Decoding the Phosphatase Code: Regulation of Cell Proliferation by Calcineurin
Abstract
:1. Introduction
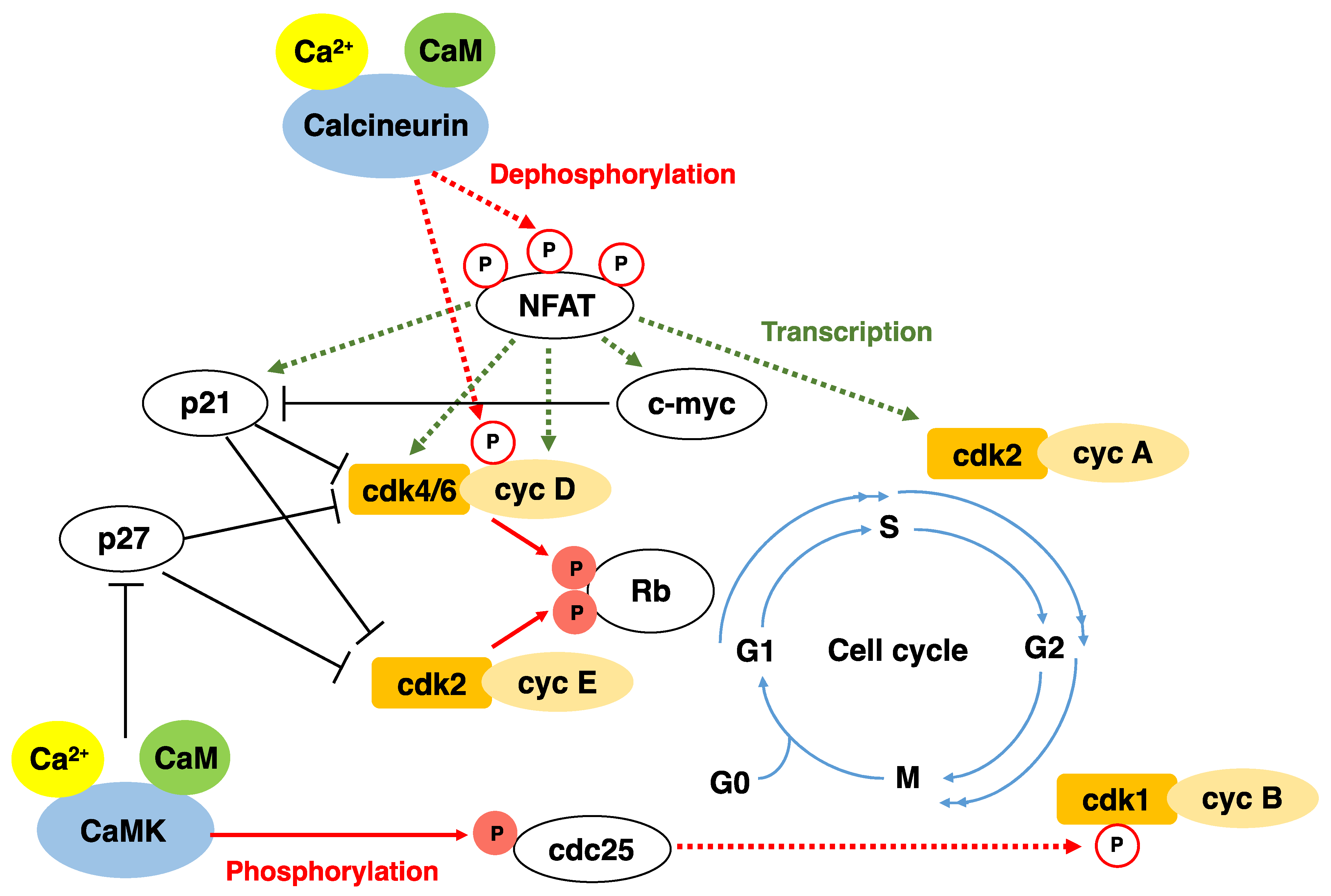
2. Cell Cycle and CDK
3. Calcium, Calmodulin, and Calmodulin-Dependent Protein Kinases in Cell Proliferation
4. Characteristics of Calcineurin
5. Mechanisms Regulating Calcineurin Activity
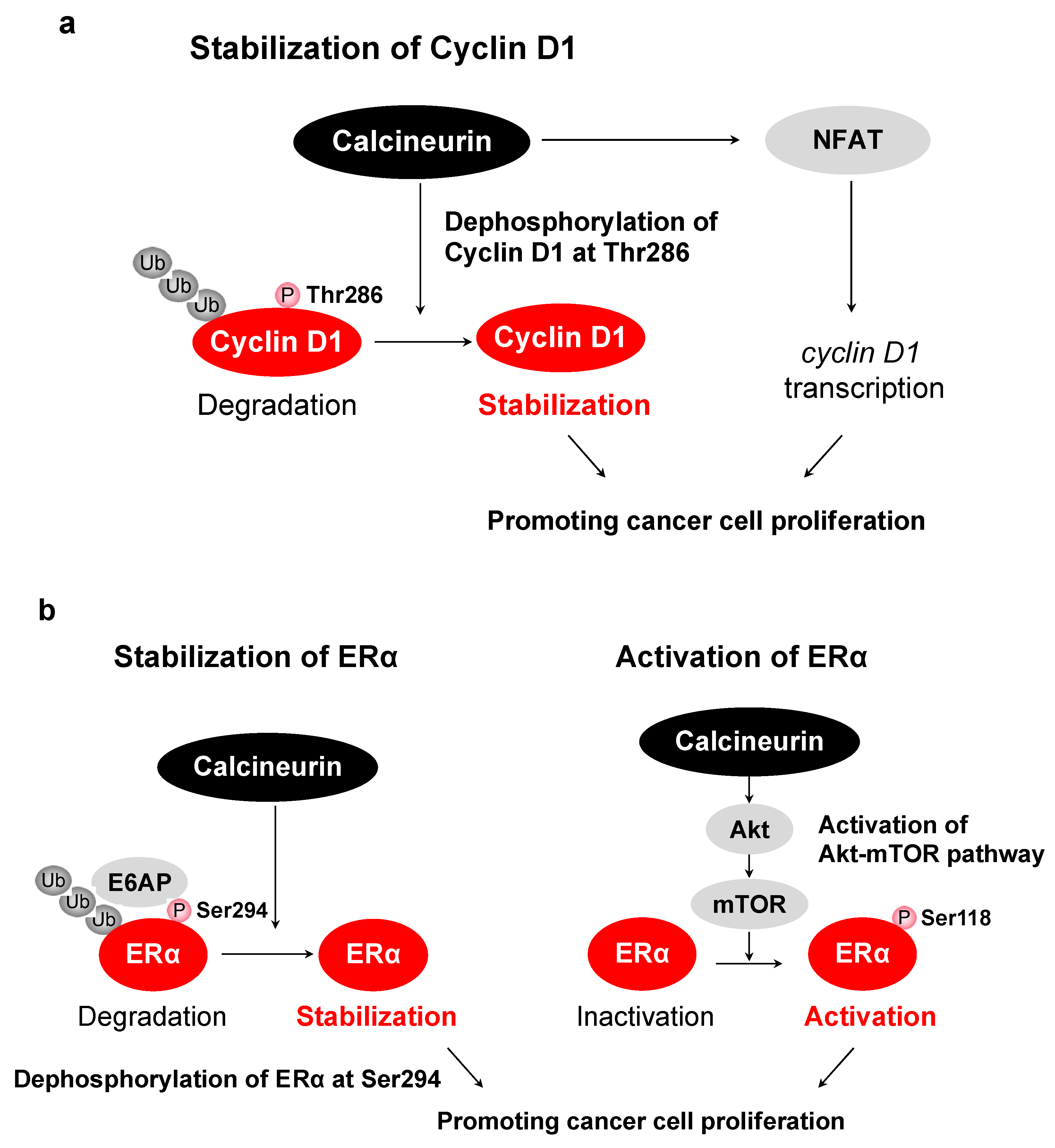
6. Functions of Calcineurin/NFAT
7. Other Substrates of Calcineurin
8. Activation of Calcineurin/NFAT Pathway in Cancer
9. A Therapeutic Perspective for Cancer
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berridge, M.J.; Lipp, P.; Bootman, M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21. [Google Scholar] [CrossRef]
- Patel, S.; Joseph, S.; Thomas, A. Molecular properties of inositol 1,4,5-trisphosphate receptors. Cell Calcium 1999, 25, 247–264. [Google Scholar] [CrossRef] [PubMed]
- Kiselyov, K.; Mignery, G.A.; Zhu, M.X.; Muallem, S. The N-Terminal Domain of the IP3 Receptor Gates Store-Operated hTrp3 Channels. Mol. Cell 1999, 4, 423–429. [Google Scholar] [CrossRef]
- Van Rossum, D.; Patterson, R.L.; Kiselyov, K.; Boehning, D.; Barrow, R.K.; Gill, D.L.; Snyder, S.H. Agonist-induced Ca2+ entry determined by inositol 1,4,5-trisphosphate recognition. Proc. Natl. Acad. Sci. USA 2004, 101, 2323–2327. [Google Scholar] [CrossRef] [Green Version]
- Venkatachalam, K.; Van Rossum, D.B.; Patterson, R.L.; Ma, H.-T.; Gill, D.L. The cellular and molecular basis of store-operated calcium entry. Nat. Cell Biol. 2002, 4, E263–E272. [Google Scholar] [CrossRef] [PubMed]
- Parkash, J.; Asotra, K. Calcium wave signaling in cancer cells. Life Sci. 2010, 87, 587–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McConkey, D.J.; Orrenius, S. The role of calcium in the regulation of apoptosis. Biochem. Biophys. Res. Commun. 1997, 239, 357–366. [Google Scholar] [CrossRef]
- Santella, L. The Role of Calcium in the Cell Cycle: Facts and Hypotheses. Biochem. Biophys. Res. Commun. 1998, 244, 317–324. [Google Scholar] [CrossRef]
- Pinto, M.C.X.; Kihara, A.; Goulart, V.A.; Tonelli, F.M.P.; Gomes, K.N.; Ulrich, H.; Resende, R.R. Calcium signaling and cell proliferation. Cell. Signal. 2015, 27, 2139–2149. [Google Scholar] [CrossRef]
- Pande, G.; Kumar, N.A.; Manogaran, P.S. Flow cytometric study of changes in the intracellular free calcium during the cell cycle. Cytometry 1996, 24, 55–63. [Google Scholar] [CrossRef]
- Smedler, E.; Uhlén, P. Frequency decoding of calcium oscillations. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 964–969. [Google Scholar] [CrossRef] [Green Version]
- Kahl, C.R.; Means, A.R. Regulation of Cell Cycle Progression by Calcium/Calmodulin-Dependent Pathways. Endocr. Rev. 2003, 24, 719–736. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, J.F. Calcium signals and cancer. Crit. Rev. Oncog. 1992, 3, 55–90. [Google Scholar]
- Cook, S.J.; Lockyer, P.J. Recent advances in Ca2+-dependent Ras regulation and cell proliferation. Cell Calcium 2006, 39, 101–112. [Google Scholar] [CrossRef]
- Morgan, D.O. Cyclin-Dependent Kinases: Engines, Clocks, and Microprocessors. Annu. Rev. Cell Dev. Biol. 1997, 13, 261–291. [Google Scholar] [CrossRef]
- Satyanarayana, A.; Kaldis, P. Mammalian cell-cycle regulation: Several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 2009, 28, 2925–2939. [Google Scholar] [CrossRef] [Green Version]
- Matsushime, H.; Ewen, M.E.; Strom, D.K.; Kato, J.Y.; Hanks, S.K.; Roussel, M.F.; Sherr, C.J. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 1992, 71, 323–334. [Google Scholar] [CrossRef]
- Connell-Crowley, L.; Harper, J.W.; Goodrich, D.W. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol. Biol. Cell 1997, 8, 287–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarkowska, T.; Mittnacht, S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J. Biol. Chem. 1997, 272, 12738–12746. [Google Scholar] [CrossRef] [Green Version]
- Bertoli, C.; Skotheim, J.M.; De Bruin, R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinberg, R.A. The retinoblastoma protein and cell cycle control. Cell 1995, 81, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Dyson, N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998, 12, 2245–2262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, B.O.; Lukas, J.; Sorensen, C.; Bartek, J.; Helin, K. Phosphorylation of mammalian CDC6 by Cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999, 18, 396–410. [Google Scholar] [CrossRef]
- Coverley, D.; Pelizon, C.; Trewick, S.; Laskey, R. Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci. 2000, 113, 1929–1938. [Google Scholar] [CrossRef] [PubMed]
- Yam, C.H.; Fung, T.K.; Poon, R. Cyclin A in cell cycle control and cancer. Cell. Mol. Life Sci. 2002, 59, 1317–1326. [Google Scholar] [CrossRef]
- Stillman, B. Cell Cycle Control of DNA Replication. Science 1996, 274, 1659–1663. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, T.; Okumura, E. In vivo regulation of the entry into M-phase: Initial activation and nuclear translocation of cyclin B/Cdc2. Prog. Cell Cycle Res. 1997, 3, 241–249. [Google Scholar] [CrossRef]
- Furuno, N.; Elzen, N.D.; Pines, J. Human Cyclin a Is Required for Mitosis until Mid Prophase. J. Cell Biol. 1999, 147, 295–306. [Google Scholar] [CrossRef] [Green Version]
- Boynton, A.L.; Whitfield, J.F.; Isaacs, R.J.; Tremblay, R. The control of human WI-38 cell proliferation by extracellular calcium and its elimination by SV-40 virus-induced proliferative transformation. J. Cell. Physiol. 1977, 92, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Russa, A.D.; Maesawa, C.; Satoh, Y. Spontaneous [Ca2+]i oscillations in G1/S phase-synchronized cells. J. Electron. Microsc. 2009, 58, 321–329. [Google Scholar] [CrossRef]
- Chin, D.; Means, A.R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000, 10, 322–328. [Google Scholar] [CrossRef]
- Colomer, J.; Lopezgirona, A.; Agell, N.; Bachs, O. Calmodulin Regulates the Expression of CDKS, Cyclins and Replicative Enzymes During Proliferative Activation of Human T Lymphocytes. Biochem. Biophys. Res. Commun. 1994, 200, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, C.D.; Means, A.R. Calmodulin is required for cell-cycle progression during G1 and mitosis. EMBO J. 1989, 8, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Husain, M. Calmodulin-Mediated Cell Cycle Regulation: New Mechanisms for Old Observations. Cell Cycle 2006, 5, 2183–2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulman, H.; Hanson, P.I. Multifunctional Ca2+/calmodulin-dependent protein kinase. Neurochem. Res. 1993, 18, 65–77. [Google Scholar] [CrossRef]
- Fujisawa, H. Regulation of the activities of multifunctional Ca2+/calmodulin-dependent protein kinases. J. Biochem. 2001, 129, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Karpurapu, M.; Wang, D.; Van Quyen, D.; Kim, T.K.; Kundumani-Sridharan, V.; Pulusani, S.; Rao, G.N. Cyclin D1 is a bona fide target gene of NFATc1 and is sufficient in the mediation of injury-induced vascular wall remodeling. J. Biol. Chem. 2010, 285, 3510–3523. [Google Scholar] [CrossRef] [Green Version]
- Goshima, T.; Habara, M.; Maeda, K.; Hanaki, S.; Kato, Y.; Shimada, M. Calcineurin regulates cyclin D1 stability through dephosphorylation at T286. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Masuo, T.; Okamura, S.; Zhang, Y.; Mori, M. Cyclosporine A inhibits colorectal cancer proliferation probably by regulating expression levels of c-Myc, p21WAF1/CIP1 and proliferating cell nuclear antigen. Cancer Lett. 2009, 285, 66–72. [Google Scholar] [CrossRef]
- Perez-Neut, M.; Rao, V.R.; Gentile, S. hERG1/Kv11.1 activation stimulates transcription of p21waf/cip in breast cancer cells via a calcineurin-dependent mechanism. Oncotarget 2016, 7, 58893–58902. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liu, Y.; Sun, X.; Li, Z.; Fang, P.; He, P.; Shi, H.; Xie, M.; Wang, X.; Zhang, D.; et al. Sildenafil inhibits calcineurin/NFATc2-mediated cyclin A expression in pulmonary artery smooth muscle cells. Life Sci. 2011, 89, 644–649. [Google Scholar] [CrossRef]
- Choi, J.; Chiang, A.; Taulier, N.; Gros, R.; Pirani, A.; Husain, M. A calmodulin-binding site on cyclin E mediates Ca2+-sensitive G1/s transitions in vascular smooth muscle cells. Circ. Res. 2006, 98, 1273–1281. [Google Scholar] [CrossRef] [Green Version]
- Kahl, C.R.; Means, A.R. Regulation of cyclin D1/Cdk4 complexes by calcium/calmodulin-dependent protein kinase I. J. Biol. Chem. 2004, 279, 15411–15419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Li, N.; Liu, X.; Zheng, Y.; Cao, X. A novel endogenous human CaMKII inhibitory protein suppresses tumor growth by inducing cell cycle arrest via p27 stabilization. J. Biol. Chem. 2008, 283, 11565–11574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, R.; Holt, M.; Philipova, R.; Moss, S.; Schulman, H.; Hidaka, H.; Whitaker, M. Calcium/calmodulin-dependent phosphorylation and activation of human Cdc25-C at the G2/M phase transition in HeLa cells. J. Biol. Chem. 1999, 274, 7958–7968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parmer, T.G.; Ward, M.D.; Hait, W.N. Effects of rottlerin, an inhibitor of calmodulin-dependent protein kinase III, on cellular proliferation, viability, and cell cycle distribution in malignant glioma cells. Cell Growth Differ. Mol. Boil. J. Am. Assoc. Cancer Res. 1997, 8, 327–334. [Google Scholar]
- Minami, H.; Inoue, S.; Hidaka, H. The effect of KN-62, Ca2+/calmodulin dependent protein kinase II inhibitor on cell cycle. Biochem. Biophys. Res. Commun. 1994, 199, 241–248. [Google Scholar] [CrossRef]
- Skelding, K.A.; Rostas, J.A.P.; Verrills, N.M. Controlling the cell cycle: The role of calcium/calmodulin-stimulated protein kinases I and II. Cell Cycle 2011, 10, 631–639. [Google Scholar] [CrossRef] [Green Version]
- House, S.J.; Ginnan, R.G.; Armstrong, S.E.; Singer, H.A. Calcium/calmodulin-dependent protein kinase II-delta isoform regulation of vascular smooth muscle cell proliferation. Am. J. Physiol. Cell Physiol. 2007, 292, C2276–C2287. [Google Scholar] [CrossRef] [Green Version]
- Tombes, R.M.; Grant, S.; Westin, E.H.; Krystal, G. G1 cell cycle arrest and apoptosis are induced in NIH 3T3 cells by KN-93, an inhibitor of CaMK-II (the multifunctional Ca2+/CaM kinase). Cell Growth Differ. Mol. Boil. J. Am. Assoc. Cancer Res. 1995, 6, 1063–1070. [Google Scholar]
- Morris, T.A.; DeLorenzo, R.J.; Tombes, R.M. CaMK-II inhibition reduces cyclin D1 levels and enhances the association of p27kip1 with Cdk2 to cause G1 arrest in NIH 3T3 cells. Exp. Cell Res. 1998, 240, 218–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, E.H.; Gorelick, F.; Czernik, A.J.; Bagaglio, D.M.; Hait, W.N. Calmodulin-dependent protein kinases in rat glioblastoma. Cell Growth Differ. Mol. Boil. J. Am. Assoc. Cancer Res. 1995, 6, 615–621. [Google Scholar]
- Si, J.; Collins, S.J. Activated Ca2+/calmodulin-dependent protein kinase IIgamma is a critical regulator of myeloid leukemia cell proliferation. Cancer Res. 2008, 68, 3733–3742. [Google Scholar] [CrossRef] [Green Version]
- Klee, C.B.; Ren, H.; Wang, X. Regulation of the Calmodulin-stimulated Protein Phosphatase, Calcineurin. J. Biol. Chem. 1998, 273, 13367–13370. [Google Scholar] [CrossRef] [Green Version]
- Hogan, P.G.; Li, H. Calcineurin. Curr. Biol. 2005, 15, R442–R443. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Rao, A.; Hogan, P.G. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011, 21, 91–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusnak, F.; Mertz, P. Calcineurin: Form and Function. Physiol. Rev. 2000, 80, 1483–1521. [Google Scholar] [CrossRef]
- Klee, C.B.; Draetta, G.F.; Hubbard, M.J. Calcineurin. Adv. Enzym. Relat. Areas Mol. Biol. 1988, 61, 149–200. [Google Scholar]
- Cohen, P.T.; Chen, M.X.; Armstrong, C.G. Novel Protein Phosphatases That May Participate in Cell Signaling. Adv. Pharmacol. 1996, 36, 67–89. [Google Scholar] [CrossRef]
- Goto, S.; Matsukado, Y.; Mihara, Y.; Inoue, N.; Miyamoto, E. Calcineurin in human brain and its relation to extrapyramidal system. Immunohistochemical study on postmortem human brains. Acta Neuropathol. 1986, 72, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Kuno, T.; Mukai, H.; Ito, A.; Chang, C.D.; Kishima, K.; Saito, N.; Tanaka, C. Distinct cellular expression of calcineurin A alpha and A beta in rat brain. J. Neurochem. 1992, 58, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Hallhuber, M.; Burkard, N.; Wu, R.; Buch, M.H.; Engelhardt, S.; Hein, L.; Neyses, L.; Schuh, K.; Ritter, O. Inhibition of nuclear import of calcineurin prevents myocardial hypertrophy. Circ. Res. 2006, 99, 626–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.-J.; Yoo, S.-A.; Kim, M.; Kim, W.-U. The Role of Calcium–Calcineurin–NFAT Signaling Pathway in Health and Autoimmune Diseases. Front. Immunol. 2020, 11, 195. [Google Scholar] [CrossRef]
- Wallace, R.W.; Tallant, E.A.; Cheung, W.Y. High levels of a heat-labile calmodulin-binding protein (CaM-BP80) in bovine neostriatum. Biochemistry 1980, 19, 1831–1837. [Google Scholar] [CrossRef]
- Usuda, N.; Arai, H.; Sasaki, H.; Hanai, T.; Nagata, T.; Muramatsu, T.; Kincaid, R.L.; Higuchi, S. Differential subcellular localization of neural isoforms of the catalytic subunit of calmodulin-dependent protein phosphatase (calcineurin) in central nervous system neurons: Immunohistochemistry on formalin-fixed paraffin sections employing antigen retrieval by microwave irradiation. J. Histochem. Cytochem. 1996, 44, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Anthony, F.A.; Winkler, M.A.; Edwards, H.H.; Cheung, W.Y. Quantitative subcellular localization of calmodulin-dependent phosphatase in chick forebrain. J. Neurosci. 1988, 8, 1245–1253. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, K.; Ness, J.; Wooge, C.H.; Janovick, J.A.; Conn, P.M. Specific Identification and Subcellular Localization of Three Calmodulin-binding Proteins in the Rat Gonadotrope: Spectrin, Caldesmon, and Calcineurin. Biol. Reprod. 1991, 44, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Shibasaki, F.; Price, E.R.; Milan, D.; McKeon, F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature 1996, 382, 370–373. [Google Scholar] [CrossRef]
- Coghlan, V.M.; Perrino, B.A.; Howard, M.; Langeberg, L.K.; Hicks, J.B.; Gallatin, W.M.; Scott, J.D. Association of Protein Kinase A and Protein Phosphatase 2B with a Common Anchoring Protein. Science 1995, 267, 108–111. [Google Scholar] [CrossRef]
- Baggott, R.R.; Mohamed, T.M.; Oceandy, D.; Holton, M.; Blanc, M.C.; Roux-Soro, S.C.; Brown, S.; Brown, J.E.; Cartwright, E.; Wang, W.; et al. Disruption of the interaction between PMCA2 and calcineurin triggers apoptosis and enhances paclitaxel-induced cytotoxicity in breast cancer cells. Carcinogenesis 2012, 33, 2362–2368. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Barber, D.L. A calcineurin homologous protein inhibits GTPase-stimulated Na-H exchange. Proc. Natl. Acad. Sci. USA 1996, 93, 12631–12636. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Sikkink, R.A.; Rusnak, F.; Barber, D.L. Inhibition of Calcineurin Phosphatase Activity by a Calcineurin B Homologous Protein. J. Biol. Chem. 1999, 274, 36125–36131. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Youn, H.-D.; Loh, C.; Stolow, M.; He, W.; Liu, J.O. Cabin 1, A Negative Regulator for Calcineurin Signaling in T Lymphocytes. Immunity 1998, 8, 703–711. [Google Scholar] [CrossRef] [Green Version]
- Lai, M.M.; Burnett, P.E.; Wolosker, H.; Blackshaw, S.; Snyder, S.H. Cain, A Novel Physiologic Protein Inhibitor of Calcineurin. J. Biol. Chem. 1998, 273, 18325–18331. [Google Scholar] [CrossRef] [Green Version]
- Rothermel, B.; Vega, R.B.; Yang, J.; Wu, H.; Bassel-Duby, R.; Williams, R.S. A Protein Encoded within the Down Syndrome Critical Region Is Enriched in Striated Muscles and Inhibits Calcineurin Signaling. J. Biol. Chem. 2000, 275, 8719–8725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuentes, J.J.; Genescà, E.; Kingsbury, T.J.; Cunningham, K.W.; Perez-Riba, M.; Estivill, X.; de la Luna, S. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum. Mol. Genet. 2000, 9, 1681–1690. [Google Scholar] [CrossRef] [Green Version]
- Kingsbury, T.J.; Cunningham, K.W. A conserved family of calcineurin regulators. Genes Dev. 2000, 14, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Shirane, M.; Nakayama, K.I. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat. Cell Biol. 2003, 5, 28–37. [Google Scholar] [CrossRef]
- Li, H.; Pink, M.D.; Murphy, J.G.; Stein, A.; Dell’Acqua, M.L.; Hogan, P.G. Balanced interactions of calcineurin with AKAP79 regulate Ca2+–calcineurin–NFAT signaling. Nat. Struct. Mol. Biol. 2012, 19, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Görlach, J.; Fox, D.S.; Cutler, N.S.; Cox, G.M.; Perfect, J.R.; Heitman, J. Identification and characterization of a highly conserved calcineurin binding protein, CBP1/calcipressin, inCryptococcus neoformans. EMBO J. 2000, 19, 3618–3629. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Martinez, S.; Genescà, E.; Rodriguez, A.; Raya, A.; Salichs, E.; Were, F.; Lopez-Maderuelo, M.D.; Redondo, J.M.; de la Luna, S. The RCAN carboxyl end mediates calcineurin docking-dependent inhibition via a site that dictates binding to substrates and regulators. Proc. Natl. Acad. Sci. USA 2009, 106, 6117–6122. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, C.; Garen, C. Activation of calmodulin-dependent enzymes can be selectively inhibited by histone H1. J. Biol. Chem. 1993, 268, 23788–23791. [Google Scholar] [CrossRef]
- Wang, X.; Culotta, V.C.; Klee, C.B. Superoxide dismutase protects calcineurin from inactivation. Nature 1996, 383, 434–437. [Google Scholar] [CrossRef] [PubMed]
- Sommer, D.; Fakata, K.L.; Swanson, S.A.; Stemmer, P. Modulation of the phosphatase activity of calcineurin by oxidants and antioxidants in vitro. JBIC J. Biol. Inorg. Chem. 2000, 267, 2312–2322. [Google Scholar] [CrossRef] [PubMed]
- Namgaladze, D.; Shcherbyna, I.; Kienhöfer, J.; Hofer, H.W.; Ullrich, V. Superoxide targets calcineurin signaling in vascular endothelium. Biochem. Biophys. Res. Commun. 2005, 334, 1061–1067. [Google Scholar] [CrossRef]
- Zhou, X.; Mester, C.; Stemmer, P.; Reid, G.E. Oxidation-Induced Conformational Changes in Calcineurin Determined by Covalent Labeling and Tandem Mass Spectrometry. Biochemistry 2014, 53, 6754–6765. [Google Scholar] [CrossRef]
- Mukerjeea, N.; McGinnis, K.M.; Gnegy, M.E.; Wang, K.K. Caspase-Mediated Calcineurin Activation Contributes to IL-2 Release during T Cell Activation. Biochem. Biophys. Res. Commun. 2001, 285, 1192–1199. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.-J.; Jo, D.-G.; Hong, G.-S.; Kim, B.J.; Lai, M.; Cho, D.-H.; Kim, K.-W.; Bandyopadhyay, A.; Hong, Y.-M.; Kim, D.H.; et al. Calpain-dependent cleavage of cain/cabin1 activates calcineurin to mediate calcium-triggered cell death. Proc. Natl. Acad. Sci. USA 2002, 99, 9870–9875. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.-Y.; Tomizawa, K.; Oda, Y.; Wei, F.-Y.; Lu, Y.-F.; Matsushita, M.; Li, S.T.; Moriwaki, A.; Matsui, H. Critical Role of Calpain-mediated Cleavage of Calcineurin in Excitotoxic Neurodegeneration. J. Biol. Chem. 2004, 279, 4929–4940. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, Y.; King, M.M.; Soderling, T.R. Regulatory interactions of calmodulin-binding proteins: Phosphorylation of calcineurin by autophosphorylated Ca2+/calmodulin-dependent protein kinase II. Proc. Natl. Acad. Sci. USA 1988, 85, 7001–7005. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, Y.; Soderling, T.R. Regulation of calcineurin by phosphorylation. Identification of the regulatory site phosphorylated by Ca2+/calmodulin-dependent protein kinase II and protein kinase C. J. Biol. Chem. 1989, 264, 16524–16529. [Google Scholar] [CrossRef]
- Martensen, T.M.; Martin, B.M.; Kincaid, R.L. Identification of the site on calcineurin phosphorylated by Ca2+/CaM-dependent kinase II: Modification of the CaM-binding domain. Biochemistry 1989, 28, 9243–9247. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Farmer, J.D.; Lane, W.S.; Friedman, J.; Weissman, I.; Schreiber, S.L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 1991, 66, 807–815. [Google Scholar] [CrossRef]
- Shaw, K.T.; Ho, A.M.; Raghavan, A.; Kim, J.; Jain, J.; Park, J.; Sharma, S.; Rao, A.; Hogan, P.G. Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells. Proc. Natl. Acad. Sci. USA 1995, 92, 11205–11209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogan, P.G.; Chen, L.; Nardone, J.; Rao, A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003, 17, 2205–2232. [Google Scholar] [CrossRef] [Green Version]
- Roy, J.; Cyert, M.S. Identifying New Substrates and Functions for an Old Enzyme: Calcineurin. Cold Spring Harb. Perspect. Biol. 2019, 12, 3. [Google Scholar] [CrossRef]
- Lally, C.; Healy, E.; Ryan, M.P. Cyclosporine A-induced cell cycle arrest and cell death in renal epithelial cells. Kidney Int. 1999, 56, 1254–1257. [Google Scholar] [CrossRef] [Green Version]
- Schneider, G.; Oswald, F.; Wahl, C.; Greten, F.R.; Adler, G.; Schmid, R.M. Cyclosporine inhibits growth through the activating transcription factor/cAMP-responsive element-binding protein binding site in the cyclin D1 promoter. J. Biol. Chem. 2002, 277, 43599–43607. [Google Scholar] [CrossRef] [Green Version]
- Kahl, C.R.; Means, A.R. Calcineurin Regulates Cyclin D1 Accumulation in Growth-stimulated Fibroblasts. Mol. Biol. Cell 2004, 15, 1833–1842. [Google Scholar] [CrossRef] [Green Version]
- Toyota, N.; Hashimoto, Y.; Matsuo, S.; Kitamura, Y.; Iizuka, H. Effects of FK506 and cyclosporin A on proliferation, histamine release and phenotype of murine mast cells. Arch. Dermatol. Res. 1996, 288, 474–480. [Google Scholar] [CrossRef]
- Khanna, A.K.; Hosenpud, J.D. Cyclosporine Induces The Expression of the Cyclin Inhibitor p21. Transplantation 1999, 67, 1262–1268. [Google Scholar] [CrossRef]
- Tomono, M.; Toyoshima, K.; Ito, M.; Amano, H.; Kiss, Z. Inhibitors of Calcineurin Block Expression of Cyclins A and E Induced by Fibroblast Growth Factor in Swiss 3T3 Fibroblasts. Arch. Biochem. Biophys. 1998, 353, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Mognol, G.P.; Carneiro, F.; Robbs, B.; Faget, D.; Viola, J.P.B. Cell cycle and apoptosis regulation by NFAT transcription factors: New roles for an old player. Cell Death Dis. 2016, 7, e2199. [Google Scholar] [CrossRef] [Green Version]
- Bueno, O.F.; Brandt, E.B.; Rothenberg, M.E.; Molkentin, J.D. Defective T cell development and function in calcineurin A beta-deficient mice. Proc. Natl. Acad. Sci. USA 2002, 99, 9398–9403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogan, P.G. Calcium–NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium 2017, 63, 66–69. [Google Scholar] [CrossRef] [Green Version]
- Vaeth, M.; Feske, S. NFAT control of immune function: New Frontiers for an Abiding Trooper. F1000Research 2018, 7, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraner, S.D.; Norris, C.M. Astrocyte Activation and the Calcineurin/NFAT Pathway in Cerebrovascular Disease. Front. Aging Neurosci. 2018, 10, 287. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.Z.A.; Hussain, T.; Zhao, D.; Yang, L. A central role for calcineurin in protein misfolding neurodegenerative diseases. Cell. Mol. Life Sci. 2016, 74, 1061–1074. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, K.; Yamaguchi, A. The Functional Role of Calcineurin in Hypertrophy, Regeneration, and Disorders of Skeletal Muscle. J. Biomed. Biotechnol. 2010, 2010, 721219. [Google Scholar] [CrossRef] [Green Version]
- Tu, M.K.; Levin, J.B.; Hamilton, A.M.; Borodinsky, L.N. Calcium signaling in skeletal muscle development, maintenance and regeneration. Cell Calcium 2016, 59, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Dewenter, M.; Von Der Lieth, A.; Katus, H.A.; Backs, J. Calcium Signaling and Transcriptional Regulation in Cardiomyocytes. Circ. Res. 2017, 121, 1000–1020. [Google Scholar] [CrossRef]
- Parra, V.; Rothermel, B.A. Calcineurin signaling in the heart: The importance of time and place. J. Mol. Cell. Cardiol. 2017, 103, 121–136. [Google Scholar] [CrossRef] [Green Version]
- Caetano, M.S.; Vieira-De-Abreu, A.; Teixeira, L.K.; Werneck, M.; Barcinski, M.A.; Viola, J. NFATC2 transcription factor regulates cell cycle progression during lymphocyte activation: Evidence of its involvement in the control of cyclin gene expression. FASEB J. 2002, 16, 1940–1942. [Google Scholar] [CrossRef]
- Baksh, S.; Widlund, H.; Frazer-Abel, A.A.; Du, J.; Fosmire, S.; Fisher, D.E.; DeCaprio, J.A.; Modiano, J.; Burakoff, S.J. NFATc2-Mediated Repression of Cyclin-Dependent Kinase 4 Expression. Mol. Cell 2002, 10, 1071–1081. [Google Scholar] [CrossRef]
- Buchholz, M.; Schatz, A.; Wagner, M.; Michl, P.; Linhart, T.; Adler, G.; Gress, T.M.; Ellenrieder, V. Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J. 2006, 25, 3714–3724. [Google Scholar] [CrossRef] [Green Version]
- Leone, G.; DeGregori, J.; Sears, R.; Jakoi, L.; Nevins, J.R. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature 1997, 387, 422–426. [Google Scholar] [CrossRef]
- Neal, J.W.; Clipstone, N.A. A Constitutively Active NFATc1 Mutant Induces a Transformed Phenotype in 3T3-L1 Fibroblasts. J. Biol. Chem. 2003, 278, 17246–17254. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Gu, J.; Ren, Q.; Shi, Y.; Xia, Q.; Wang, J.; Wang, S.; Wang, Y.; Wang, J. NFATC1 promotes cell growth and tumorigenesis in ovarian cancer up-regulating c-Myc through ERK1/2/p38 MAPK signal pathway. Tumour. Biol. 2016, 37, 4493–4500. [Google Scholar] [CrossRef]
- Köenig, A.; Linhart, T.; Schlengemann, K.; Reutlinger, K.; Wegele, J.; Adler, G.; Singh, G.; Hofmann, L.; Kunsch, S.; Büch, T.; et al. NFAT-Induced Histone Acetylation Relay Switch Promotes c-Myc-Dependent Growth in Pancreatic Cancer Cells. Gastroenterology 2010, 138, 1189–1199.e2. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, N.; Kaminuma, O. Isoform-Selective NFAT Inhibitor: Potential Usefulness and Development. Int. J. Mol. Sci. 2021, 22, 2725. [Google Scholar] [CrossRef]
- Sheftic, S.R.; Page, R.; Peti, W. Investigating the human Calcineurin Interaction Network using the piLxVP SLiM. Sci. Rep. 2016, 6, 38920. [Google Scholar] [CrossRef]
- Pallen, C.; Wang, J.H. A multifunctional calmodulin-stimulated phosphatase. Arch. Biochem. Biophys. 1985, 237, 281–291. [Google Scholar] [CrossRef]
- Morioka, M.; Hamada, J.-I.; Ushio, Y.; Miyamoto, E. Potential role of calcineurin for brain ischemia and traumatic injury. Prog. Neurobiol. 1999, 58, 1–30. [Google Scholar] [CrossRef]
- Wei, Q.; Holzer, M.; Brueckner, M.K.; Liu, Y.; Arendt, T. Dephosphorylation of tau protein by calcineurin triturated into neural living cells. Cell. Mol. Neurobiol. 2002, 22, 13–24. [Google Scholar] [CrossRef]
- Liu, Q.; Wilkins, B.J.; Lee, Y.J.; Ichijo, H.; Molkentin, J.D. Direct Interaction and Reciprocal Regulation between ASK1 and Calcineurin-NFAT Control Cardiomyocyte Death and Growth. Mol. Cell. Biol. 2006, 26, 3785–3797. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.-G.; Pathan, N.; Ethell, I.M.; Krajewski, S.; Yamaguchi, Y.; Shibasaki, F.; McKeon, F.; Bobo, T.; Franke, T.F.; Reed, J.C. Ca2+-Induced Apoptosis Through Calcineurin Dephosphorylation of BAD. Science 1999, 284, 339–343. [Google Scholar] [CrossRef]
- Woolfrey, K.M.; Dell’Acqua, M.L. Coordination of Protein Phosphorylation and Dephosphorylation in Synaptic Plasticity. J. Biol. Chem. 2015, 290, 28604–28612. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-C.; Wang, J.-M.; Kikkawa, U.; Mukai, H.; Shen, M.-R.; Morita, I.; Chen, B.-K.; Chang, W.-C. Calcineurin-mediated dephosphorylation of c-Jun Ser-243 is required for c-Jun protein stability and cell transformation. Oncogene 2007, 27, 2422–2429. [Google Scholar] [CrossRef] [Green Version]
- Michod, D.; Bartesaghi, S.; Khelifi, A.; Bellodi, C.; Berliocchi, L.; Nicotera, P.; Salomoni, P. Calcium-Dependent Dephosphorylation of the Histone Chaperone DAXX Regulates H3.3 Loading and Transcription upon Neuronal Activation. Neuron 2012, 74, 122–135. [Google Scholar] [CrossRef] [Green Version]
- Cereghetti, G.M.; Stangherlin, A.; de Brito, O.M.; Chang, C.-R.; Blackstone, C.; Bernardi, P.; Scorrano, L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. USA 2008, 105, 15803–15808. [Google Scholar] [CrossRef] [Green Version]
- Bodmer, D.; Ascaño, M.; Kuruvilla, R. Isoform-Specific Dephosphorylation of Dynamin1 by Calcineurin Couples Neurotrophin Receptor Endocytosis to Axonal Growth. Neuron 2011, 70, 1085–1099. [Google Scholar] [CrossRef] [Green Version]
- Masaki, T.; Habara, M.; Sato, Y.; Goshima, T.; Maeda, K.; Hanaki, S.; Shimada, M. Calcineurin regulates the stability and activity of estrogen receptor α. Proc. Natl. Acad. Sci. USA 2021, 118, 44. [Google Scholar] [CrossRef]
- Sanderson, J.L.; Gorski, J.A.; Dell’Acqua, M.L. NMDA Receptor-Dependent LTD Requires Transient Synaptic Incorporation of Ca2+-Permeable AMPARs Mediated by AKAP150-Anchored PKA and Calcineurin. Neuron 2016, 89, 1000–1015. [Google Scholar] [CrossRef] [Green Version]
- Dougherty, M.K.; Ritt, D.A.; Zhou, M.; Specht, S.I.; Monson, D.M.; Veenstra, T.D.; Morrison, D.K. KSR2 Is a Calcineurin Substrate that Promotes ERK Cascade Activation in Response to Calcium Signals. Mol. Cell 2009, 34, 652–662. [Google Scholar] [CrossRef] [Green Version]
- Goto, S.; Yamamoto, H.; Fukunaga, K.; Iwasa, T.; Matsukado, Y.; Miyamoto, E. Dephosphorylation of Microtubule-Associated Protein 2? Factor, and Tubulin by Calcineurin. J. Neurochem. 1985, 45, 276–283. [Google Scholar] [CrossRef]
- Flavell, S.W.; Cowan, C.W.; Kim, T.-K.; Greer, P.L.; Lin, Y.; Paradis, S.; Griffith, E.C.; Hu, L.S.; Chen, C.; Greenberg, M.E. Activity-Dependent Regulation of MEF2 Transcription Factors Suppresses Excitatory Synapse Number. Science 2006, 311, 1008–1012. [Google Scholar] [CrossRef] [Green Version]
- Kolozsvári, B.; Bakó, É.; Bécsi, B.; Kiss, A.; Czikora, Á.; Tóth, A.; Vámosi, G.; Gergely, P.; Erdődi, F. Calcineurin regulates endothelial barrier function by interaction with and dephosphorylation of myosin phosphatase. Cardiovasc. Res. 2012, 96, 494–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brun, M.; Glubrecht, D.D.; Baksh, S.; Godbout, R. Calcineurin Regulates Nuclear Factor I Dephosphorylation and Activity in Malignant Glioma Cell Lines. J. Biol. Chem. 2013, 288, 24104–24115. [Google Scholar] [CrossRef] [Green Version]
- Chow, C.-W.; Dong, C.; Flavell, R.A.; Davis, R.J. c-Jun NH2-Terminal Kinase Inhibits Targeting of the Protein Phosphatase Calcineurin to NFATc1. Mol. Cell. Biol. 2000, 20, 5227–5234. [Google Scholar] [CrossRef] [Green Version]
- Okamura, H.; Aramburu, J.; Garcia-Rodriguez, C.; Viola, J.; Raghavan, A.; Tahiliani, M.; Zhang, X.; Qin, J.; Hogan, P.G.; Rao, A. Concerted Dephosphorylation of the Transcription Factor NFAT1 Induces a Conformational Switch that Regulates Transcriptional Activity. Mol. Cell 2000, 6, 539–550. [Google Scholar] [CrossRef]
- Kim, H.B.; Kumar, A.; Wang, L.; Liu, G.H.; Keller, S.R.; Lawrence, J.C.; Finck, B.N.; Harris, T.E. Lipin 1 represses NFATc4 transcriptional activity in adipocytes to inhibit secretion of inflammatory factors. Mol. Cell. Biol. 2010, 30, 3126–3139. [Google Scholar] [CrossRef] [Green Version]
- Hendus-Altenburger, R.; Wang, X.; Sjogaard-Frich, L.M.; Pedraz-Cuesta, E.; Sheftic, S.R.; Bendsoe, A.H.; Page, R.; Kragelund, B.B.; Pedersen, S.F.; Peti, W. Molecular basis for the binding and selective dephosphorylation of Na(+)/H(+) exchanger 1 by calcineurin. Nat. Commun. 2019, 10, 3489. [Google Scholar] [CrossRef]
- Blumenthal, D.K.; Takio, K.; Hansen, R.S.; Krebs, E.G. Dephosphorylation of cAMP-dependent protein kinase regulatory subunit (type II) by calmodulin-dependent protein phosphatase. Determinants of substrate specificity. J. Biol. Chem. 1986, 261, 8140–8145. [Google Scholar] [CrossRef]
- Li, Y.; Sheftic, S.R.; Grigoriu, S.; Schwieters, C.D.; Page, R.; Peti, W. The structure of the RCAN1:CN complex explains the inhibition of and substrate recruitment by calcineurin. Sci. Adv. 2020, 6, eaba3681. [Google Scholar] [CrossRef]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R.; Montefusco, S.; Scotto-Rosato, A.; Prezioso, C.; Forrester, A.; et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czirják, G.; Tóth, Z.E.; Enyedi, P. The Two-pore Domain K+ Channel, TRESK, Is Activated by the Cytoplasmic Calcium Signal through Calcineurin. J. Biol. Chem. 2004, 279, 18550–18558. [Google Scholar] [CrossRef] [Green Version]
- Nishi, A.; Snyder, G.L.; Nairn, A.; Greengard, P. Role of Calcineurin and Protein Phosphatase-2A in the Regulation of DARPP-32 Dephosphorylation in Neostriatal Neurons. J. Neurochem. 2008, 72, 2015–2021. [Google Scholar] [CrossRef]
- Sugimoto, T.; Stewart, S.; Guan, K.-L. The Calcium/Calmodulin-dependent Protein Phosphatase Calcineurin Is the Major Elk-1 Phosphatase. J. Biol. Chem. 1997, 272, 29415–29418. [Google Scholar] [CrossRef] [Green Version]
- Dawson, T.M.; Steiner, J.P.; Dawson, V.L.; Dinerman, J.L.; Uhl, G.R.; Snyder, S.H. Immunosuppressant FK506 enhances phosphorylation of nitric oxide synthase and protects against glutamate neurotoxicity. Proc. Natl. Acad. Sci. USA 1993, 90, 9808–9812. [Google Scholar] [CrossRef] [Green Version]
- Alao, J.P. The regulation of cyclin D1 degradation: Roles in cancer development and the potential for therapeutic invention. Mol. Cancer 2007, 6, 24. [Google Scholar] [CrossRef] [Green Version]
- Russell, A.F.; Thompson, M.A.; Hendley, J.; Trute, L.; Armes, J.E.; Germain, D.S. Cyclin D1 and D3 associate with the SCF complex and are coordinately elevated in breast cancer. Oncogene 1999, 18, 1983–1991. [Google Scholar] [CrossRef] [Green Version]
- Lara-Pezzi, E.; Winn, N.; Paul, A.; Mccullagh, K.; Slominsky, E.; Santini, M.P.; Mourkioti, F.; Sarathchandra, P.; Fukushima, S.; Suzuki, K.; et al. A naturally occurring calcineurin variant inhibits FoxO activity and enhances skeletal muscle regeneration. J. Cell Biol. 2007, 179, 1205–1218. [Google Scholar] [CrossRef]
- Quang, C.T.; Leboucher, S.; Passaro, D.; Fuhrmann, L.; Nourieh, M.; Vincent-Salomon, A.; Ghysdael, J. The calcineurin/NFAT pathway is activated in diagnostic breast cancer cases and is essential to survival and metastasis of mammary cancer cells. Cell Death Dis. 2015, 6, e1658. [Google Scholar] [CrossRef]
- Minami, T.; Jiang, S.; Schadler, K.; Suehiro, J.; Osawa, T.; Oike, Y.; Miura, M.; Naito, M.; Kodama, T.; Ryeom, S. The Calcineurin-NFAT-Angiopoietin-2 Signaling Axis in Lung Endothelium Is Critical for the Establishment of Lung Metastases. Cell Rep. 2013, 4, 709–723. [Google Scholar] [CrossRef] [Green Version]
- Manda, K.R.; Tripathi, P.; His, A.C.; Ning, J.; Ruzinova, M.B.; Liapis, H.; Bailey, M.H.; Zhang, H.; Maher, C.A.; Humphrey, P.A.; et al. NFATc1 promotes prostate tumorigenesis and overcomes PTEN loss-induced senescence. Oncogene 2016, 35, 3282–3292. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Kang, X.; Cao, S.; Cheng, H.; Wang, D.; Geng, J. Calcineurin/NFATc1 Pathway Contributes to Cell Proliferation in Hepatocellular Carcinoma. Dig. Dis. Sci. 2012, 57, 3184–3188. [Google Scholar] [CrossRef]
- Tie, X.; Han, S.; Meng, L.; Wang, Y.; Wu, A. NFAT1 Is Highly Expressed in, and Regulates the Invasion of, Glioblastoma Multiforme Cells. PLoS ONE 2013, 8, e66008. [Google Scholar] [CrossRef] [Green Version]
- Shoshan, E.; Braeuer, R.R.; Kamiya, T.; Mobley, A.K.; Huang, L.; Vasquez, M.E.; Velazquez-Torres, G.; Chakravarti, N.; Ivan, C.; Prieto, V.; et al. NFAT1 Directly Regulates IL8 and MMP3 to Promote Melanoma Tumor Growth and Metastasis. Cancer Res. 2016, 76, 3145–3155. [Google Scholar] [CrossRef] [Green Version]
- Gachet, S.; Genescà, E.; Passaro, D.; Irigoyen, M.; Alcalde, H.; Clemenson, C.; Poglio, S.; Pflumio, F.; Janin, A.; Lasgi, C.; et al. Leukemia-initiating cell activity requires calcineurin in T-cell acute lymphoblastic leukemia. Leukemia 2013, 27, 2289–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medyouf, H.; Alcalde, H.; Berthier, C.; Guillemin, M.C.; dos Santos, N.R.; Janin, A.; Decaudin, D.; De Thé, H.; Ghysdael, J. Targeting calcineurin activation as a therapeutic strategy for T-cell acute lymphoblastic leukemia. Nat. Med. 2007, 13, 736–741. [Google Scholar] [CrossRef]
- Marafiot, T.; Pozzobon, M.; Hansmann, M.-L.; Ventura, R.; Pileri, S.A.; Roberton, H.; Gesk, S.; Gaulard, P.; Barth, T.F.E.; Du, M.Q.; et al. The NFATc1 transcription factor is widely expressed in white cells and translocates from the cytoplasm to the nucleus in a subset of human lymphomas. Br. J. Haematol. 2005, 128, 333–342. [Google Scholar] [CrossRef]
- Pham, L.V.; Tamayo, A.T.; Yoshimura, L.C.; Lin-Lee, Y.C.; Ford, R.J. Constitutive NF-kappaB and NFAT activation in aggressive B-cell lymphomas synergistically activates the CD154 gene and maintains lymphoma cell survival. Blood 2005, 106, 3940–3947. [Google Scholar] [CrossRef]
- Xin, B.; Ji, K.Q.; Liu, Y.S.; Zhao, X.D. NFAT Overexpression Correlates with CA72-4 and Poor Prognosis of Ovarian Clear-Cell Carcinoma Subtype. Reprod. Sci. 2021, 28, 745–756. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Liu, L.; Cui, Z.; Shi, R.; Hou, J.; Liu, Z.; Yang, L.; Wang, L.; Li, Y. NFAT2 overexpression suppresses the malignancy of hepatocellular carcinoma through inducing Egr2 expression. BMC Cancer 2020, 20, 966. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Shu, P.; Zou, S.; Shen, X.; Qu, Y.; Zhang, Y.; Sun, K.; Zhang, J. NFATc1 is a tumor suppressor in hepatocellular carcinoma and induces tumor cell apoptosis by activating the FasL-mediated extrinsic signaling pathway. Cancer Med. 2018, 7, 4701–4717. [Google Scholar] [CrossRef] [Green Version]
- Akimzhanov, A.; Krenacs, L.; Schlegel, T.; Klein-Hessling, S.; Bagdi, E.; Stelkovics, E.; Kondo, E.; Chuvpilo, S.; Wilke, P.; Avots, A.; et al. Epigenetic Changes and Suppression of the Nuclear Factor of Activated T Cell 1 (NFATC1) Promoter in Human Lymphomas with Defects in Immunoreceptor Signaling. Am. J. Pathol. 2008, 172, 215–224. [Google Scholar] [CrossRef] [Green Version]
- Fruman, D.A.; Pai, S.Y.; Burakoff, S.J.; Bierer, B.E. Characterization of a mutant calcineurin A alpha gene expressed by EL4 lymphoma cells. Mol. Cell. Biol. 1995, 15, 3857–3863. [Google Scholar] [CrossRef] [Green Version]
- Gross, K.L.; Cioffi, E.A.; Scammell, J.G. Increased Activity of the Calcineurin–Nuclear Factor of Activated T Cells Pathway In Squirrel Monkey B-Lymphoblasts Identified By Powerblot™. In Vitro Cell. Dev. Biol. Anim. 2004, 40, 57–63. [Google Scholar] [CrossRef]
- Lehen’Kyi, V.; Flourakis, M.; Skryma, R.; Prevarskaya, N. TRPV6 channel controls prostate cancer cell proliferation via Ca2+/NFAT-dependent pathways. Oncogene 2007, 26, 7380–7385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fixemer, T.; Wissenbach, U.; Flockerzi, V.; Bonkhoff, H. Expression of the Ca2+-selective cation channel TRPV6 in human prostate cancer: A novel prognostic marker for tumor progression. Oncogene 2003, 22, 7858–7861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Abel, M.; Hoenderop, J.G.J.; Bindels, R.J.M. The epithelial calcium channels TRPV5 and TRPV6: Regulation and implications for disease. Naunyn-Schmiedebergs Arch. Exp. Pathol. Pharmakol. 2005, 371, 295–306. [Google Scholar] [CrossRef] [Green Version]
- Furman, J.L.; Norris, C.M. Calcineurin and glial signaling: Neuroinflammation and beyond. J. Neuroinflamm. 2014, 11, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musson, R.E.; Smit, N.P. Regulatory mechanisms of calcineurin phosphatase activity. Curr. Med. Chem. 2011, 18, 301–315. [Google Scholar] [CrossRef]
- Kawahara, T.; Kashiwagi, E.; Ide, H.; Li, Y.; Zheng, Y.; Ishiguro, H.; Miyamoto, H. The role of NFATc1 in prostate cancer progression: Cyclosporine A and tacrolimus inhibit cell proliferation, migration, and invasion. Prostate 2015, 75, 573–584. [Google Scholar] [CrossRef]
- Peuker, K.; Muff, S.; Wang, J.; Kunzel, S.; Bosse, E.; Zeissig, Y.; Luzzi, G.; Basic, M.; Strigli, A.; Ulbricht, A.; et al. Epithelial calcineurin controls microbiota-dependent intestinal tumor development. Nat. Med. 2016, 22, 506–515. [Google Scholar] [CrossRef] [Green Version]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Nag, S.; Wang, W.; Zhou, J.; Zhang, W.-D.; Wang, H.; Zhang, R. NFAT as cancer target: Mission possible? Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1846, 297–311. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, T.; Kashiwagi, E.; Ide, H.; Li, Y.; Zheng, Y.; Miyamoto, Y.; Netto, G.J.; Ishiguro, H.; Miyamoto, H. Cyclosporine A and tacrolimus inhibit bladder cancer growth through down-regulation of NFATc1. Oncotarget 2015, 6, 1582–1593. [Google Scholar] [CrossRef] [Green Version]
- Siamakpour-Reihani, S.; Caster, J.; Bandhu Nepal, D.; Courtwright, A.; Hilliard, E.; Usary, J.; Ketelsen, D.; Darr, D.; Shen, X.J.; Patterson, C.; et al. The Role of Calcineurin/NFAT in SFRP2 Induced Angiogenesis—A Rationale for Breast Cancer Treatment with the Calcineurin Inhibitor Tacrolimus. PLoS ONE 2011, 6, e20412. [Google Scholar] [CrossRef] [Green Version]
- Hojo, M.; Morimoto, T.; Maluccio, M.; Asano, T.; Morimoto, K.; Lagman, M.; Shimbo, T.; Suthanthiran, M. Cyclosporine induces cancer progression by a cell-autonomous mechanism. Nature 1999, 397, 530–534. [Google Scholar] [CrossRef]
- Datta, D.; Contreras, A.; Basu, A.; Dormond, O.; Flynn, E.; Briscoe, D.; Pal, S. Calcineurin Inhibitors Activate the Proto-Oncogene Ras and Promote Protumorigenic Signals in Renal Cancer Cells. Cancer Res. 2009, 69, 8902–8909. [Google Scholar] [CrossRef] [Green Version]
- Han, W.; Ming, M.; He, T.-C.; He, Y.-Y. Immunosuppressive Cyclosporin A Activates AKT in Keratinocytes through PTEN Suppression: Implications In Skin Carcinogenesis. J. Biol. Chem. 2010, 285, 11369–11377. [Google Scholar] [CrossRef] [Green Version]
- Cui, C.; Merritt, R.; Fu, L.; Pan, Z. Targeting calcium signaling in cancer therapy. Acta Pharm. Sin. B 2017, 7, 3–17. [Google Scholar] [CrossRef]
- Pusl, T.; Wu, J.J.; Zimmerman, T.L.; Zhang, L.; Ehrlich, B.E.; Berchtold, M.W.; Hoek, J.B.; Karpen, S.J.; Nathanson, M.H.; Bennett, A.M. Epidermal Growth Factor-mediated Activation of the ETS Domain Transcription Factor Elk-1 Requires Nuclear Calcium. J. Biol. Chem. 2002, 277, 27517–27527. [Google Scholar] [CrossRef] [Green Version]
- Andrade, L.M.; Geraldo, J.M. Nucleoplasmic Calcium Buffering Sensitizes Human Squamous Cell Carcinoma to Anticancer Therapy. J. Cancer Sci. Ther. 2012, 4, 5. [Google Scholar] [CrossRef]
| Calcineurin Substrates | Dephosphorylation Sites | Reaction by Dephosphorylation | Reference |
|---|---|---|---|
| 12E8 | Ser262, Ser356 | ND | [124] |
| ASK1 | Ser967 | Promotes the dissociation of ASK1 from the 14-3-3 protein, resulting in the activation of ASK1 | [125] |
| AT270 | Thr181 | ND | [124] |
| BAD | Ser155 | Promotes heterodimerization of BAD and Bcl-xL, which induces apoptosis | [126] |
| CaMKIIγ | Ser334 | Translocates CaMKIIγ to the nucleus | [127] |
| c-Jun | Ser243 | Stabilizes c-Jun, promotes the interaction between c-Jun and Sp1 | [128] |
| Cyclin D1 | Thr286 | Stabilizes cyclin D1, inducing G1/S progression | [38] |
| DAXX | Ser669 | Promotes H3.3 uptake by DAXX | [129] |
| DRP1 | Ser637 | Splites the organelle by Drp1 | [130] |
| Dynamin 1 | Ser774, Ser778 | Promotes endocytosis of TrkA receptors and axonal growth | [131] |
| ERα | Ser294 | Stabilizes ERα and promotes the activity of ERα | [132] |
| GluA1 | Ser845 | Promotes removal of AMPARs from synapses and endocytosis | [133] |
| KSR2 | Ser198, Thr287, Ser310 | Activates ERK and induces membrane localization of KSR2 | [134] |
| MAP2 | ND | [135] | |
| MEF2A | Ser221, Ser255, Ser408 | Activates MEF2A and promotes the change from sumoylation to acetylation of Leu403 | [136] |
| MYPT1 | Thr696 | Affects actin polymerization by activating MP and improves endothelial barrier function | [137] |
| NF1 | ND | Activates transcription | [138] |
| NFATC1 | Ser172 | Promotes nuclear transfer of NFATC1 | [139] |
| NFATC2 * | Five residues among following sites, Ser170, Ser173, Ser174, Ser176, Ser177, Ser179, Ser182, in SRR-1 domain | Promotes nuclear transfer of NFATC2 | [140] |
| Ser215, Ser219, Ser223 in SP-2 domain | |||
| Ser270, Ser276, Ser278, Ser282 in SP-3 domain | |||
| Ser328 in KTS motif | |||
| NFATC4 | Ser170 | Promotes nuclear transfer of NFATC4 | [141] |
| NHE1 | Thr779 | Inhibits NHE1 activity | [142] |
| PHF1 | Ser396, Ser404 | ND | [124] |
| RIIα | Ser95 | ND | [143] |
| RCAN1 | Ser108, Ser112, Thr124, Thr192 | ND | [144] |
| Tau1 | Ser199, Ser202 | ND | [124] |
| TFEB | Ser142, Ser211 | Promotes nuclear transfer of TFEB | [145] |
| TRESK | Ser276 | Increases K+ current, decreases channel responsiveness to calcium signals | [146] |
| Factor | Alterations in Cancer | Types of Cancer | Reference |
|---|---|---|---|
| Calcineurin (CnA) | Overexpression | glioma (malignant gliomas, including grades III and IV astrocytomas) | [138] |
| breast cancer (ER-α–positive) | [132] | ||
| Activation | lymphomas (lymphoid malignancies) | [160] | |
| Overexpression, activation | colon cancer | [39] | |
| NFATc1 | Overexpression | ovarian cancer (clear-cell carcinoma) | [118,163] |
| liver cancer (hepatocellular carcinoma) | [156] | ||
| prostate cancer | [155] | ||
| lymphomas (large B-cell lymphoma) | [162] | ||
| Nuclear localization | lymphomas (diffuse large B-cell lymphomas) | [161] | |
| breast cancer (triple-negative) | [153] | ||
| Suppression | liver cancer (hepatocellular carcinoma) | [164,165] | |
| lymphomas (anaplastic large cell lymphomas and classical Hodgkin’s lymphomas) | [166] | ||
| Overexpression, nuclear localization | pancreatic cancer (pancreatic adenocarcinoma) | [115] | |
| NFATc2 | Overexpression | melanoma | [158] |
| glioma (glioblastoma) | [157] | ||
| Nuclear localization | lung cancer | [154] | |
| NFATc1, NFATc3 | Dephosphorylation | leukemia (T-cell acute lymphoblastic leukemia) | [159] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masaki, T.; Shimada, M. Decoding the Phosphatase Code: Regulation of Cell Proliferation by Calcineurin. Int. J. Mol. Sci. 2022, 23, 1122. https://doi.org/10.3390/ijms23031122
Masaki T, Shimada M. Decoding the Phosphatase Code: Regulation of Cell Proliferation by Calcineurin. International Journal of Molecular Sciences. 2022; 23(3):1122. https://doi.org/10.3390/ijms23031122
Chicago/Turabian StyleMasaki, Takahiro, and Midori Shimada. 2022. "Decoding the Phosphatase Code: Regulation of Cell Proliferation by Calcineurin" International Journal of Molecular Sciences 23, no. 3: 1122. https://doi.org/10.3390/ijms23031122
APA StyleMasaki, T., & Shimada, M. (2022). Decoding the Phosphatase Code: Regulation of Cell Proliferation by Calcineurin. International Journal of Molecular Sciences, 23(3), 1122. https://doi.org/10.3390/ijms23031122






