Abiotic Stress and Belowground Microbiome: The Potential of Omics Approaches
Abstract
1. Introduction
2. Plant Responses to Abiotic Stresses
3. Role of Soil Microbes in Interaction with Plants under Abiotic Stress Conditions
3.1. Arbuscular Mycorrhizal Fungi
3.2. Actinomycetes
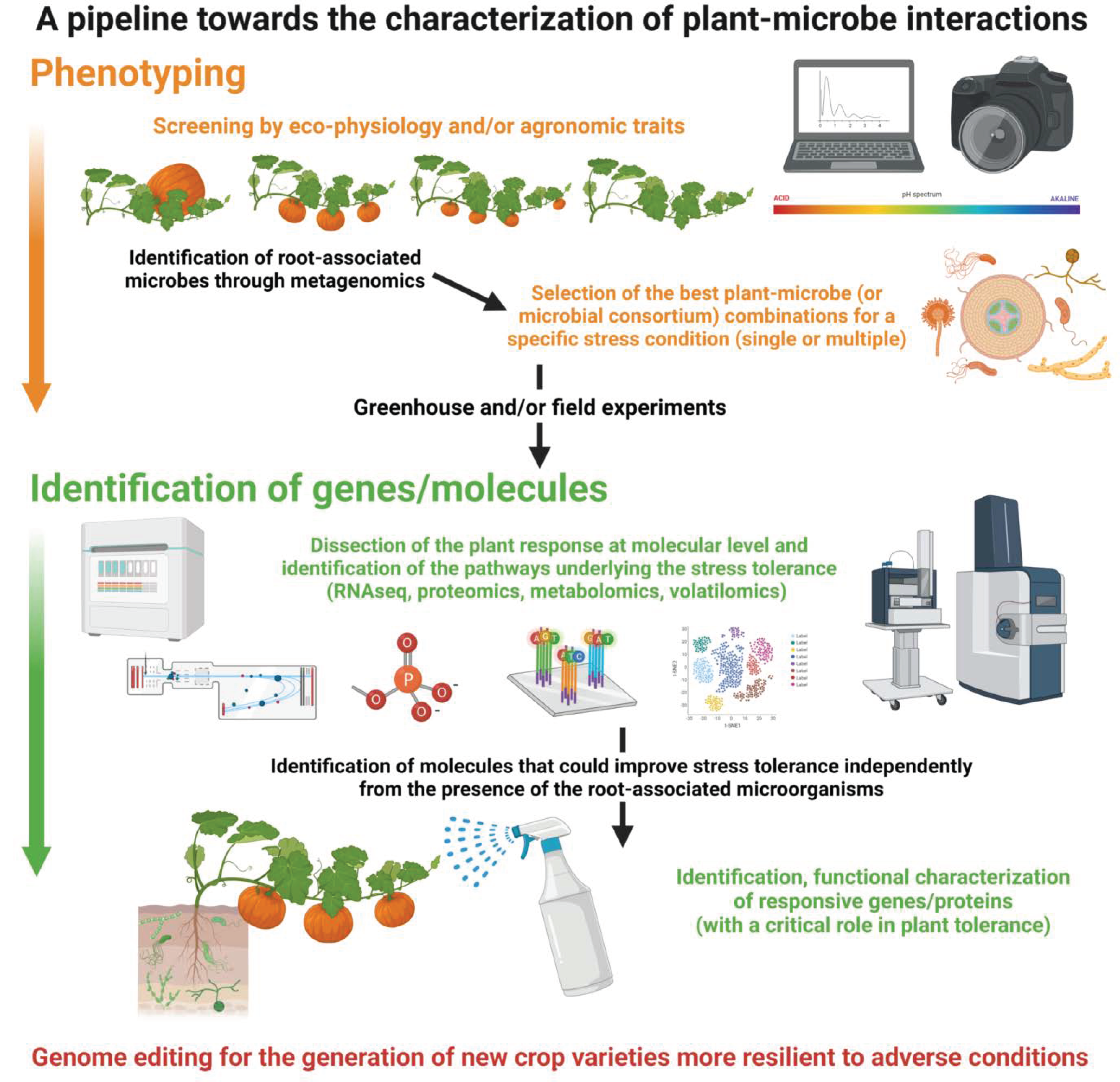
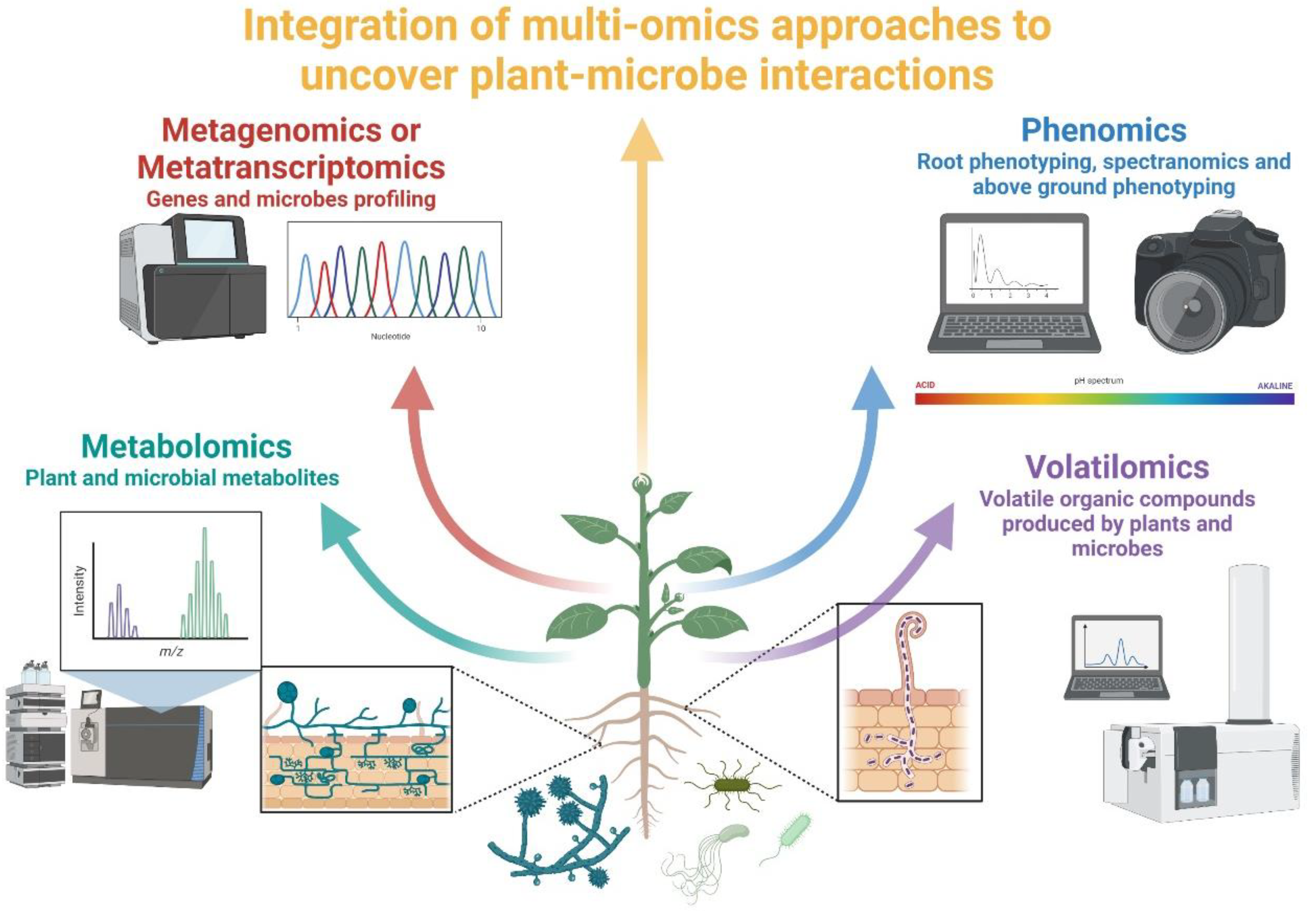
4. Diversity and Complementarity of Omics Approaches in Studying Plant–Microbe Interactions
4.1. Metagenomics vs. Metabarcoding
4.2. Meta-Transcriptomics and Meta-Proteomics
4.3. Metabolomics
4.4. Non-Destructive Approaches (Phenomics)
4.4.1. Volatilomics
4.4.2. Shovelomics
4.4.3. Spectranomics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berg, G.; Rybakova, D.; Grube, M.; Köberl, M. The plant microbiome explored: Implications for experimental botany. J. Exp. Bot. 2016, 67, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Sudheer, S.; Usmani, Z.; Rani, R.; Gupta, P. Deciphering the omics of plant-microbe interaction: Perspectives and new insights. Curr. Genom. 2020, 21, 343–362. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S. Disentangling the factors shaping microbiota composition across the plant holobiont. New Phytol. 2016, 209, 454–457. [Google Scholar] [CrossRef]
- Beattie, G.A. Microbiomes: Curating communities from plants. Nature 2015, 528, 340–341. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Campisano, A.; Antonielli, L.; Pancher, M.; Yousaf, S.; Pindo, M.; Pertot, I. Bacterial endophytic communities in the grapevine depend on pest management. PLoS ONE 2014, 9, e112763. [Google Scholar] [CrossRef]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Chialva, M.; Salvioli, A.; Daghino, S.; Ghignone, S.; Bagnaresi, P.; Chiapello, M.; Novero, M.; Spadaro, D.; Perotto, S.; Bonfante, P. Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytol. 2018, 220, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Gouda, S.; Kerry, R.G.; Das, G.; Paramithiotis, S.; Shin, H.S.; Patra, J.K. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 2018, 206, 131–140. [Google Scholar] [CrossRef]
- Berruti, A.; Lumini, E.; Balestrini, R.; Bianciotto, V. Arbuscular mycorrhizal fungi as natural biofertilizers: Let’s benefit from past successes. Front. Microbiol. 2016, 6, 1559. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis; Academic Press: London, UK, 2008. [Google Scholar]
- Balestrini, R.; Lumini, E. Focus on mycorrhizal symbioses. Appl. Soil Ecol. 2018, 123, 299–304. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef] [PubMed]
- Olanrewaju, O.S.; Babalola, O.O. Streptomyces: Implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 2019, 103, 1179–1188. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Chukwuneme, C.F.; Babalola, O.O.; Kutu, F.R.; Ojuederie, O.B. Characterization of actinomycetes isolates for plant growth promoting traits and their effects on drought tolerance in maize. J. Plant Interact. 2020, 15, 93–105. [Google Scholar] [CrossRef]
- Nimnoi, P.; Pongsilp, N.; Lumyong, S. Endophytic actinomycetes isolated from Aquilaria crassna Pierre ex Lec and screening of plant growth promoters production. World J. Microbiol. Biotechnol. 2010, 26, 193–203. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Lohani, N.; Jain, D.; Singh, M.B.; Bhalla, P.L. Engineering multiple abiotic Stress tolerance in canola, Brassica napus. Front. Plant Sci. 2020, 11, 3. [Google Scholar] [CrossRef]
- Trenberth, K.E.; Dai, A.; Van Der Schrier, G.; Jones, P.D.; Barichivich, J.; Briffa, K.R.; Sheffield, J. Global warming and changes in drought. Nat. Clim. Chang. 2014, 4, 17. [Google Scholar] [CrossRef]
- Evelin, H.; Devi, T.S.; Gupta, S.; Kapoor, R. Mitigation of salinity stress in plants by arbuscular mycorrhizal symbiosis: Current understanding and new challenges. Front. Plant Sci. 2019, 10, 470. [Google Scholar] [CrossRef] [PubMed]
- Nephali, L.; Piater, L.A.; Dubery, I.A.; Patterson, V.; Huyser, J.; Burgess, K.; Tugizimana, F. Biostimulants for plant growth and mitigation of abiotic stresses: A metabolomics perspective. Metabolites 2020, 10, 505. [Google Scholar] [CrossRef]
- Westman, S.M.; Kloth, K.J.; Hanson, J.; Ohlsson, A.B.; Albrectsen, B.R. Defense priming in Arabidopsis—A Meta-Analysis. Sci. Rep. 2019, 9, 13309. [Google Scholar] [CrossRef]
- Ruelland, E.; Zachowski, A. How plants sense temperature. Environ. Exp. Bot. 2010, 69, 225–232. [Google Scholar] [CrossRef]
- Martinez-Medina, A.; Flors, V.; Heil, M.; Mauch-mani, B.; Pieterse, C.M.J. Recognizing plant defense priming. Trends Microbiol. 2016, 21, 818–822. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Nahar, K.; Hasanuzzaman, M.; Fujita, M. Roles of osmolytes in plant adaptation to drought and salinity. In Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies; Iqbal, N., Nazar, R., Eds.; Springer: Chennai, India, 2015; pp. 1–170. ISBN 9788132226161. [Google Scholar]
- Plett, J.M.; Martin, F.M. Know your enemy, embrace your friend: Using omics to understand how plants respond differently to pathogenic and mutualistic microorganisms. Plant J. 2018, 93, 729–746. [Google Scholar] [CrossRef] [PubMed]
- Balestrini, R.; Chitarra, W.; Fotopoulos, V.; Ruocco, M. Potential Role of Beneficial Soil Microorganisms in Plant Tolerance to Abiotic Stress Factors; Lukac, M., Grenni, P., Gamboni, M., Eds.; Springer: Cham, Switzerland, 2017; Chapter 12. [Google Scholar] [CrossRef]
- Antoine, S.; Hériché, M.; Boussageon, R.; Noceto, P.A.; van Tuinen, D.; Wipf, D.; Courty, P.E. A historical perspective on mycorrhizal mutualism emphasizing arbuscular mycorrhizas and their emerging challenges. Mycorrhiza 2021, 31, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Qin, C.; Ahanger, M.A.; Raza, S.; Khan, M.I.; Ashraf, M.; Ahmed, N.; Zhang, L. Role of arbuscular mycorrhizal fungi in plant growth regulation: Implications in abiotic stress tolerance. Front. Plant Sci. 2019, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, M.; Boopathi, T.; Manivannan, P. Comprehensive assessment of ameliorative effects of AMF in alleviating abiotic stress in tomato plants. J. Fungi 2021, 7, 303. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T. Plant proteome responses to salinity stress—Comparison of glycophytes and halophytes. Funct. Plant Biol. 2013, 40, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Peng, F.; Tedeschi, A.; Xue, X.; Wang, T.; Liao, J.; Zhang, W.; Huang, C. Do halophytes and glycophytes differ in their interactions with arbuscular mycorrhizal fungi under salt stress? A meta-analysis. Bot. Stud. 2020, 61, 13. [Google Scholar] [CrossRef]
- Augé, R.M.; Toler, H.D.; Saxton, A.M. Arbuscular mycorrhizal symbiosis and osmotic adjustment in response to NaCl stress: A meta-analysis. Front. Plant Sci. 2014, 5, 562. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, M.; Boughattas, S.; Hu, S.; Oh, S.H.; Sa, T. A meta-analysis of arbuscular mycorrhizal effects on plants grown under salt stress. Mycorrhiza 2014, 24, 611–625. [Google Scholar] [CrossRef]
- Evelin, H.; Giri, B.; Kapoor, R. Ultrastructural evidence for AMF mediated salt stress mitigation in Trigonella foenum-graecum. Mycorrhiza 2013, 23, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Evelin, H.; Kapoor, R.; Giri, B. Arbuscular mycorrhizal fungi in alleviation of salt stress: A review. Ann. Bot. 2009, 104, 1263–1280. [Google Scholar] [CrossRef]
- Eroğlu, Ç.G.; Cabral, C.; Ravnskov, S.; Bak Topbjerg, H.; Wollenweber, B. Arbuscular mycorrhiza influences carbon-use efficiency and grain yield of wheat grown under pre- and post-anthesis salinity stress. Plant Biol. 2020, 22, 863–871. [Google Scholar] [CrossRef]
- Pollastri, S.; Savvides, A.; Pesando, M.; Lumini, E.; Volpe, M.G.; Ozudogru, E.A.; Faccio, A.; De Cunzo, F.; Michelozzi, M.; Lambardi, M.; et al. Impact of two arbuscular mycorrhizal fungi on Arundo donax L. response to salt stress. Planta 2018, 247, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Chitarra, W.; Pagliarani, C.; Maserti, B.; Lumini, E.; Siciliano, I.; Cascone, P.; Schubert, A.; Gambino, G.; Balestrini, R.; Guerrieri, E. Insights on the impact of arbuscular mycorrhizal symbiosis on tomato tolerance to water stress. Plant Physiol. 2016, 171, 1009–1023. [Google Scholar] [CrossRef]
- Volpe, V.; Chitarra, W.; Cascone, P.; Volpe, M.G.; Bartolini, P.; Moneti, G.; Pieraccini, G.; Di Serio, C.; Maserti, B.; Guerrieri, E.; et al. The association with two different arbuscular mycorrhizal fungi differently affects water stress tolerance in tomato. Front. Plant Sci. 2018, 9, 1480. [Google Scholar] [CrossRef] [PubMed]
- Rivero, J.; Álvarez, D.; Flors, V.; Azcón-Aguilar, C.; Pozo, M.J. Root metabolic plasticity underlies functional diversity in mycorrhiza-enhanced stress tolerance in tomato. New Phytol. 2018, 220, 1322–1336. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Ahanger, M.A.; Su, Y.; Lei, Y.; Mustafa, N.S.A.; Ahmad, P.; Zhang, L. Improved drought tolerance by AMF inoculation in maize (Zea mays) involves physiological and biochemical implications. Plants 2019, 8, 579. [Google Scholar] [CrossRef] [PubMed]
- Balestrini, R.; Brunetti, C.; Chitarra, W.; Nerva, L. Photosynthetic traits and nitrogen uptake in crops: Which is the role of arbuscular mycorrhizal fungi? Plants 2020, 9, 1105. [Google Scholar] [CrossRef] [PubMed]
- Mathur, S.; Tomar, R.S.; Jajoo, A. Arbuscular mycorrhizal fungi (AMF) protects photosynthetic apparatus of wheat under drought stress. Photosynth. Res. 2019, 139, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.N.; Wu, Q.S.; Kuča, K. Unravelling the role of arbuscular mycorrhizal fungi in mitigating the oxidative burst of plants under drought stress. Plant Biol. 2021, 23, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Druzhinina, I.S.; Pan, X.; Yuan, Z. Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 2016, 34, 1245–1259. [Google Scholar] [CrossRef]
- Güimil, S.; Chang, H.S.; Zhu, T.; Sesma, A.; Osbourn, A.; Roux, C.; Ioannidis, V.; Oakeley, E.J.; Docquier, M.; Descombes, P.; et al. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Natl. Acad. Sci. USA 2005, 102, 8066–8070. [Google Scholar] [CrossRef]
- Fiorilli, V.; Vallino, M.; Biselli, C.; Faccio, A.; Bagnaresi, P.; Bonfante, P. Host and non-host roots in rice: Cellular and molecular approaches reveal differential responses to arbuscular mycorrhizal fungi. Front. Plant Sci. 2015, 6, 636. [Google Scholar] [CrossRef]
- Fiorilli, V.; Catoni, M.; Miozzi, L.; Novero, M.; Accotto, G.P.; Lanfranco, L. Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol. 2009, 184, 975–987. [Google Scholar] [CrossRef]
- Balestrini, R.; Salvioli, A.; Dal Molin, A.; Novero, M.; Gabelli, G.; Papparelli, E.; Marroni, F.; Bonfante, P. Impact of an arbuscular mycorrhizal fungus versus a mixed microbial inoculum on the transcriptome reprogramming of grapevine roots. Mycorrhiza 2017, 27, 417–430. [Google Scholar] [CrossRef]
- Fiorilli, V.; Vannini, C.; Ortolani, F.; Garcia-Seco, D.; Chiapello, M.; Novero, M.; Domingo, G.; Terzi, V.; Morcia, C.; Bagnaresi, P.; et al. Omics approaches revealed how arbuscular mycorrhizal symbiosis enhances yield and resistance to leaf pathogen in wheat. Sci. Rep. 2018, 8, 9625. [Google Scholar] [CrossRef]
- Balestrini, R.; Rosso, L.C.; Veronico, P.; Melillo, M.T.; De Luca, F.; Fanelli, E.; Colagiero, M.; Salvioli, A.; Ciancio, A.; Pentimone, I. Transcriptomic responses to water deficit and nematode infection in mycorrhizal tomato roots. Front. Microbiol. 2019, 10, 1807. [Google Scholar] [CrossRef]
- Grover, M.; Bodhankar, S.; Maheswari, M.; Srinivasarao, C. Actinomycetes as mitigators of climate change and abiotic stress. In Plant Growth Promoting Actinobacteria; Subramaniam, G., Arumugam, S., Rajendran, V., Eds.; Springer: Singapore, 2016; pp. 203–212. [Google Scholar] [CrossRef]
- Yandigeri, M.S.; Meena, K.K.; Singh, D.; Malviya, N.; Singh, D.P.; Solanki, M.K.; Yadav, A.K.; Arora, D.K. Drought-tolerant endophytic Actinobacteria promote growth of wheat (Triticum aestivum) under water stress conditions. Plant Growth Regul. 2012, 68, 411–420. [Google Scholar] [CrossRef]
- Aly, M.M.; El-Sabbagh, S.M.; El-Shouny, W.A.; Ebrahim, M.K.H. Physiological response of Zea mays to NaCl stress with respect to Azotobacter chroococcum and Streptomyces niveus. Pak. J. Biol. Sci. 2003, 6, 2073–2080. [Google Scholar] [CrossRef]
- Palaniyandi, S.A.; Damodharan, K.; Yang, S.H.; Suh, J.W. Streptomyces sp. Strain PGPA39 alleviates salt stress and promotes growth of ‘Micro Tom’ tomato plants. J. Appl. Microbiol. 2014, 117, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, S.; Meguro, A.; Toyoda, K.; Nishimura, T.; Kunoh, H. Drought tolerance of tissue-cultured seedlings of mountain laurel (Kalmia latifolia L.) induced by an endophytic actinomycete II. Acceleration of callose accumulation and lignification. Actinomycetologica 2005, 19, 13–17. [Google Scholar] [CrossRef][Green Version]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The soil microbiome influences grapevine-associated microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Monje, D.; Lundberg, D.S.; Lazarovits, G.; Reis, V.M.; Raizada, M.N. Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil 2016, 405, 337–355. [Google Scholar] [CrossRef]
- White, R.A., III; Rivas-Ubach, A.; Borkum, M.I.; Köberl, M.; Bilbao, A.; Colby, S.M.; Hoyt, D.W.; Bingol, K.; Kim, Y.M.; Wendler, J.P.; et al. The state of rhizospheric science in the era of multi-omics: A practical guide to omics technologies. Rhizosphere 2017, 3, 212–221. [Google Scholar] [CrossRef]
- Alawiye, T.T.; Babalola, O.O. Bacterial diversity and community structure in typical plant rhizosphere. Diversity 2019, 11, 179. [Google Scholar] [CrossRef]
- Priya, P.; Aneesh, B.; Harikrishnan, K. Genomics as a potential tool to unravel the rhizosphere microbiome interactions on plant health. J. Microbiol. Methods 2021, 185, 106215. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.T.; Song, B. Plant adaptation to climate change—Where are we? J. Syst. Evol. 2020, 58, 533–545. [Google Scholar] [CrossRef]
- Palit, P.; Kudapa, H.; Zougmore, R.; Kholova, J.; Whitbread, A.; Sharma, M.; Varshney, R.K. An integrated research framework combining genomics, systems biology, physiology, modelling and breeding for legume improvement in response to elevated CO2 under climate change scenario. Curr. Plant Biol. 2020, 22, 100149. [Google Scholar] [CrossRef]
- Li, Q.; Yan, J. Sustainable agriculture in the era of omics: Knowledge-driven crop breeding. Genome Biol. 2020, 21, 154. [Google Scholar] [CrossRef]
- Levy, A.; Conway, J.M.; Dangl, J.L.; Woyke, T. Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe 2018, 24, 475–485. [Google Scholar] [CrossRef]
- Khatabi, B.; Gharechahi, J.; Ghaffari, M.R.; Liu, D.; Haynes, P.A.; McKay, M.J.; Mirzaei, M.; Salekdeh, G.H. Plant-microbe symbiosis: What has proteomics taught us? Proteomics 2019, 19, e1800105. [Google Scholar] [CrossRef]
- Chaston, J.; Douglas, A.E. Making the most of “omics” for symbiosis research. Biol. Bull. 2012, 223, 21–29. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; Vera-Ponce de León, A.; Sanchez-Flores, A. The road to metagenomics: From microbiology to DNA sequencing technologies and bioinformatics. Front. Genet. 2015, 6, 348. [Google Scholar] [CrossRef]
- Singer, E.; Bushnell, B.; Coleman-Derr, D.; Bowman, B.; Bowers, R.M.; Levy, A.; Gies, E.A.; Cheng, J.F.; Copeland, A.; Klenk, H.P.; et al. High-resolution phylogenetic microbial community profiling. ISME J. 2016, 10, 2020–2032. [Google Scholar] [CrossRef]
- Vogel, T.M.; Hirsch, P.R.; Simonet, P.; Jansson, J.K.; Tiedje, J.M.; van Elsas, J.D.; Nalin, R.; Philippot, L.; Bailey, M.J. Advantages of the metagenomic approach for soil exploration: Reply from Vogel et al. Nat. Rev. Microbiol. 2009, 7, 756–757. [Google Scholar] [CrossRef]
- Solden, L.; Lloyd, K.; Wrighton, K. The bright side of microbial dark matter: Lessons learned from the uncultivated majority. Curr. Opin. Microbiol. 2016, 31, 217–226. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Fungal DNA barcoding. Genome 2016, 59, 913–932. [Google Scholar] [CrossRef] [PubMed]
- Öpik, M.; Metsis, M.; Daniell, T.J.; Zobel, M.; Moora, M. Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol. 2009, 184, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Lumini, E.; Orgiazzi, A.; Borriello, R.; Bonfante, P.; Bianciotto, V. Disclosing arbuscular mycorrhizal fungal biodiversity in soil through a land-use gradient using a pyrosequencing approach. Environ. Microbiol. 2010, 12, 2165–2179. [Google Scholar] [CrossRef]
- Victorino, Í.M.M.; Berruti, A.; Orgiazzi, A.; Voyron, S.; Bianciotto, V.; Lumini, E. High-throughput DNA sequence-based analysis of AMF communities. In Arbuscular Mycorrhizal Fungi. Methods in Molecular Biology; Ferrol, N., Lanfranco, L., Eds.; Humana: New York, NY, USA, 2020; Volume 2146. [Google Scholar] [CrossRef]
- Zepeda Mendoza, M.L.; Sicheritz-Pontén, T.; Gilbert, M.T. Environmental genes and genomes: Understanding the differences and challenges in the approaches and software for their analyses. Brief. Bioinform. 2015, 16, 745–758. [Google Scholar] [CrossRef]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Põlme, S.; Riit, T.; Liiv, I.; Kõljalg, U.; Kisand, V.; Nilsson, R.H.; Hildebrand, F.; et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Crandall, S.G.; Gold, K.M.; Jiménez-Gasco, M.D.M.; Filgueiras, C.C.; Willett, D.S. A multi-omics approach to solving problems in plant disease ecology. PLoS ONE 2020, 15, e0237975. [Google Scholar] [CrossRef]
- Kuske, C.R.; Hesse, C.N.; Challacombe, J.F.; Cullen, D.; Herr, J.R.; Mueller, R.C.; Tsang, A.; Vilgalys, R. Prospects and challenges for fungal metatranscriptomics of complex communities. Fungal Ecol. 2015, 14, 133–137. [Google Scholar] [CrossRef]
- Bastida, F.; Hernández, T.; García, C. Metaproteomics of soils from semiarid environment: Functional and phylogenetic information obtained with different protein extraction methods. J. Proteom. 2014, 101, 31–42. [Google Scholar] [CrossRef]
- Wilmes, P.; Bond, P.L. The application of twodimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ. Microbiol. 2004, 6, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Marcelino, V.R.; Irinyi, L.; Eden, J.S.; Meyer, W.; Holmes, E.C.; Sorrell, T.C. Metatranscriptomics as a tool to identify fungal species and subspecies in mixed communities—A proof of concept under laboratory conditions. IMA Fungus 2019, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Chialva, M.; Ghignone, S.; Novero, M.; Hozzein, W.N.; Lanfranco, L.; Bonfante, P. Tomato RNA-seq data mining reveals the taxonomic and functional diversity of root-associated microbiota. Microorganisms 2019, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Chiapello, M.; Zampieri, E.; Mello, A. A small effort for researchers, a big gain for soil metaproteomics. Front. Microbiol. 2020, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Alonso, E.; Pena-Perez, S.; Serrano, S.; Garcia-Lopez, E.; Alcazar, A.; Cid, C. Taxonomic and functional characterization of a microbial community from a volcanic englacial ecosystem in Deception Island, Antarctica. Sci. Rep. 2019, 9, 12158. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, E.; Chiapello, M.; Daghino, S.; Bonfante, P.; Mello, A. Soil metaproteomics reveals an inter-kingdom stress response to the presence of black truffles. Sci. Rep. 2016, 6, 25773. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Jarnuczak, A.F.; Ternent, T.; Brazma, A.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Liu, X.; Locasale, J.W. Metabolomics: A primer. Trends Biochem. Sci. 2017, 42, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 30–32. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies—Challenges and emerging directions. J. Am. Soc. Mass. Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Rivero, J.; Gamir, J.; Pozo, M.J.; Flors, V. Metabolic transition in mycorrhizal tomato roots. Front. Microbiol. 2015, 6, 598. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Metabolomics in plant priming research: The way forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef]
- Bernardo, L.; Carletti, P.; Badeck, F.W.; Rizza, F.; Morcia, C.; Ghizzoni, R.; Rouphael, Y.; Colla, G.; Terzi, V.; Lucini, L. Metabolomic responses triggered by arbuscular mycorrhiza enhance tolerance to water stress in wheat cultivars. Plant Physiol. Biochem. 2019, 137, 203–212. [Google Scholar] [CrossRef]
- Rivero, J.; Lidoy, J.; Llopis-Giménex, A.; Herrero, S.; Flors, V.; Pozo, M.J. Mycorrhizal symbiosis primes the accumulation of antiherbivore compounds and enhances herbivore mortality in tomato. J. Exp. Bot. 2021, 72, 5038–5050. [Google Scholar] [CrossRef]
- Escudero-Martinez, C.; Bulgarelli, D. Tracing the evolutionary routes of plant-microbiota interactions. Curr. Opin. Microbiol. 2019, 49, 34–40. [Google Scholar] [CrossRef]
- Iannucci, A.; Fragasso, M.; Beleggia, R.; Nigro, F.; Papa, R. Evolution of the crop rhizosphere: Impact of domestication on root exudates in tetraploid wheat (Triticum turgidum L.). Front. Plant Sci. 2017, 8, 2124. [Google Scholar] [CrossRef]
- Fiorani, F.; Schurr, U. Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef] [PubMed]
- Tardieu, F.; Cabrera-Bosquet, L.; Pridmore, T.; Bennett, M. Plant phenomics, from sensors to knowledge. Curr. Biol. 2017, 27, R770–R783. [Google Scholar] [CrossRef]
- Costa, C.; Schurr, U.; Loreto, F.; Menesatti, P.; Carpentier, S. Plant phenotyping research trends, a science mapping approach. Front. Plant Sci. 2019, 9, 1933. [Google Scholar] [CrossRef]
- Costa, J.M.; Marques da Silva, J.; Pinheiro, C.; Barón, M.; Mylona, P.; Centritto, M.; Haworth, M.; Loreto, F.; Uzilday, B.; Turkan, I.; et al. Opportunities and limitations of crop phenotyping in Southern European countries. Front. Plant Sci. 2019, 10, 1125. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef] [PubMed]
- Catola, S.; Centritto, M.; Cascone, P.; Ranieri, A.; Loreto, F.; Calamai, L.; Balestrini, R.; Guerrieri, E. Effects of single or combined water deficit and aphid attack on tomato volatile organic compound (VOC) emission and plant-plant communication. Environ. Exp. Bot. 2018, 153, 54–62. [Google Scholar] [CrossRef]
- Brilli, F.; Loreto, F.; Baccelli, I. Exploiting plant volatile organic compounds (VOCs) in agriculture to improve sustainable defense strategies and productivity of crops. Front. Plant Sci. 2019, 10, 264. [Google Scholar] [CrossRef]
- Bailly, A.; Weisskopf, L. Mining the volatilomes of plant-associated microbiota for new biocontrol solutions. Front. Microbiol. 2017, 8, 1638. [Google Scholar] [CrossRef] [PubMed]
- Fincheira, P.; Quiroz, A.; Tortella, G.; Diez, M.C.; Rubilar, O. Current advances in plant-microbe communication via volatile organic compounds as an innovative strategy to improve plant growth. Microb. Res. 2021, 247, 126726. [Google Scholar] [CrossRef]
- Kanchiswamy, C.N.; Malnoy, M.; Maffei, M.E. Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Sci. 2015, 6, 151. [Google Scholar] [CrossRef]
- Romano, A.; Hanna, G.B. Identification and quantification of VOCs by proton transfer reaction time of flight mass spectrometry: An experimental workflow for the optimization of specificity, sensitivity, and accuracy. J. Mass Spectrom. 2018, 53, 287–295. [Google Scholar] [CrossRef]
- Arifuzzaman, M.; Oladzadabbasabadi, A.; McClean, P.; Rahman, M. Shovelomics for phenotyping root architectural traits of rapeseed/canola (Brassica napus L.) and genome-wide association mapping. Mol. Genet. Genom. 2019, 294, 985–1000. [Google Scholar] [CrossRef]
- Lynch, J.P. Roots of the second green revolution. Aust. J. Bot. 2007, 55, 493–512. [Google Scholar] [CrossRef]
- Kuijken, R.C.; van Eeuwijk, F.A.; Marcelis, L.F.; Bouwmeester, H.J. Root phenotyping: From component trait in the lab to breeding. J. Exp. Bot. 2015, 66, 5389–5401. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.A.; Pound, M.P.; Bennett, M.J.; Wells, D.M. Uncovering the hidden half of plants using new advances in root phenotyping. Curr. Opin. Biotechnol. 2019, 55, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, P.H.R.; de Lima, I.P.; de Castro, A.P.; Lanna, A.C.; Guimarães Santos Melo, P.; de Raïssac, M. Phenotyping root systems in a set of Japonica rice accessions: Can structural traits predict the response to drought? Rice 2020, 13, 67. [Google Scholar] [CrossRef] [PubMed]
- McGrail, R.K.; Van Sanford, D.A.; McNear, D.H., Jr. Trait-based root phenotyping as a necessary tool for crop selection and improvement. Agronomy 2020, 10, 1328. [Google Scholar] [CrossRef]
- Takahashi, H.; Pradal, C. Root phenotyping: Important and minimum information required for root modeling in crop plants. Breed. Sci. 2021, 71, 109–116. [Google Scholar] [CrossRef]
- Trachsel, S.; Kaeppler, S.M.; Brown, K.M.; Lynch, J.P. Shovelomics: High throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 2011, 341, 75–87. [Google Scholar] [CrossRef]
- Aguilar, J.J.; Moore, M.; Johnson, L.; Greenhut, R.F.; Rogers, E.; Walker, D.; O’Neil, F.; Edwards, J.L.; Thystrup, J.; Farrow, S.; et al. Capturing in-field root system dynamics with RootTracker. Plant Physiol. 2021, 187, 1117–1130. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop phenomics and high-throughput phenotyping: Past decades, current challenges, and future perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef] [PubMed]
- Fine, P.V.A.; Salazar, D.; Martin, R.E.; Metz, M.R.; Misiewicz, T.M.; Asner, G.P. Exploring the links between secondary metabolites and leaf spectral reflectance in a diverse genus of Amazonian trees. Ecosphere 2021, 12, e03362. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Spectranomics: Emerging science and conservation opportunities at the interface of biodiversity and remote sensing. Glob. Ecol. Conserv. 2016, 8, 212–219. [Google Scholar] [CrossRef]
- Fisher, J.B.; Sweeney, S.; Brzostek, E.R.; Evans, T.P.; Johnson, D.J.; Myers, J.A.; Bourg, N.A.; Wolf, A.T.; Howe, R.W.; Phillips, R.P. Tree-mycorrhizal associations detected remotely from canopy spectral properties. Glob. Chang. Biol. 2016, 22, 2596–2607. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandrini, M.; Nerva, L.; Sillo, F.; Balestrini, R.; Chitarra, W.; Zampieri, E. Abiotic Stress and Belowground Microbiome: The Potential of Omics Approaches. Int. J. Mol. Sci. 2022, 23, 1091. https://doi.org/10.3390/ijms23031091
Sandrini M, Nerva L, Sillo F, Balestrini R, Chitarra W, Zampieri E. Abiotic Stress and Belowground Microbiome: The Potential of Omics Approaches. International Journal of Molecular Sciences. 2022; 23(3):1091. https://doi.org/10.3390/ijms23031091
Chicago/Turabian StyleSandrini, Marco, Luca Nerva, Fabiano Sillo, Raffaella Balestrini, Walter Chitarra, and Elisa Zampieri. 2022. "Abiotic Stress and Belowground Microbiome: The Potential of Omics Approaches" International Journal of Molecular Sciences 23, no. 3: 1091. https://doi.org/10.3390/ijms23031091
APA StyleSandrini, M., Nerva, L., Sillo, F., Balestrini, R., Chitarra, W., & Zampieri, E. (2022). Abiotic Stress and Belowground Microbiome: The Potential of Omics Approaches. International Journal of Molecular Sciences, 23(3), 1091. https://doi.org/10.3390/ijms23031091









