Unlocking Prognostic Genes and Multi-Targeted Therapeutic Bioactives from Herbal Medicines to Combat Cancer-Associated Cachexia: A Transcriptomics and Network Pharmacology Approach
Abstract
1. Introduction
2. Results
2.1. Selection of Phytocompounds
2.2. Selection of Unique CAC Genes and Human Targets
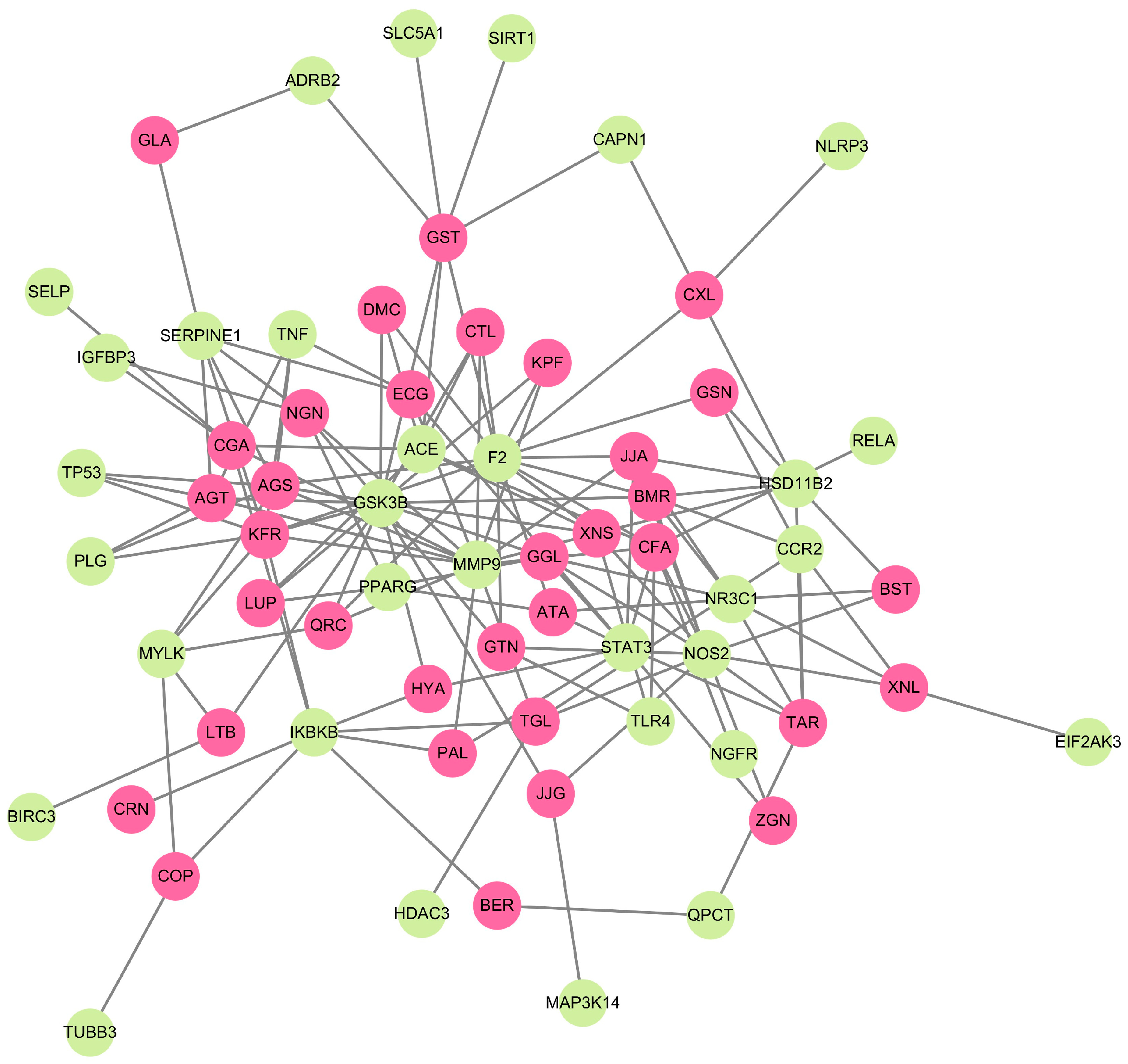
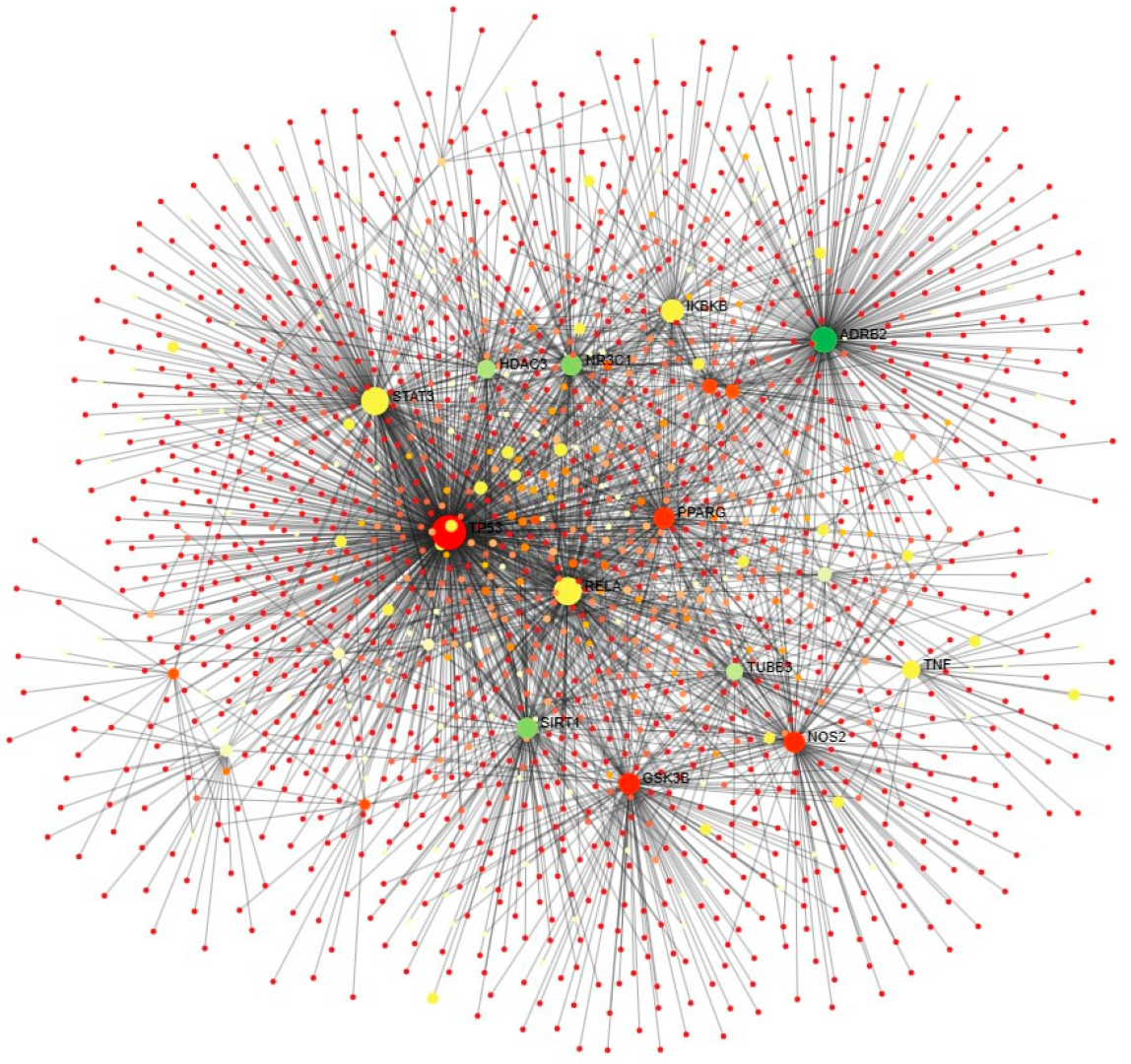
2.3. Molecular Network of Human Targets and Phytocompounds
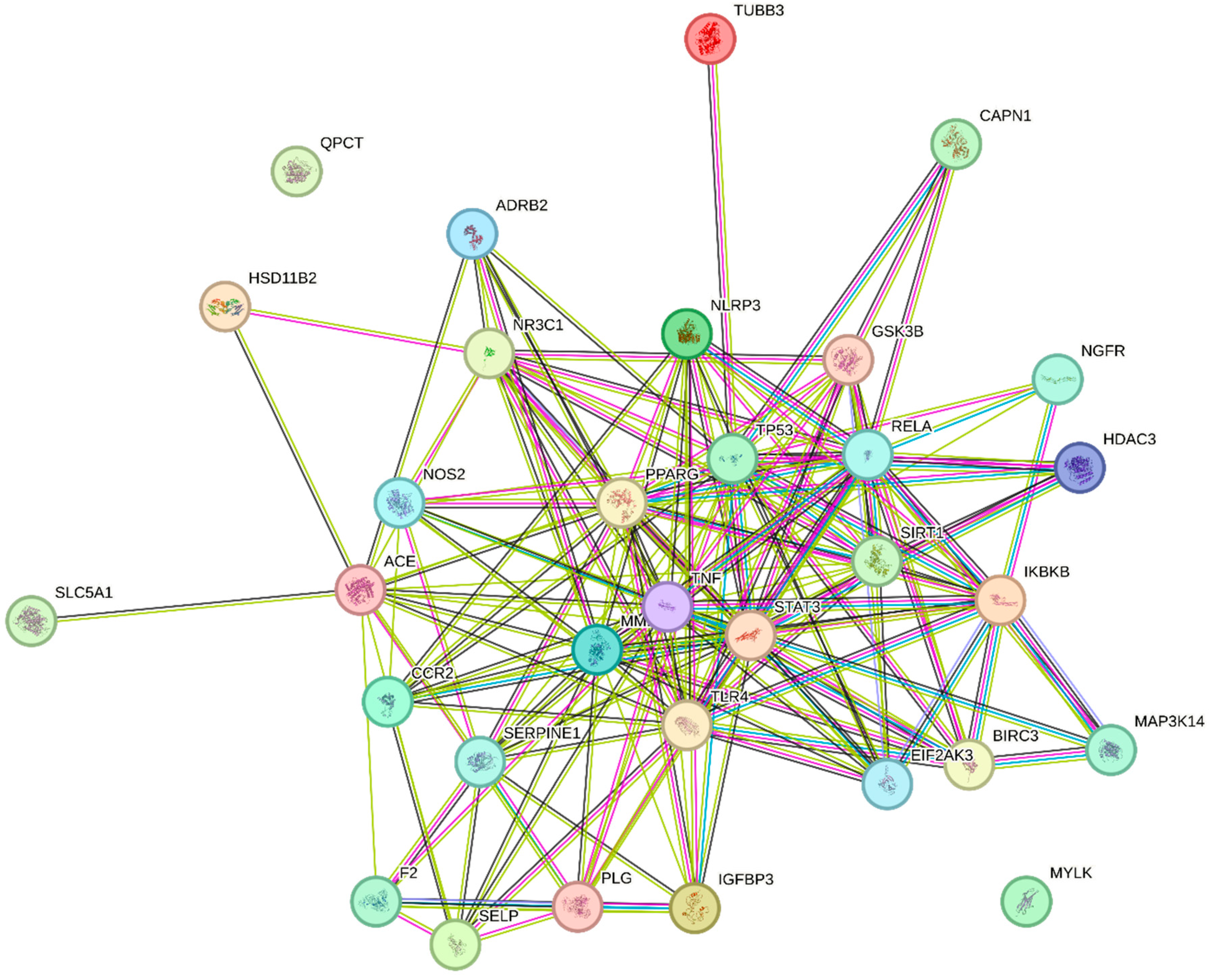
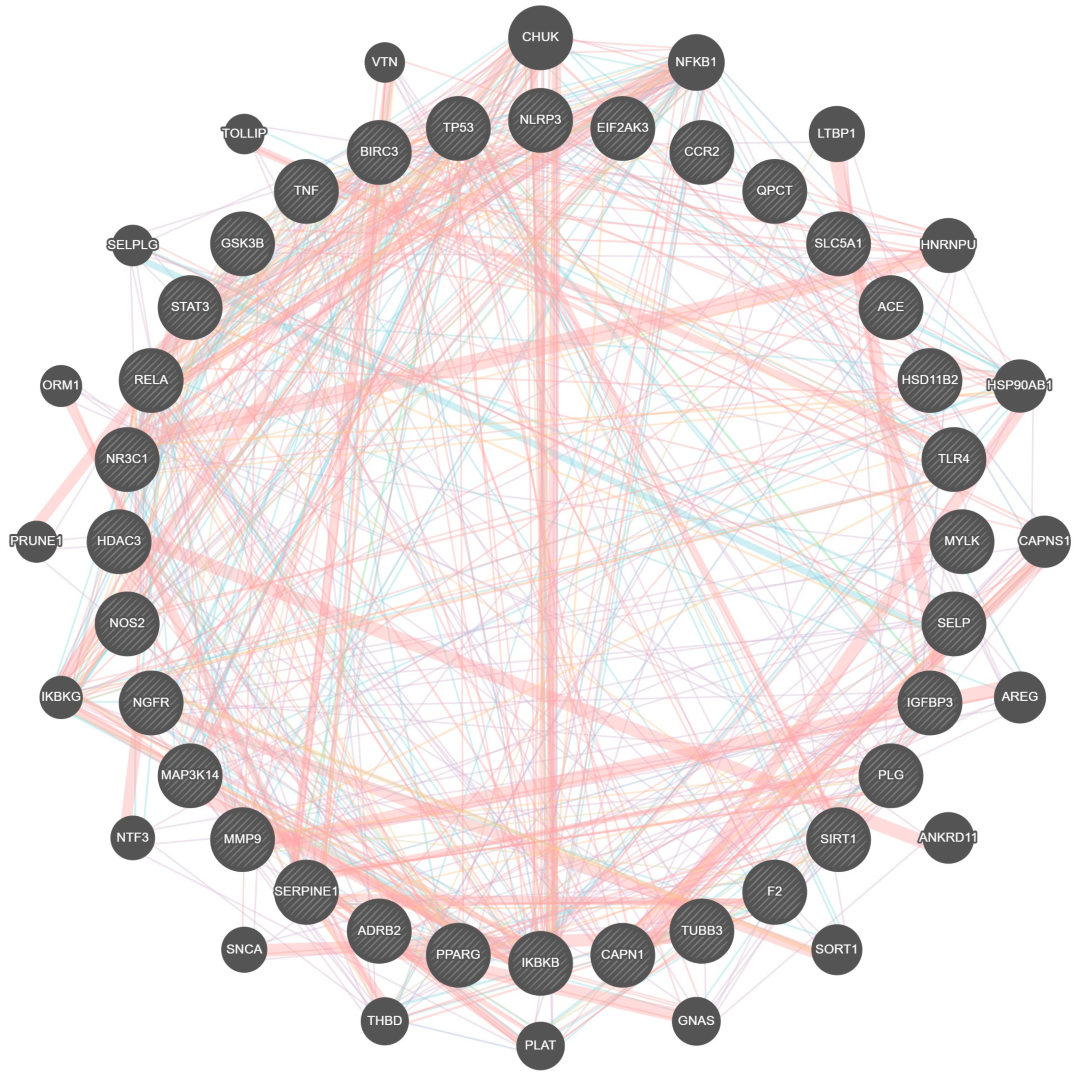
2.4. Protein Interaction and Pathway Analysis
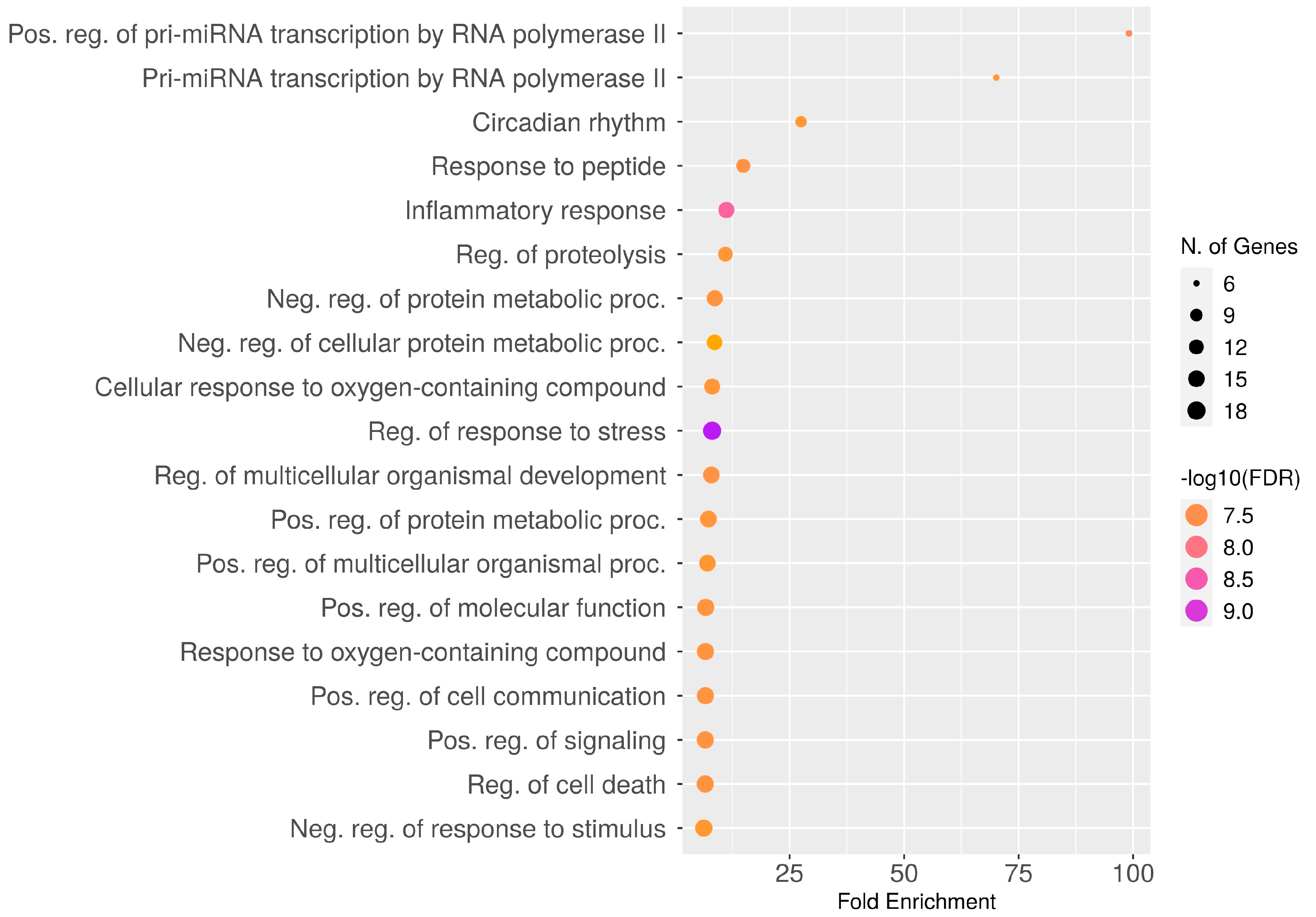
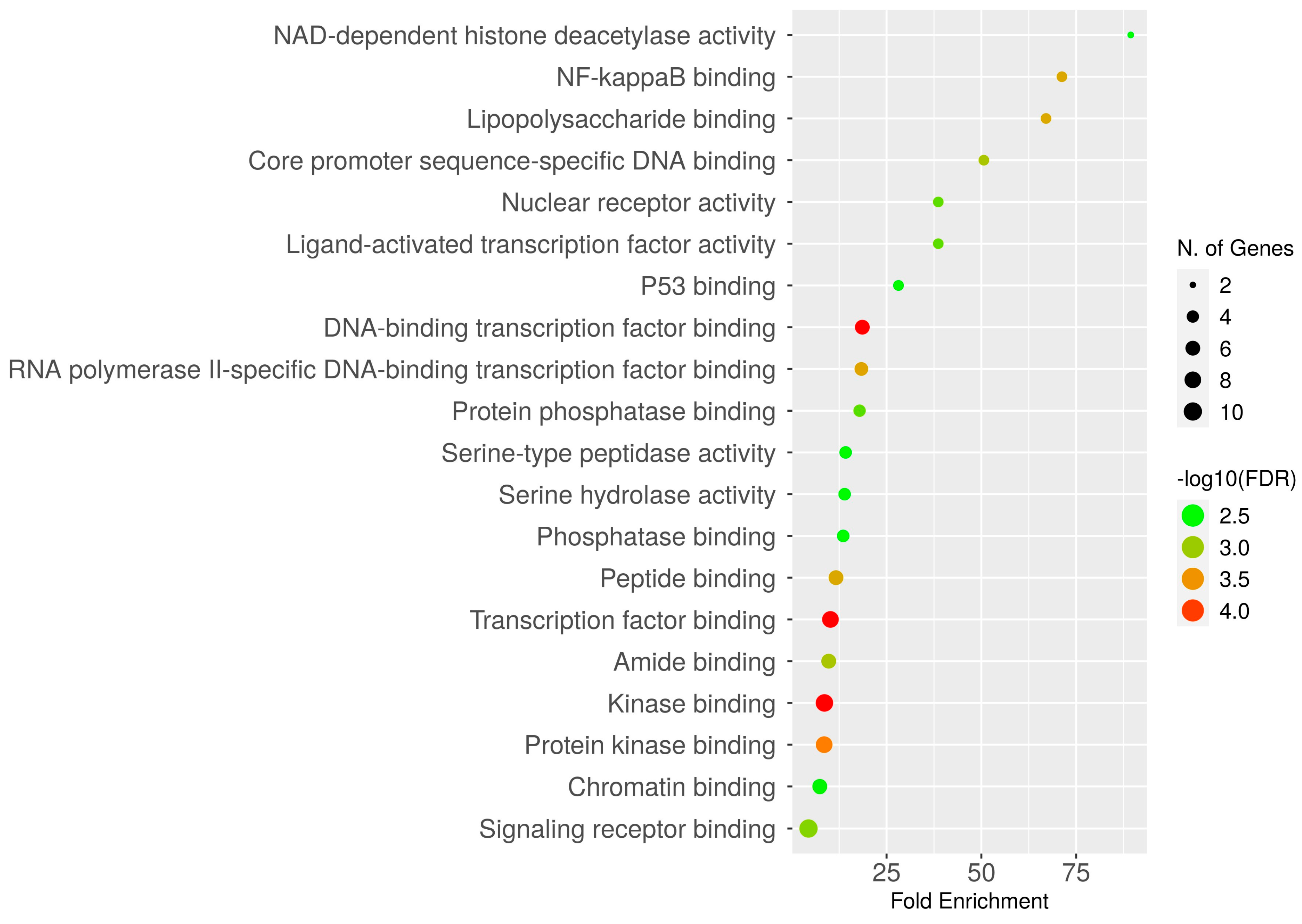
2.5. Gene Ontology Enrichment Analysis
2.6. Over-Representation Analysis (ORA)
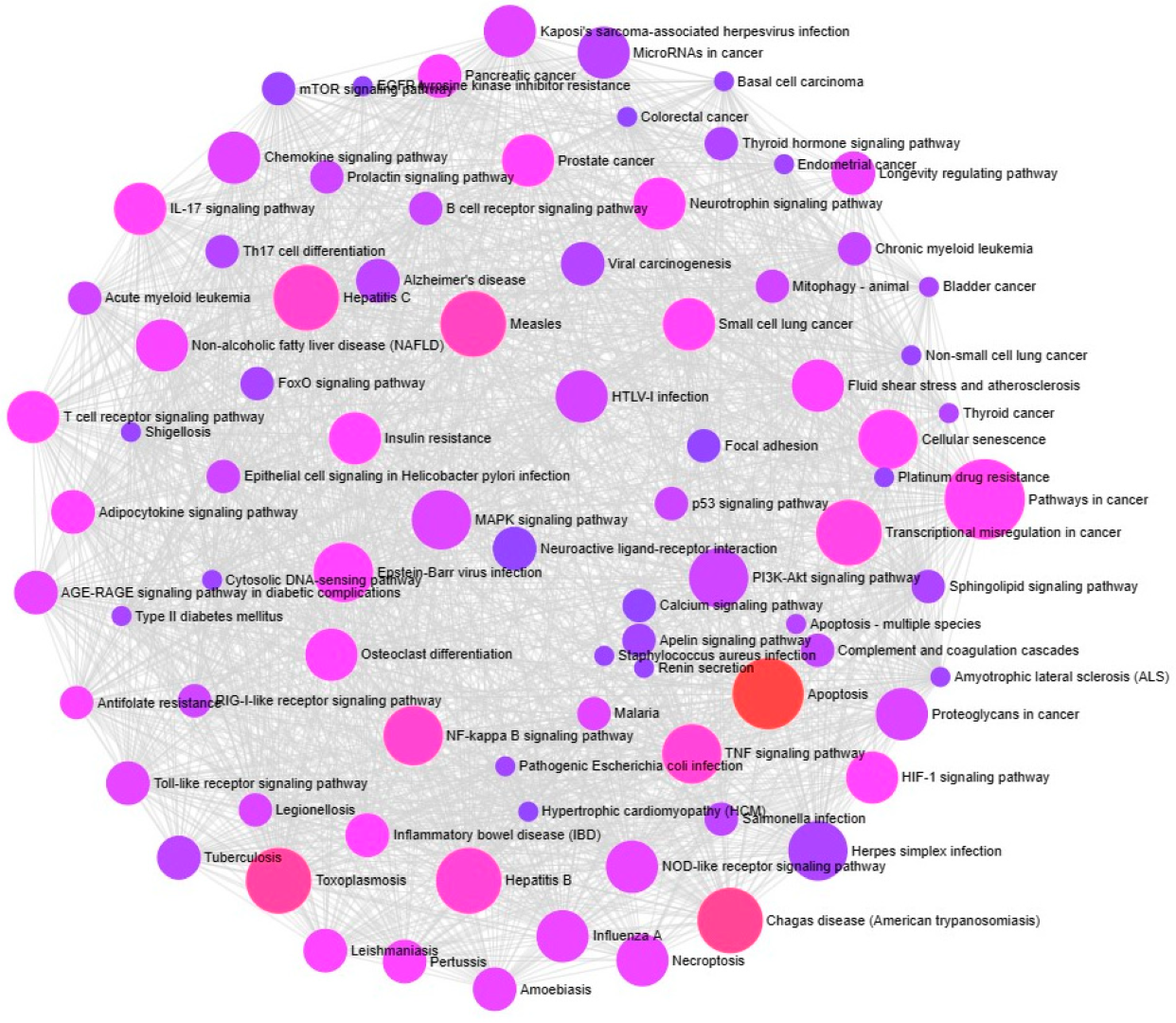
2.7. KEGG Pathway Enrichment Analysis
2.8. Unique Genes Survival Analysis
2.9. Pharmacological Properties of Phytocompounds
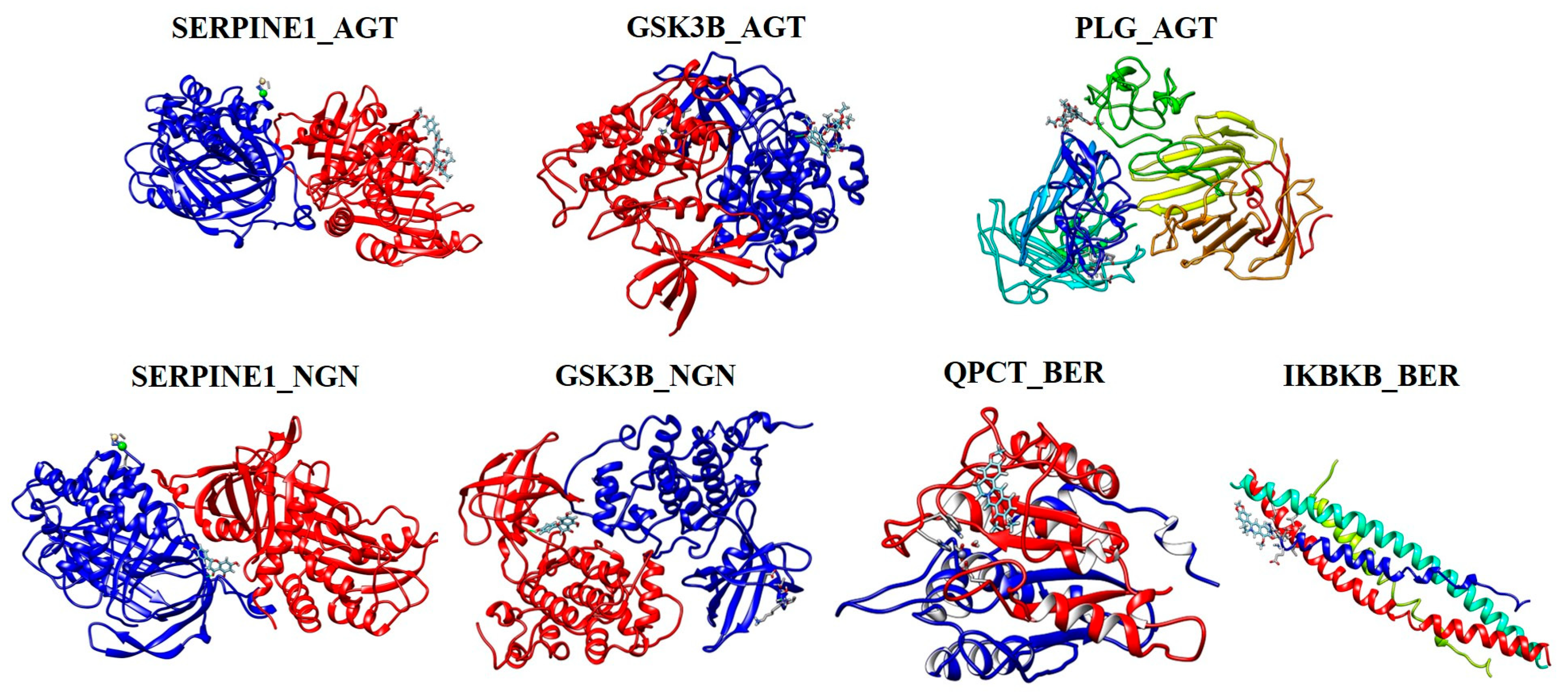
2.10. Molecular Docking Analysis
2.11. Effect of the Bioactive Compounds on 3T3-L1 Cell Viability
3. Discussion
4. Materials and Methods
4.1. Screening of Herbal Medicines and Phytocompounds
4.2. Retrieval of Phytocompounds and Pharmacological Properties
4.3. Checking of Human Targets and Curation of Unique Genes
4.4. Compound-Target Network (CTN) Analysis
4.5. Interactome and Gene Enrichment Analyses
4.6. Over-Representation Analysis (ORA)
4.7. KEGG Pathway Enrichment Analysis
4.8. Overall Survival Heat MAP Analysis
4.9. Molecular Docking
4.10. Cell Culture and Cachectic Condition Media
4.11. Cell Viability Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fearon, K.C.; Glass, D.J.; Guttridge, D.C. Cancer cachexia: Mediators, signaling, and metabolic pathways. Cell Metab. 2012, 16, 153–166. [Google Scholar] [CrossRef] [PubMed]
- van der Ende, M.; Plas, R.L.; van Dijk, M.; Dwarkasing, J.T.; van Gemerden, F.; Sarokhani, A.; Swarts, H.J.M.; van Schothorst, E.M.; Grefte, S.; Witkamp, R.F.; et al. Effects of whole-body vibration training in a cachectic C26 mouse model. Sci. Rep. 2021, 11, 21563. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Li, L.; Su, Z.; Wei, L.; Pu, W.; Zhao, C.; Ding, Y.; Wazir, J.; Cao, W.; Wang, H.; et al. An integrative transcriptome study reveals Ddit4/Redd1 as a key regulator of cancer cachexia in rodent models. Cell Death Dis. 2021, 12, 652. [Google Scholar] [CrossRef] [PubMed]
- Muthamil, S.; Kim, H.Y.; Jang, H.J.; Lyu, J.H.; Shin, U.C.; Go, Y.; Park, S.-H.; Lee, H.G.; Park, J.H. Understanding the relationship between cancer associated cachexia and hypoxia-inducible factor-1. Biomed. Pharmacother. 2023, 163, 114802. [Google Scholar] [CrossRef] [PubMed]
- Argilés, J.M.; López-Soriano, F.J.; Stemmler, B.; Busquets, S. Cancer-associated cachexia—Understanding the tumour macroenvironment and microenvironment to improve management. Nat. Rev. Clin. Oncol. 2023, 20, 250–264. [Google Scholar] [CrossRef] [PubMed]
- von Haehling, S.; Anker, S.D. Prevalence, incidence and clinical impact of cachexia: Facts and numbers—Update 2014. J. Cachexia Sarcopenia Muscle 2014, 5, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Baracos, V.E.; Martin, L.; Korc, M.; Guttridge, D.C.; Fearon, K.C. Cancer-associated cachexia. Nat. Rev. Dis. Primers 2018, 4, 1–18. [Google Scholar] [CrossRef]
- Ni, J.; Zhang, L. Cancer cachexia: Definition, staging, and emerging treatments. Cancer Manag. Res. 2020, 12, 5597–5605. [Google Scholar] [CrossRef]
- Wakabayashi, H.; Arai, H.; Inui, A. The regulatory approval of anamorelin for treatment of cachexia in patients with non-small cell lung cancer, gastric cancer, pancreatic cancer, and colorectal cancer in Japan: Facts and numbers. J. Cachexia Sarcopenia Muscle 2021, 12, 14–16. [Google Scholar] [CrossRef]
- Adarshan, S.; Akassh, S.; Avinash, K.; Bharathkumar, M.; Muthuramalingam, P.; Shin, H.; Baskar, V.; Chen, J.-T.; Bhuvaneshwari, V.; Ramesh, M. Transcriptomics, cheminformatics, and systems pharmacology strategies unveil the potential bioactives to combat COVID-19. Molecules 2022, 27, 5955. [Google Scholar] [CrossRef]
- Muthamil, S.; Prasath, K.G.; Priya, A.; Precilla, P.; Pandian, S.K. Global proteomic analysis deciphers the mechanism of action of plant derived oleic acid against Candida albicans virulence and biofilm formation. Sci. Rep. 2020, 10, 5113. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Gouvinhas, I.; Rocha, J.; Barros, A.I. Phytochemical and antioxidant analysis of medicinal and food plants towards bioactive food and pharmaceutical resources. Sci. Rep. 2021, 11, 10041. [Google Scholar] [CrossRef] [PubMed]
- Oelkrug, C.; Lange, C.M.; Wenzel, E.; Fricke, S.; Hartke, M.; Simasi, J.; Schubert, A. Analysis of the tumoricidal and anti-cachectic potential of curcumin. Anticancer Res. 2014, 34, 4781–4788. [Google Scholar] [PubMed]
- Sohn, S.I.; Priya, A.; Balasubramaniam, B.; Muthuramalingam, P.; Sivasankar, C.; Selvaraj, A.; Valliammai, A.; Jothi, R.; Pandian, S. Biomedical applications and bioavailability of curcumin—An updated overview. Pharmaceutics 2021, 13, 2102. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wei, L.; Wazir, J.; Li, L.; Song, S.; Lin, K.; Pu, W.; Zhao, C.; Su, Z.; Wang, H.; et al. Curcumin treatment suppresses cachexia-associated adipose wasting in mice by blocking the cAMP/PKA/CREB signaling pathway. Phytomedicine 2023, 109, 154563. [Google Scholar] [CrossRef] [PubMed]
- Kuchta, K.; Cameron, S. Phytotherapy for cachexia: Where do we stand? Front. Pharmacol. 2020, 11, 917. [Google Scholar] [CrossRef]
- Li, W.; Swiderski, K.; Murphy, K.T.; Lynch, G.S. Role for plant-derived antioxidants in attenuating cancer cachexia. Antioxidants 2022, 11, 183. [Google Scholar] [CrossRef]
- Kim, C.; Hwang, J.K. Flavonoids: Nutraceutical potential for counteracting muscle atrophy. Food Sci. Biotechnol. 2020, 29, 1619–1640. [Google Scholar] [CrossRef]
- Abusaliya, A.; Ha, S.E.; Bhosale, P.B.; Kim, H.H.; Park, M.Y.; Vetrivel, P.; Kim, G.S. Glycosidic flavonoids and their potential applications in cancer research: A review. Mol. Cell. Toxicol. 2022, 18, 9–16. [Google Scholar] [CrossRef]
- Prado, B.L.; Qian, Y. Anti-cytokines in the treatment of cancer cachexia. Ann. Palliat. Med. 2019, 8, 67–79. [Google Scholar] [CrossRef]
- Yaghobi, N.; Mehrzad, V.; Badri, S.; Yegdaneh, A.; Moghaddas, A. Combination of traditional herbal medicine for the treatment of cancer-induced Anorexia/Cachexia: A pilot, randomized, double-blinded and placebo-controlled clinical trial. J. Herb. Med. 2021, 29, 100499. [Google Scholar] [CrossRef]
- Lee, C.; Kang, M.J.; Kim, S.; Han, I.H.; Bae, H. Screening of phytochemicals effective on relieving cancer cachexia in cisplatin-induced in vitro sarcopenia model. Mol. Cell. Toxicol. 2022, 18, 111–120. [Google Scholar] [CrossRef]
- Wang, X.; Hu, Y.; Zhou, X.; Li, S. Network pharmacology and traditional medicine: Setting the new standards by combining in silico and experimental work. Front. Pharmacol. 2022, 13, 1002537. [Google Scholar] [CrossRef]
- Noor, F.; Tahir ul Qamar, M.; Ashfaq, U.A.; Albutti, A.; Alwashmi, A.S.; Aljasir, M.A. Network pharmacology approach for medicinal plants: Review and assessment. Pharmaceuticals 2022, 15, 572. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.H.; Wu, F.; Cao, W.Y.; Wu, Z.G.; Chao, Y.C.; Peng, F.; Liang, C. Network pharmacology for the identification of phytochemicals in traditional Chinese medicine for COVID-19 that may regulate interleukin-6. Biosci. Rep. 2021, 41, BSR20202583. [Google Scholar] [CrossRef] [PubMed]
- Muthuramalingam, P.; Akassh, S.; Rithiga, S.B.; Prithika, S.; Gunasekaran, R.; Shin, H.; Kumar, R.; Baskar, V.; Kim, J. Integrated omics profiling and network pharmacology uncovers the prognostic genes and multi-targeted therapeutic bioactives to combat lung cancer. Eur. J. Pharmacol. 2023, 940, 175479. [Google Scholar] [CrossRef] [PubMed]
- Muthuramalingam, P.; Jeyasri, R.; Valliammai, A.; Selvaraj, A.; Karthika, C.; Gowrishankar, S.; Pandian, S.K.; Ramesh, M.; Chen, J.T. Global multi-omics and systems pharmacological strategy unravel the multi-targeted therapeutic potential of natural bioactive molecules against COVID-19: An in silico approach. Genomics 2020, 112, 4486–4504. [Google Scholar] [CrossRef]
- Yuan, C.; Wang, M.H.; Wang, F.; Chen, P.Y.; Ke, X.G.; Yu, B.; Yang, Y.-F.; You, P.-T.; Wu, H.Z. Network pharmacology and molecular docking reveal the mechanism of Scopoletin against non-small cell lung cancer. Life Sci. 2021, 270, 119105. [Google Scholar] [CrossRef]
- Aarthy, M.; Muthuramalingam, P.; Ramesh, M.; Singh, S.K. Unraveling the multi-targeted curative potential of bioactive molecules against cervical cancer through integrated omics and systems pharmacology approach. Sci. Rep. 2022, 12, 14245. [Google Scholar] [CrossRef]
- Muthuramalingam, P.; Govindasamy, R.; Venkidasamy, B.; Krishnan, M.; Shin, H. Network pharmacology: A systems perspective possible underpinning approach for oral cancer treatment. DARU J. Pharm. Sci. 2023, 31, 273–275. [Google Scholar] [CrossRef]
- Liu, X.G.; Lv, M.C.; Huang, M.Y.; Sun, Y.Q.; Gao, P.Y.; Li, D.Q. A network pharmacology study on the triterpene saponins from medicago sativa l. For the treatment of neurodegenerative diseases. J. Food Biochem. 2019, 43, e12955. [Google Scholar] [CrossRef] [PubMed]
- Jeyasri, R.; Muthuramalingam, P.; Suba, V.; Ramesh, M.; Chen, J.T. Bacopa monnieri and their bioactive compounds inferred multi-target treatment strategy for neurological diseases: A cheminformatics and system pharmacology approach. Biomolecules 2020, 10, 536. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ren, R.; Zhang, Q.; Wu, J.; Zhang, Y.; Xue, M.; Yin, D.; Yang, Y. Coptis chinensis Franch polysaccharides provide a dynamically regulation on intestinal microenvironment, based on the intestinal flora and mucosal immunity. J. Ethnopharmacol. 2021, 267, 113542. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Huo, J.; Wang, T.; Sun, G.; Wang, W. Chemical profiling of Coptis rootlet and screening of its bioactive compounds in inhibiting Staphylococcus aureus by UPLC-Q-TOF/MS. J. Pharm. Biomed. Anal. 2020, 180, 113089. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, L.; Lou, G.H.; Zeng, H.R.; Hu, J.; Huang, Q.W.; Peng, W.; Yang, X.B. Coptidis Rhizoma: A comprehensive review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. Pharm. Biol. 2019, 57, 193–225. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Su, B. Chemical Constituents of Coptis chinensis. Chem. Nat. Compd. 2022, 58, 1131–1133. [Google Scholar] [CrossRef]
- Jeong, D.; Dong, G.Z.; Lee, H.J.; Ryu, J.H. Anti-inflammatory compounds from Atractylodes macrocephala. Molecules 2019, 24, 1859. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, C.; Yin, L.; Si, J.; Yu, M.; Li, J.; Li, L.; Zhang, T.; Zou, Z. Bioactive constituents from the rhizomes of Atractylodes macrocephala. Fitoterapia 2023, 165, 105431. [Google Scholar] [CrossRef]
- Ratan, Z.A.; Haidere, M.F.; Hong, Y.H.; Park, S.H.; Lee, J.O.; Lee, J.; Cho, J.Y. Pharmacological potential of ginseng and its major component ginsenosides. J. Ginseng Res. 2021, 45, 199–210. [Google Scholar] [CrossRef]
- Piao, X.; Zhang, H.; Kang, J.P.; Yang, D.U.; Li, Y.; Pang, S.; Jin, Y.; Yang, D.C.; Wang, Y. Advances in saponin diversity of Panax ginseng. Molecules 2020, 25, 3452. [Google Scholar] [CrossRef]
- Li, D.; Yue, D.; Liu, D.; Zhang, L.; Song, S. Phytochemical and chemotaxonomic study on Ziziphus jujuba Mill. (Rhamnaceae). Biochem. Syst. Ecol. 2020, 91, 104058. [Google Scholar] [CrossRef]
- Abdoul-Azize, S. Potential benefits of jujube (Zizyphus Lotus L.) bioactive compounds for nutrition and health. J. Nutr. Metab. 2016, 2016, 2867470. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M.; Ali, A.; Ali, M.; Mir, S.R. Chromatographic isolation and spectroscopic identification of phytoconstituents of jujuba seeds (Zizyphus jujuba Mill.). J. Pharm. Bioallied Sci. 2017, 9, 26. [Google Scholar]
- Devaraj, R.D.; Jeepipalli, S.P.; Xu, B. Phytochemistry and health promoting effects of Job’s tears (Coix lacryma-jobi)—A critical review. Food Biosci. 2020, 34, 100537. [Google Scholar] [CrossRef]
- Diningrat, D.S.; Risfandi, M.; Harahap, N.S.; Sari, A.N. Phytochemical screening and antibacterial activity Coix lacryma-jobi oil. J. Plant Biotechnol. 2020, 47, 100–106. [Google Scholar] [CrossRef]
- Salam, S.G.A.; Rashed, M.M.; Ibrahim, N.A.; Rahim, E.A.A.; Aly, T.A.; Al-Farga, A. Phytochemical screening and in-vitro biological properties of unprocessed and household processed fenugreek (Trigonella foenum-graecum Linn.) seeds and leaves. Sci. Rep. 2023, 13, 7032. [Google Scholar] [CrossRef]
- Singh, N.; Yadav, S.S.; Kumar, S.; Narashiman, B. Ethnopharmacological, phytochemical and clinical studies on Fenugreek (Trigonella foenum-graecum L.). Food Biosci. 2022, 46, 101546. [Google Scholar] [CrossRef]
- Wani, S.A.; Kumar, P. Fenugreek: A review on its nutraceutical properties and utilization in various food products. J. Saudi Soc. Agric. Sci. 2018, 17, 97–106. [Google Scholar] [CrossRef]
- Zilani, M.N.H.; Sultana, T.; Asabur Rahman, S.M.; Anisuzzman, M.; Islam, M.A.; Shilpi, J.A.; Hossain, M.G. Chemical composition and pharmacological activities of Pisum sativum. BMC Complement. Altern. Med. 2017, 17, 1–9. [Google Scholar] [CrossRef]
- Castaldo, L.; Izzo, L.; Gaspari, A.; Lombardi, S.; Rodríguez-Carrasco, Y.; Narváez, A.; Grosso, M.; Ritieni, A. Chemical Composition of green pea (Pisum sativum L.) pods extracts and their potential exploitation as ingredients in nutraceutical formulations. Antioxidants 2021, 11, 105. [Google Scholar] [CrossRef]
- Mukjerjee, S.; Karati, D. A mechanistic view on phytochemistry, pharmacognostic properties, and pharmacological activities of phytocompounds present in zingiber officinale: A comprehensive review. Pharmacol. Res. Mod. Chin. Med. 2022, 5, 100173. [Google Scholar] [CrossRef]
- Mao, Q.Q.; Xu, X.Y.; Cao, S.Y.; Gan, R.Y.; Corke, H.; Beta, T.; Li, H.B. Bioactive compounds and bioactivities of ginger (Zingiber officinale Roscoe). Foods 2019, 8, 185. [Google Scholar] [CrossRef] [PubMed]
- Nigam, M.; Atanassova, M.; Mishra, A.P.; Pezzani, R.; Devkota, H.P.; Plygun, S.; Salehi, B.; Setzer, W.N.; Sharifi-Rad, J. Bioactive compounds and health benefits of Artemisia species. Nat. Prod. Commun. 2019, 14, 1–17. [Google Scholar]
- Ganie, S.A.; Gulla, S.S.; Yadav, S.S. Xanthium strumerium L.: An ethnomedicinal and phytochemical review. Int. J. Phytomedicine 2014, 6, 471–476. [Google Scholar]
- Sahoo, R.K.; Tamuli, K.J.; Lhouvum, N.; Dutta, D.; Bordoloi, M.; Sharma, H.K.; Gogoi, K.; Bhattacharyya, D.R. Phytochemical constituents from Xanthium strumarium L. and evaluation of their in vitro antimalarial activities. S. Afr. J. Bot. 2020, 135, 35–40. [Google Scholar] [CrossRef]
- Hao, D.C.; Song, Y.; Xiao, P.; Zhong, Y.; Wu, P.; Xu, L. The genus Chrysanthemum: Phylogeny, biodiversity, phytometabolites, and chemodiversity. Front. Plant Sci. 2022, 13, 973197. [Google Scholar] [CrossRef] [PubMed]
- Lal, M.; Chandraker, S.K.; Shukla, R. Antimicrobial Properties of Selected Plants Used in Traditional Chinese Medicine. In Functional and Preservative Properties of Phytochemicals; Academic Press: Cambridge, MA, USA, 2020; pp. 119–143. [Google Scholar]
- Zhang, L.L.; Tian, K.; Tang, Z.H.; Chen, X.J.; Bian, Z.X.; Wang, Y.T.; Lu, J.J. Phytochemistry and Pharmacology of Carthamus Tinctorius L. Am. J. Chin. Med. 2016, 44, 197–226. [Google Scholar] [CrossRef]
- Poisson, J.; Martinez-Tapia, C.; Heitz, D.; Geiss, R.; Albrand, G.; Falandry, C.; Gisselbrecht, M.; Couderc, A.; Boulahssass, R.; Paillaud, E.; et al. Prevalence and prognostic impact of cachexia among older patients with cancer: A nationwide cross-sectional survey (NutriAgeCancer). J. Cachexia Sarcopenia Muscle 2021, 12, 1477–1488. [Google Scholar] [CrossRef]
- Prigerson, H.G.; Bao, Y.; Shah, M.A.; Paulk, M.E.; LeBlanc, T.W.; Schneider, B.J.; Garrido, M.M.; Reid, C.; Berlin, D.A.; Maciejewski, P.K.; et al. Chemotherapy use, performance status, and quality of life at the end of life. JAMA Oncol. 2015, 1, 778–784. [Google Scholar] [CrossRef]
- Arends, J.; Muscaritoli, M.; Anker, S.; Audisio, R.; Barazzoni, R.; Bosnjak, S.; Bossi, P.; Bowman, J.; Gijssels, S.; Aapro, M.; et al. Overcoming barriers to timely recognition and treatment of cancer cachexia: Sharing Progress in Cancer Care Task Force Position Paper and Call to Action. Crit. Rev. Oncol. Hematol. 2023, 185, 103965. [Google Scholar] [CrossRef]
- Segal, R.; Zwaal, C.; Green, E.; Tomasone, J.R.; Loblaw, A.; Petrella, T. Exercise for people with cancer: A clinical practice guideline. Curr. Oncol. 2017, 24, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.C.; Cook, J.; Maddocks, M.; Skipworth, R.J.; Fallon, M.; Laird, B.J. Combined exercise and nutritional rehabilitation in outpatients with incurable cancer: A systematic review. Support. Care Cancer 2019, 27, 2371–2384. [Google Scholar] [CrossRef] [PubMed]
- Arends, J.; Strasser, F.; Gonella, S.; Solheim, T.S.; Madeddu, C.; Ravasco, P.; Buonaccorso, L.; de van der Schueren, M.; Baldwin, C.; Ripamonti, C.I.; et al. Cancer cachexia in adult patients: ESMO Clinical Practice Guidelines. ESMO Open 2021, 6, 100092. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-García, V.; López-Briz, E.; Carbonell-Sanchis, R.; Bort-Martí, S.; Gonzálvez-Perales, J.L. Megestrol acetate for cachexia–anorexia syndrome. A systematic review. J. Cachexia Sarcopenia Muscle 2018, 9, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.L.; Teoh, S.E.; Yaow, C.Y.L.; Lin, D.J.; Masuda, Y.; Han, M.X.; Yeo, W.S.; Ng, Q.X. A systematic review and meta-analysis of the clinical use of megestrol acetate for cancer-related anorexia/cachexia. J. Clin. Med. 2022, 11, 3756. [Google Scholar] [CrossRef] [PubMed]
- Zarifi, S.H.; Bagherniya, M.; Banach, M.; Johnston, T.P.; Sahebkar, A. Phytochemicals: A potential therapeutic intervention for the prevention and treatment of cachexia. Clin. Nutr. 2022, 41, 2843–2857. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, W.; Fayazuddin, M.; Shariq, S.; Singh, O.; Moin, S.; Akhtar, K.; Kumar, A. Anti-inflammatory activity of roots of Cichorium intybus due to its inhibitory effect on various cytokines and antioxidant activity. Anc. Sci. Life 2014, 34, 44. [Google Scholar] [CrossRef]
- Yadav, U.C.; Baquer, N.Z. Pharmacological effects of Trigonella foenum-graecum L. in health and disease. Pharm. Biol. 2014, 52, 243–254. [Google Scholar] [CrossRef]
- Park, B.; You, S.; Cho, W.C.; Choi, J.Y.; Lee, M.S. A systematic review of herbal medicines for the treatment of cancer cachexia in animal models. J. Zhejiang Univ. Sci. B 2019, 20, 9. [Google Scholar] [CrossRef]
- Aquila, G.; Re Cecconi, A.D.; Brault, J.J.; Corli, O.; Piccirillo, R. Nutraceuticals and exercise against muscle wasting during cancer cachexia. Cells 2020, 9, 2536. [Google Scholar] [CrossRef]
- Penedo-Vázquez, A.; Duran, X.; Mateu, J.; López-Postigo, A.; Barreiro, E. Curcumin and resveratrol improve muscle function and structure through attenuation of proteolytic markers in experimental cancer-induced cachexia. Molecules 2021, 26, 4904. [Google Scholar] [CrossRef] [PubMed]
- VanderVeen, B.N.; Cardaci, T.D.; Cunningham, P.; McDonald, S.J.; Bullard, B.M.; Fan, D.; Murphy, E.A.; Velázquez, K.T. Quercetin improved muscle mass and mitochondrial content in a murine model of cancer and chemotherapy-induced cachexia. Nutrients 2022, 15, 102. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lai, Y.J.; Chan, Y.L.; Li, T.L.; Wu, C.J. Epigallocatechin-3-gallate effectively attenuates skeletal muscle atrophy caused by cancer cachexia. Cancer Lett. 2011, 305, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.; Nahashon, S.; Taka, E.; Adinew, G.M.; Soliman, K.F. Epigallocatechin-3-Gallate (EGCG): New therapeutic perspectives for neuroprotection, aging, and neuroinflammation for the modern age. Biomolecules 2022, 12, 371. [Google Scholar] [CrossRef] [PubMed]
- Baazim, H.; Antonio-Herrera, L.; Bergthaler, A. The interplay of immunology and cachexia in infection and cancer. Nat. Rev. Immunol. 2022, 22, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E. Understanding cachexia as a cancer metabolism syndrome. Oncogenesis 2016, 5, e200. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Xi, Y.; Chen, J.; Zhu, P.; Kang, J.; Zou, Z.; Wang, F.; Bu, S. STAT3 stimulates adipogenic stem cell proliferation and cooperates with HMGA2 during the early stage of differentiation to promote adipogenesis. Biochem. Biophys. Res. Commun. 2017, 482, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Wang, X.; Lu, J. Advances in IKBKE as a potential target for cancer therapy. Cancer Med. 2020, 9, 247–258. [Google Scholar] [CrossRef]
- Ray, P.D.; Maclellan, R.A.; He, J.; Liu, Z.; Wu, J. Downregulation of RelA (p65) by rapamycin inhibits murine adipocyte differentiation and reduces fat mass of C57BL/6J mice despite high fat diet. Int. Sch. Res. Not. 2014, 2014, 540582. [Google Scholar] [CrossRef][Green Version]
- You, K.; Gu, H.; Yuan, Z.; Xu, X. Tumor necrosis factor alpha signaling and organogenesis. Front. Cell Dev. Biol. 2021, 9, 727075. [Google Scholar] [CrossRef]
- Yang, Y.; Han, L.; Yuan, Y.; Li, J.; Hei, N.; Liang, H. Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat. Commun. 2014, 5, 3231. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Cheng, X.; Zhang, Z.; Han, L.; Wu, K.; Li, Y.; Lin, X. TGF-Beta Induced Key Genes of Osteogenic and Adipogenic Differentiation in Human Mesenchymal Stem Cells and MiRNA–mRNA Regulatory Networks. Front. Genet. 2021, 12, 759596. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; Meng, Y.; Zhang, C.; Di, L. GSK3-activated STAT5 regulates expression of SFRPs to modulate adipogenesis. FASEB J. 2018, 32, 4714–4726. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Bhosale, P.B.; Abusaliya, A.; Kim, H.H.; Ha, S.E.; Park, M.Y.; Jeong, S.H.; Vetrivel, P.; Heo, J.D.; Kim, J.-A.; Kim, G.S.; et al. Apigetrin promotes TNFα-induced apoptosis, necroptosis, G2/M phase cell cycle arrest, and ROS generation through inhibition of NF-κB pathway in Hep3B liver cancer cells. Cells 2022, 11, 2734. [Google Scholar] [CrossRef] [PubMed]
- Hadrich, F.; Sayadi, S. Apigetrin inhibits adipogenesis in 3T3-L1 cells by downregulating PPARγ and CEBP-α. Lipids Health Dis. 2018, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Neag, M.A.; Mocan, A.; Echeverría, J.; Pop, R.M.; Bocsan, C.I.; Crişan, G.; Buzoianu, A.D. Berberine: Botanical occurrence, traditional uses, extraction methods, and relevance in cardiovascular, metabolic, hepatic, and renal disorders. Front. Pharmacol. 2018, 9, 557. [Google Scholar] [CrossRef]
- Salehi, B.; Fokou, P.V.T.; Sharifi-Rad, M.; Zucca, P.; Pezzani, R.; Martins, N.; Sharifi-Rad, J. The therapeutic potential of naringenin: A review of clinical trials. Pharmaceuticals 2019, 12, 11. [Google Scholar] [CrossRef]
- Song, J.G.; Liu, L. Naringenin alleviates bone cancer pain in rats via down-regulating spinal P2X7R/PI3K/AKT signaling: Involving suppression in spinal inflammation. Mol. Cell. Toxicol. 2021, 17, 475–484. [Google Scholar] [CrossRef]
- Kim, S.H.; Namkoong, S.; Ha, C.W.; Jang, S.; Hong, S.; Kim, M.J.; Koo, H.J.; Hadiwidjaja, M.; Lee, S.R.; Sohn, E.H. Korean red ginseng extract exploits NF-κB to promote wound repair and protein expression in keratinocytes. Mol. Cell. Toxicol. 2022, 8, 213–223. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.H.; Ross, J.A.; Kaasa, S.; Skorpen, F.; Fearon, K.C.; European Palliative Care Research Collaborative. Identification of possible genetic polymorphisms involved in cancer cachexia: A systematic review. J. Genet. 2011, 90, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, A.; Zhong, X.; Au, E.P.; Ceppa, E.P.; Nakeeb, A.; House, M.G.; Zyromski, N.J.; Schmidt, C.M.; Schloss, K.N.H.; Zimmers, T.A.; et al. Profiling of adipose and skeletal muscle in human pancreatic cancer cachexia reveals distinct gene profiles with convergent pathways. Cancers 2021, 13, 1975. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, T.; Sari, I.N.; Wijaya, Y.T.; Julianto, N.M.; Muhammad, J.A.; Lee, H.; Chae, J.H.; Kwon, H.Y. Cancer cachexia: Molecular mechanisms and treatment strategies. J. Hematol. Oncol. 2023, 16, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Freire, P.P.; Fernandez, G.J.; Cury, S.S.; De Moraes, D.; Oliveira, J.S.; De Oliveira, G.; Dal-Pai-Silva, M.; Reis, P.P.D.; Carvalho, R.F. The pathway to cancer cachexia: microRNA-regulated networks in muscle wasting based on integrative meta-analysis. Int. J. Mol. Sci. 2019, 20, 1962. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Syahrir, N.H.A.; Hasbi, M. Compound-Target Prediction and Network-Target Analysis on Jamu Formula. J. Phys. Conf. Ser. 2021, 1752, 012028. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Karp, P.D.; Midford, P.E.; Caspi, R.; Khodursky, A. Pathway size matters: The influence of pathway granularity on over-representation (enrichment analysis) statistics. BMC Genom. 2021, 22, 1–11. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.U.; Ezeh, E.M.; Ofoke, I.H.; Ogbu, C.O.; Ugwuja, E.I.; Aja, P.M. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Suppl. 2), W270–W277. [Google Scholar] [CrossRef]
- Irwin, J.J.; Tang, K.G.; Young, J.; Dandarchuluun, C.; Wong, B.R.; Khurelbaatar, M.; Moroz, Y.S.; Mayfield, J.; Sayle, R.A. ZINC20—A free ultralarge-scale chemical database for ligand discovery. J. Chem. Inf. Model. 2020, 60, 6065–6073. [Google Scholar] [CrossRef]
- Wang, S.M.; Wu, H.E.; Yasui, Y.; Geva, M.; Hayden, M.; Maurice, T.; Cozzolino, M.; Su, T.P. Nucleoporin POM121 signals TFEB-mediated autophagy via activation of SIGMAR1/sigma-1 receptor chaperone by pridopidine. Autophagy 2023, 19, 126–151. [Google Scholar] [CrossRef]



| S. No | Phytocompounds | Abbreviation | References | |
|---|---|---|---|---|
| 1. | Coptis chinensis | Berberine Palmatine Coptisine | BER PAL COP | [33,34,35,36] |
| 2. | Atractylodesmacrocephala | Atractylenolide I Taraxeryl acetate Lupeol | ATA TAR LUP | [37,38] |
| 3. | Panax ginseng C. A. Meyer | Ginsenosides Ginseng Tetrapeptide | GSN GST | [39,40] |
| 4. | Zizyphus jujuba | Lotusine B Jujubogenin Jujuboside A | LTB JJG JJA | [41,42,43] |
| 5. | Coix lacryma-jobi | Coixenolide Βetasitosterol Caffeic acid Naringenin | CXL BST CFA NGN | [44,45] |
| 6. | Trigonella foenum-graecum L. | Trigonelline Gentianine Quercetin | TGL GTN QRC | [46,47,48] |
| 7. | Pisum sativum | Gallicacid Epicatechin gallate Kaempferol | GLA ECG KPF | [49,50] |
| 8. | Zingiber officinale | 6-gingerol Zingerone Citral | GGL ZGN CTL | [51,52] |
| 9. | Artemisia japonica Thunb. | Beta-amyrin Β-sitosterol 7,8- dimethoxycoumarin | BMR BSL DMC | [53] |
| 10. | Xanthium strumarium L. | Xanthinosin Caffeic acid Xanthanol | XNS CFA XNL | [54,55] |
| 11. | Chrysanthemum morifolium Ramatuelle | Apigetrin Astragaloside Chlorogenic acid Kaemferol-3-O-rutinoside | AGT AGS CGA KFR | [56,57] |
| 12. | Carthamus tinctorius L. | Carthamin Hydroxysafflor yellow A | CRN HYA | [58] |
| S. No | Phytocompounds | Genes |
|---|---|---|
| 1 | ATA, LUP, NGN, GGL | PPARG |
| 2 | PAL, ATA, TAR, JJA, CFA, ECG, GGL, ZGN, HYA | STAT3 |
| 3 | GST, GLA | ADRB2 |
| 4 | GST | SIRT1 |
| 5 | CXL | NLRP |
| 6 | NGN, CGA | IGFBP3 |
| 7 | ECG, AGA, AGT, KFR | TNF |
| 8 | CFA, GTN, XNS | TLR4 |
| 9 | PAL, JJA, CFA, NGN, TGL, QRC, ECG, KPF, CTL, XNX, AGA, AGT, CGA, KFR | MMP9 |
| 10 | TAR, JJG, JJA, BST, CFA, TGL, GTN, GGL, ZGN, BMR, XNS, XNL, | NOS2 |
| 11 | BER, TAR | QPCT |
| 12 | BER, PAL, COP, TGL, CGA, KFR, CRN, HYA | IKBKB |
| 13 | COP | TUBB3 |
| 14 | COP, LTB, QRC, AGS, KFR | MYLK |
| 15 | GSN, GST, JJA, CXL, CFA, GTN, QRC, KPF, CTL, BMR, DMA, XNS, XNL, AGS, KFR, ATA | F2 |
| 16 | TAR, JJA, BST, GGL, BMR, XNL | NR3C1 |
| 17 | GSN, TGL, BMR, XNL | CCR2 |
| 18 | GSN, JJA, CXL, BST, BMR, XNS | HSD11B2 |
| 19 | LUP, GST, CFA, CTL, DMC, XNS, CGA, | ACE |
| 20 | LUP, GST, LTB, JJG, NGN, GTN, QRC, KPF, CTL, GGL, BMR, DMC, XNS, AGT, AGS, KFR, HYA | GSK3B |
| 21 | GST, CXL | CAPN1 |
| 22 | GST | SLC5A1 |
| 23 | LTB | BIRC3 |
| 24 | JJG | MAP3K14 |
| 25 | CFA | NGFR |
| 26 | CFA | RELA |
| 27 | NGN, GLA, ECG, AGT, AGS, KFR, | SERPINE1 |
| 28 | TGL | HDAC3 |
| 29 | XNL | EIF2AK3 |
| 30 | AGT, AGS, KFR | PLG |
| 31 | AGT, AGS, KFR | TP53 |
| 32 | CGA | SELP |
| S. No | Compounds | GPCR Ligand (GPCR) | Kinase Inhibitor (Ki) | Nuclear Receptor Ligand (Ncr) | Protease Inhibitor (Pi) | Enzyme Inhibitor (Ei) | No. of Violations (nVio) |
|---|---|---|---|---|---|---|---|
| 1. | BER | −0.11 | −0.27 | −0.78 | −0.35 | 0.82 | 1 |
| 2. | PAL | −0.10 | −0.22 | −0.65 | −0.29 | 0.81 | 1 |
| 3. | COP | −0.06 | −0.22 | −0.82 | −0.33 | 0.89 | 1 |
| 4. | ATA | −0.63 | −0.72 | 0.27 | −0.28 | 0.50 | 0 |
| 5. | TAR | 0.10 | −0.31 | 0.44 | −0.04 | 0.41 | 1 |
| 6. | LUP | 0.27 | −0.42 | 0.85 | 0.15 | 0.52 | 1 |
| 7. | GSN | 0.28 | −0.28 | 0.71 | 0.12 | 0.65 | 1 |
| 8. | GST | 0.49 | −0.10 | −0.08 | 0.95 | 0.41 | 2 |
| 9. | LTB | −0.01 | −0.53 | −0.68 | 0.44 | −0.35 | 1 |
| 10. | JJG | 0.22 | −0.31 | 0.79 | 0.06 | 0.67 | 1 |
| 11. | JJA | −3.83 | −3.92 | −3.87 | −3.8 | −3.79 | 3 |
| 12. | CXL | −0.02 | −0.25 | −0.14 | 0.04 | −0.07 | 2 |
| 13. | BST | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| 14. | CFA | −0.48 | −0.81 | −0.10 | −0.79 | −0.09 | 0 |
| 15. | NGN | 0.03 | −0.26 | 0.42 | −0.12 | 0.21 | 0 |
| 16. | TGL | −0.82 | −1.75 | −3.35 | −1.05 | −0.09 | 0 |
| 17. | GTN | −0.53 | −0.58 | −0.81 | −0.92 | −0.09 | 0 |
| 18. | QRC | −0.06 | 0.28 | 0.36 | −0.25 | 0.28 | 0 |
| 19. | GLA | −0.77 | −0.88 | −0.52 | −0.94 | −0.17 | 0 |
| 20. | ECG | 0.17 | 0.05 | 0.34 | 0.13 | 0.25 | 1 |
| 21. | KPF | −0.10 | 0.21 | 0.32 | −0.27 | 0.26 | 0 |
| 22. | GGL | 0.16 | −0.33 | 0.20 | 0.15 | 0.38 | 0 |
| 23. | ZGN | −0.58 | −1.15 | −0.59 | −0.72 | −0.07 | 0 |
| 24. | CTL | −0.86 | −1.29 | −0.42 | −0.57 | 0.02 | 0 |
| 25. | BMR | 0.22 | −0.31 | 0.67 | 0.11 | 0.56 | 1 |
| 26. | DMC | −0.83 | −0.76 | −0.77 | −0.95 | −0.08 | 0 |
| 27. | XNS | 0.00 | −0.85 | 0.96 | −0.31 | 0.68 | 0 |
| 28. | XNL | −0.31 | −0.34 | −0.05 | −0.54 | 0.10 | 0 |
| 29. | AGT | 0.10 | 0.14 | 0.31 | 0.02 | 0.43 | 1 |
| 30. | AGS | −0.14 | −0.28 | −0.42 | −0.15 | −0.05 | 3 |
| 31. | CGA | 0.29 | −0.00 | 0.74 | 0.27 | 0.62 | 1 |
| 32. | KFR | −0.01 | −0.09 | −0.17 | −0.04 | 0.18 | 3 |
| 33. | CRN | −3.36 | −3.64 | −3.59 | −2.83 | −3.35 | 3 |
| 34. | HYA | −0.11 | −0.32 | −0.06 | −0.02 | 0.03 | 3 |
| Phytocompound (Ligand) | Gene (Target) | Binding Energy ∆G (kcal/mol) |
|---|---|---|
| Apigetrin (AGT) | SERPINE1 | −8.54 |
| GSK3B | −9.36 | |
| PLG | −8.14 | |
| Naringenin (NGN) | SERPINE1 | −7.02 |
| GSK3B | −7.04 | |
| Berberine (BER) | IKBKB | −6.11 |
| QPCT | −6.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muthamil, S.; Muthuramalingam, P.; Kim, H.-Y.; Jang, H.-J.; Lyu, J.-H.; Shin, U.C.; Go, Y.; Park, S.-H.; Lee, H.G.; Shin, H.; et al. Unlocking Prognostic Genes and Multi-Targeted Therapeutic Bioactives from Herbal Medicines to Combat Cancer-Associated Cachexia: A Transcriptomics and Network Pharmacology Approach. Int. J. Mol. Sci. 2024, 25, 156. https://doi.org/10.3390/ijms25010156
Muthamil S, Muthuramalingam P, Kim H-Y, Jang H-J, Lyu J-H, Shin UC, Go Y, Park S-H, Lee HG, Shin H, et al. Unlocking Prognostic Genes and Multi-Targeted Therapeutic Bioactives from Herbal Medicines to Combat Cancer-Associated Cachexia: A Transcriptomics and Network Pharmacology Approach. International Journal of Molecular Sciences. 2024; 25(1):156. https://doi.org/10.3390/ijms25010156
Chicago/Turabian StyleMuthamil, Subramanian, Pandiyan Muthuramalingam, Hyun-Yong Kim, Hyun-Jun Jang, Ji-Hyo Lyu, Ung Cheol Shin, Younghoon Go, Seong-Hoon Park, Hee Gu Lee, Hyunsuk Shin, and et al. 2024. "Unlocking Prognostic Genes and Multi-Targeted Therapeutic Bioactives from Herbal Medicines to Combat Cancer-Associated Cachexia: A Transcriptomics and Network Pharmacology Approach" International Journal of Molecular Sciences 25, no. 1: 156. https://doi.org/10.3390/ijms25010156
APA StyleMuthamil, S., Muthuramalingam, P., Kim, H.-Y., Jang, H.-J., Lyu, J.-H., Shin, U. C., Go, Y., Park, S.-H., Lee, H. G., Shin, H., & Park, J. H. (2024). Unlocking Prognostic Genes and Multi-Targeted Therapeutic Bioactives from Herbal Medicines to Combat Cancer-Associated Cachexia: A Transcriptomics and Network Pharmacology Approach. International Journal of Molecular Sciences, 25(1), 156. https://doi.org/10.3390/ijms25010156








