pehau-Arnaudet, G.; Kauffmann, B.; Loquet, A.; Brûlé, S.; Cabral, V.; d’Enfert, C.; Bachellier-Bassi, S. The Pga59 cell wall protein is an amyloid forming protein involved in adhesion and biofilm establishment in the pathogenic yeast Candida albicans. NPJ Biofilms Microbiomes 2023, 9, 6. [Google Scholar] [CrossRef]
- Klebl, F.; Tanner, W. Molecular cloning of a cell wall exo-beta-1,3-glucanase from Saccharomyces cerevisiae. J. Bacteriol. 1989, 171, 6259–6264. [Google Scholar] [CrossRef]
- Mrsa, V.; Klebl, F.; Tanner, W. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-beta-1,3-glucanase. J. Bacteriol. 1993, 175, 2102–2106. [Google Scholar] [CrossRef] [PubMed]
- Goldman, R.C.; Sullivan, P.A.; Zakula, D.; Capobianco, J.O. Kinetics of beta-1,3-glucan interaction at the donor and acceptor sites of the fungal glucosyltransferase encoded by the BGL2 gene. Eur. J. Biochem. 1995, 227, 372–378. [Google Scholar] [CrossRef]
- Sarthy, A.V.; McGonigal, T.; Coen, M.; Frost, D.J.; Meulbroek, J.A.; Goldman, R.C. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-β-glucosyltransferase. Microbiology 1997, 143, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Jeng, H.W.; Holmes, A.R.; Cannon, R.D. Characterization of two Candida albicans surface mannoprotein adhesins that bind immobilized saliva components. Med. Mycol. 2005, 43, 209–217. [Google Scholar] [CrossRef]
- Kalebina, T.S.; Plotnikova, T.A.; Gorkovskii, A.A.; Selyakh, I.O.; Galzitskaya, O.V.; Bezsonov, E.E.; Gellissen, G.; Kulaev, I.S. Amyloid–like properties of Saccharomyces cerevisiae cell wall glucantransferase Bgl2p. Prion 2008, 2, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Bezsonov, E.E.; Groenning, M.; Galzitskaya, O.V.; Gorkovskii, A.A.; Semisotnov, G.V.; Selyakh, I.O.; Ziganshin, R.H.; Rekstina, V.V.; Kudryashova, I.B.; Kuznetsov, S.A.; et al. Amyloidogenic peptides of yeast cell wall glucantransferase Bgl2p as a model for the investigation of its pH-dependent fibril formation. Prion 2013, 7, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Kalebina, T.S.; Rekstina, V.V.; Pogarskaia, E.E.; Kulakovskaya, T.V. Importance of Non-Covalent Interactions in Yeast Cell Wall Molecular Organization. Int. J. Mol. Sci. 2024, 25, 2496. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; Boorsma, A.; De Groot, P.W. Cell wall construction in Saccharomyces cerevisiae. Yeast 2006, 23, 185–202. [Google Scholar] [CrossRef]
- Chaffin, W.L. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 2008, 72, 495–544. [Google Scholar] [CrossRef]
- Millet, N.; Latgé, J.P.; Mouyna, I. Members of glycosyl-hydrolase family 17 of A. fumigatus differentially affect morphogenesis. J. Fungi 2018, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Teparic, R.; Lozancic, M.; Mrsa, V. Evolutionary overview of molecular interactions and enzymatic activities in the yeast cell walls. Int. J. Mol. Sci. 2020, 21, 8996. [Google Scholar] [CrossRef]
- Henrissat, B.; Davies, G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997, 7, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, T.A.; Selyakh, I.O.; Kalebina, T.S.; Kulaev, I.S. Bgl2p and Gas1p are the major glucantransferases forming the molecular ensemble of yeast cell wall. Dokl. Biochem. Biophys. 2006, 409, 244–247. [Google Scholar] [CrossRef]
- Sabirzyanov, F.A.; Sabirzyanova, T.A.; Rekstina, V.V.; Adzhubei, A.A.; Kalebina, T.S. C–terminal sequence is involved in the incorporation of bgl2p glucanosyltransglycosylase in the cell wall of Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, fox093. [Google Scholar] [CrossRef]
- Moreno-Garcia, J.; Mauricio, J.; Moreno, J.; Garcia-Martinez, T. Differential proteome analysis of a flor yeast strain under biofilm formation. Int. J. Mol. Sci. 2017, 18, 720. [Google Scholar] [CrossRef]
- Pitarch, A.; Jiménez, A.; Nombela, C.; Gil, C. Decoding serological response to Candida cell wall immunome into novel diagnostic, prognostic, and therapeutic candidates for systemic candidiasis by proteomic and bioinformatic analyses. Mol. Cell. Proteom. 2006, 5, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Gorkovskii, A.A.; Bezsonov, E.E.; Plotnikova, T.A.; Kalebina, T.S.; Kulaev, I.S. Revealing of Saccharomyces cerevisiae yeast cell wall proteins capable of binding thioflavin T, a fluorescent dye specifically interacting with amyloid fibrils. Biochemistry 2009, 74, 1219–1224. [Google Scholar] [CrossRef]
- Bezsonov, E.E.; Kalebina, T.S.; Gorkovskii, A.A.; Kudryashova, I.B.; Semisotnov, G.V.; Kulaev, I.S. Temperature–induced conformational transitions of the glucantransferase Bgl2p isolated from Saccharomyces cerevisiae cell walls. Mol. Biol. 2010, 44, 488–491. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.B.; Yagi, H.; Manno, M.; Martorana, V.; Ban, T.; Christiansen, G.; Otzen, D.E.; Goto, Y.; Rischel, C. Branching in amyloid fibril growth. Biophys. J. 2009, 96, 1529–1536. [Google Scholar] [CrossRef]
- Rekstina, V.V.; Sabirzyanova, T.A.; Sabirzyanov, F.A.; Adzhubei, A.A.; Tkachev, Y.V.; Kudryashova, I.B.; Snalina, N.E.; Bykova, A.A.; Alessenko, A.V.; Ziganshin, R.H.; et al. The post-translational modifications, localization, and mode of attachment of noncovalently bound glucanosyltransglycosylases of yeast cell wall as a key to understanding their functioning. Int. J. Mol. Sci. 2020, 21, 8304. [Google Scholar] [CrossRef] [PubMed]
- Kalebina, T.S.; Laurinavichiute, D.K.; Packeiser, A.N.; Morenkov, O.S.; Ter-Avanesyan, M.D.; Kulaev, I.S. Correct GPI-anchor synthesis is required for the incorporation of endoglucanase/glucanosyltransferase Bgl2p into the Saccharomyces cerevisiae cell wall. FEMS Microbiol. Lett. 2002, 210, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Wojtkowiak, A.; Witek, K.; Hennig, J.; Jaskolski, M. Structures of an active–site mutant of a plant 1,3-β-glucanase in complex with oligosaccharide products of hydrolysis. Acta Crystallogr. D Biol. Crystallogr. 2012, 69, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Avinery, R.; Kornreich, M.; Beck, R. Universal and accessible entropy estimation using a compression algorithm. Phys. Rev. Lett. 2019, 123, 178102. [Google Scholar] [CrossRef]
- Tsai, P.C.; Fox, N.; Bigley, A.N.; Harvey, S.P.; Barondeau, D.P.; Raushel, F.M. Enzymes for the homeland defense: Optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents. Biochemistry 2012, 51, 6463–6475. [Google Scholar] [CrossRef] [PubMed]
- Madej, T.; Lanczycki, C.J.; Zhang, D.; Thiessen, P.A.; Geer, R.C.; Marchler-Bauer, A.; Bryant, S.H. MMDB and VAST+: Tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2013, 42, D297–D303. [Google Scholar] [CrossRef]
- Qin, Z.; Yan, Q.; Lei, J.; Yang, S.; Jiang, Z.; Wu, S. The first crystal structure of a glycoside hydrolase family 17 β-1,3-glucanosyltransferase displays a unique catalytic cleft. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 1714–1724. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.N.; Garrett, T.P.; Colman, P.M.; Chen, L.; Hoj, P.B.; Fincher, G.B. Three-dimensional structures of two plant beta-glucan endohydrolases with distinct substrate specificities. Proc. Natl. Acad. Sci. USA 1994, 91, 2785–2789. [Google Scholar] [CrossRef] [PubMed]
- Receveur-Brechot, V.; Czjzek, M.; Barre, A.; Roussel, A.; Peumans, W.J.; Damme, E.J.W.; Rougé, P. Crystal structure at 1.45 Å resolution of the major allergen endo–β–1,3–glucanase of banana as a molecular basis for the latex–fruit syndrome. Proteins 2006, 63, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Romero, A.; Hernandez-Santoyo, A.; Fuentes-Silva, D.; Palomares, L.A.; Mũnoz-Cruz, S.; Yépez-Mulia, L.; Orozco-Martinez, S. Structural analysis of the endogenous glycoallergen Hev b 2 (endo–β–1,3–glucanase) from Hevea brasiliensis and its recognition by human basophils. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Louros, N.; Rousseau, F.; Schymkowitz, J. CORDAX web server: An online platform for the prediction and 3D visualization of aggregation motifs in protein sequences. Bioinformatics 2024, 40, btae279. [Google Scholar] [CrossRef]
- Paiz, E.A.; Allen, J.H.; Correia, J.J.; Fitzkee, N.C.; Hough, L.E.; Whitten, S.T. Beta turn propensity and a model polymer scaling exponent identify intrinsically disordered phase-separating proteins. J. Biol. Chem. 2021, 297, 101343. [Google Scholar] [CrossRef]
- Xing, Y.; Nandakumar, A.; Kakinen, A.; Sun, Y.; Davis, T.P.; Ke, P.C.; Ding, F. Amyloid Aggregation under the Lens of Liquid-Liquid Phase Separation. J. Phys. Chem. Lett. 2021, 12, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.A.; Hughes, M.P.; Hu, C.J.; Sawaya, M.R.; Salwinski, L.; Pan, H.; French, S.W.; Seidler, P.M.; Eisenberg, D.S. Identifying Amyloid-Related Diseases by Mapping Mutations in Low-Complexity Protein Domains to Pathologies. Nat. Struct. Mol. Biol. 2022, 29, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef]
- Glucan 1,3–beta–glucosidase. Alphafold Structure Prediction. Available online: https://alphafold.ebi.ac.uk/entry/P15703 (accessed on 9 September 2021).
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Pall, S.; Smith, J.C.; Hess, B.; Lindahl, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Crystal structure and pair potentials: A molecular-dynamics study. Phys. Rev. Lett. 1980, 45, 1196–1199. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Strain fluctuations and elastic constants. J. Chem. Phys. 1982, 76, 2662–2666. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Yao, D.; Yu, F.; Xu, B.; Zhou, R.; Bao, X.; Lu, Z. Immobilization of Antibodies on Ultraflat Polystyrene Surfaces. Clin. Chem. 2000, 46, 1456–1463. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H.; Willems, A.R.; Woods, R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 1995, 11, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef] [PubMed]


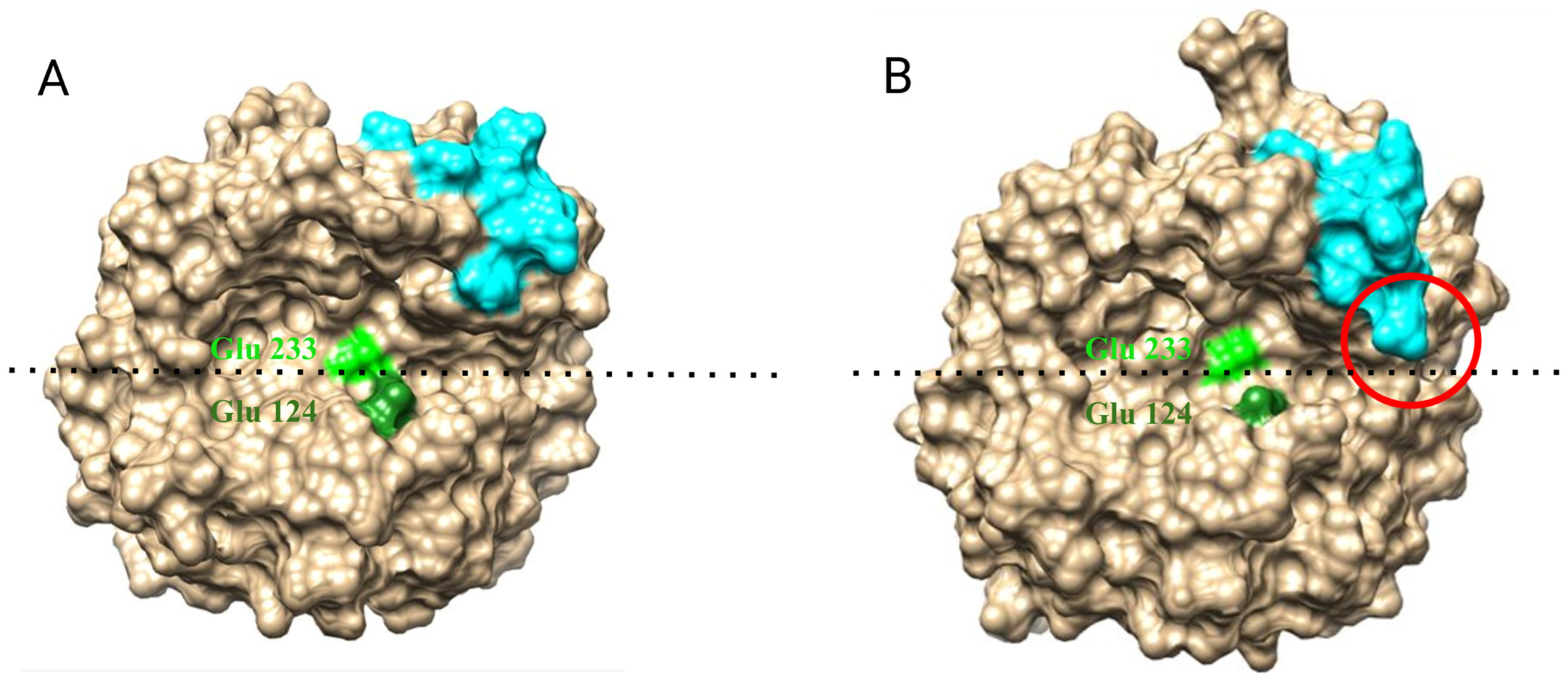
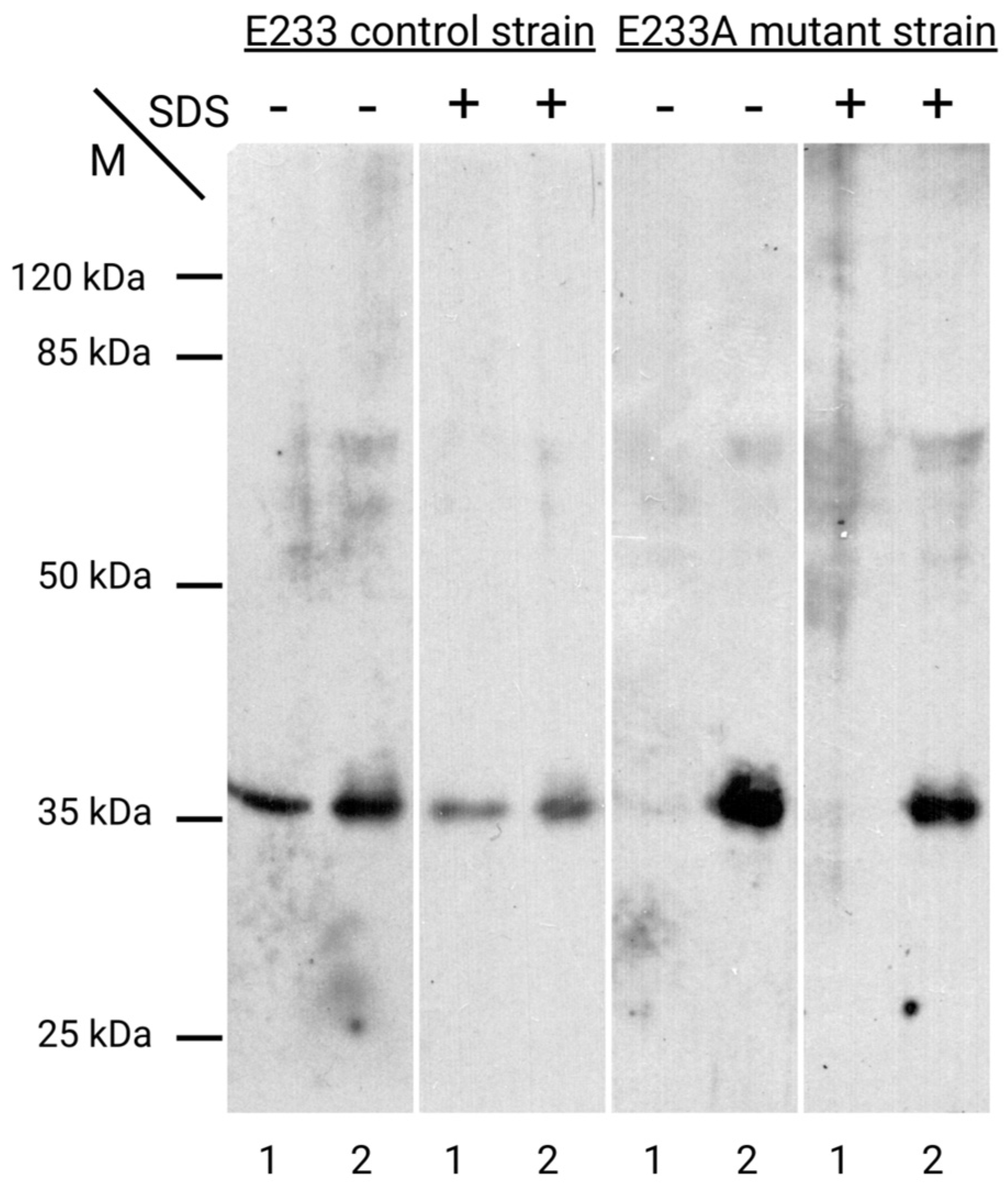
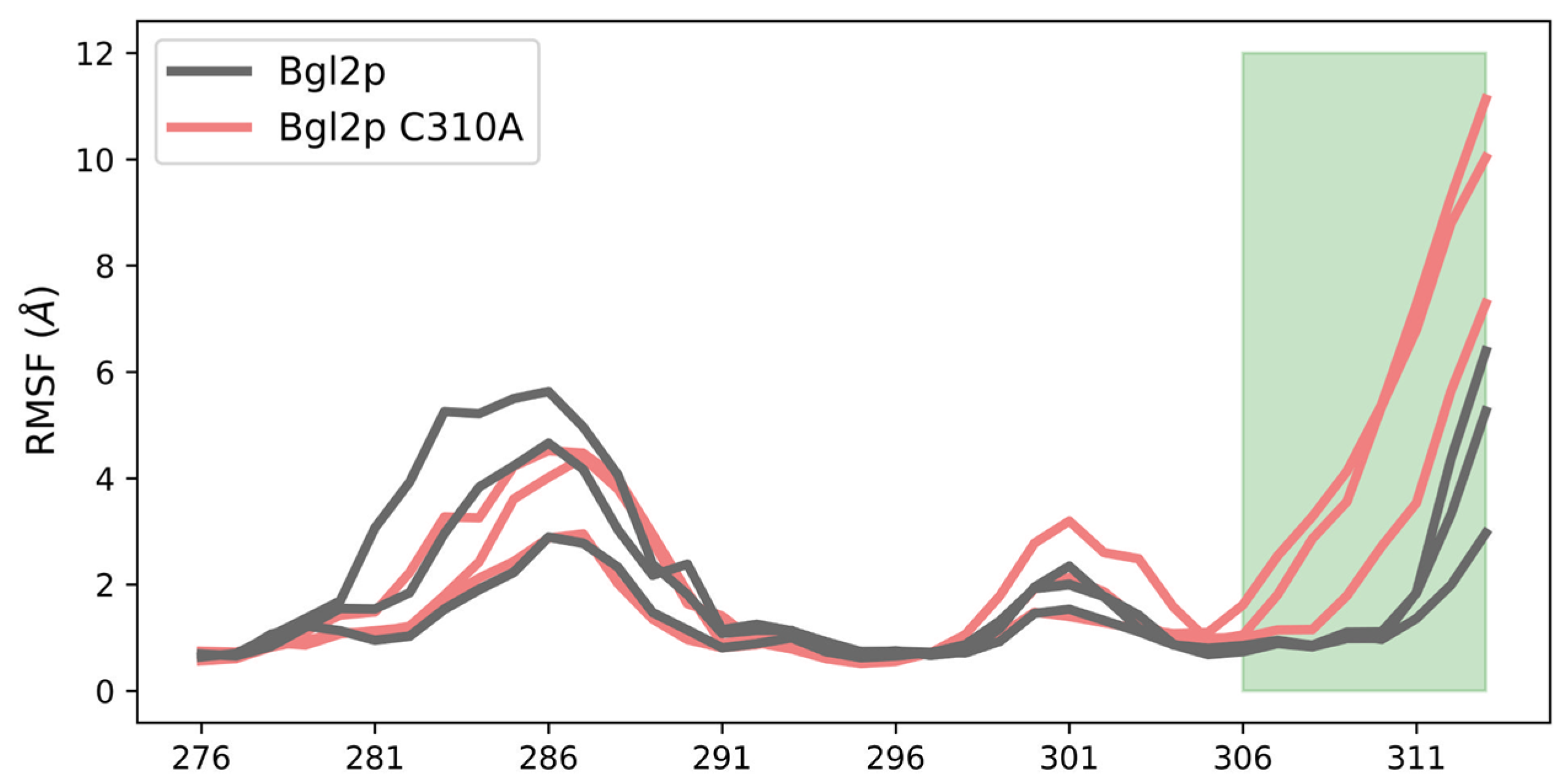
| PDB ID | Organism | Resolution, Å | Reference |
|---|---|---|---|
| 3URA | Brevundimonas diminuta | 1.88 | [30] |
| 6JMS | Cryptomeria japonica | 1.50 | [31] |
| 4WTP | Rhizomucor miehei CAU432 | 1.30 | [32] |
| 1GHS | Hordeum vulgare | 2.30 | [33] |
| 2CYG | Musa acuminata | 1.45 | [34] |
| 4IIS | Hevea brasiliensis | 2.67 | [35] |
| 4HPG | 2.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Motorin, N.A.; Makarov, G.I.; Rekstina, V.V.; Evtushenko, E.G.; Sabirzyanov, F.A.; Ziganshin, R.H.; Shaytan, A.K.; Kalebina, T.S. Yeast Glucan Remodeling Protein Bgl2p: Amyloid Properties and the Mode of Attachment in Cell Wall. Int. J. Mol. Sci. 2024, 25, 13703. https://doi.org/10.3390/ijms252413703
Motorin NA, Makarov GI, Rekstina VV, Evtushenko EG, Sabirzyanov FA, Ziganshin RH, Shaytan AK, Kalebina TS. Yeast Glucan Remodeling Protein Bgl2p: Amyloid Properties and the Mode of Attachment in Cell Wall. International Journal of Molecular Sciences. 2024; 25(24):13703. https://doi.org/10.3390/ijms252413703
Chicago/Turabian StyleMotorin, Nikita A., Gennady I. Makarov, Valentina V. Rekstina, Evgeniy G. Evtushenko, Fanis A. Sabirzyanov, Rustam H. Ziganshin, Alexey K. Shaytan, and Tatyana S. Kalebina. 2024. "Yeast Glucan Remodeling Protein Bgl2p: Amyloid Properties and the Mode of Attachment in Cell Wall" International Journal of Molecular Sciences 25, no. 24: 13703. https://doi.org/10.3390/ijms252413703
APA StyleMotorin, N. A., Makarov, G. I., Rekstina, V. V., Evtushenko, E. G., Sabirzyanov, F. A., Ziganshin, R. H., Shaytan, A. K., & Kalebina, T. S. (2024). Yeast Glucan Remodeling Protein Bgl2p: Amyloid Properties and the Mode of Attachment in Cell Wall. International Journal of Molecular Sciences, 25(24), 13703. https://doi.org/10.3390/ijms252413703






