Individual and Combined Treatments with Methylated Resveratrol Analogue DMU-214 and Gefitinib Inhibit Tongue Cancer Cells Growth via Apoptosis Induction and EGFR Inhibition
Abstract
1. Introduction
2. Results
2.1. Effect of DMU-214, Gef, and the Combination of DMU-214 and Gef on the Viability of CAL-27 and SCC-25 Tongue Cancer Cell Lines
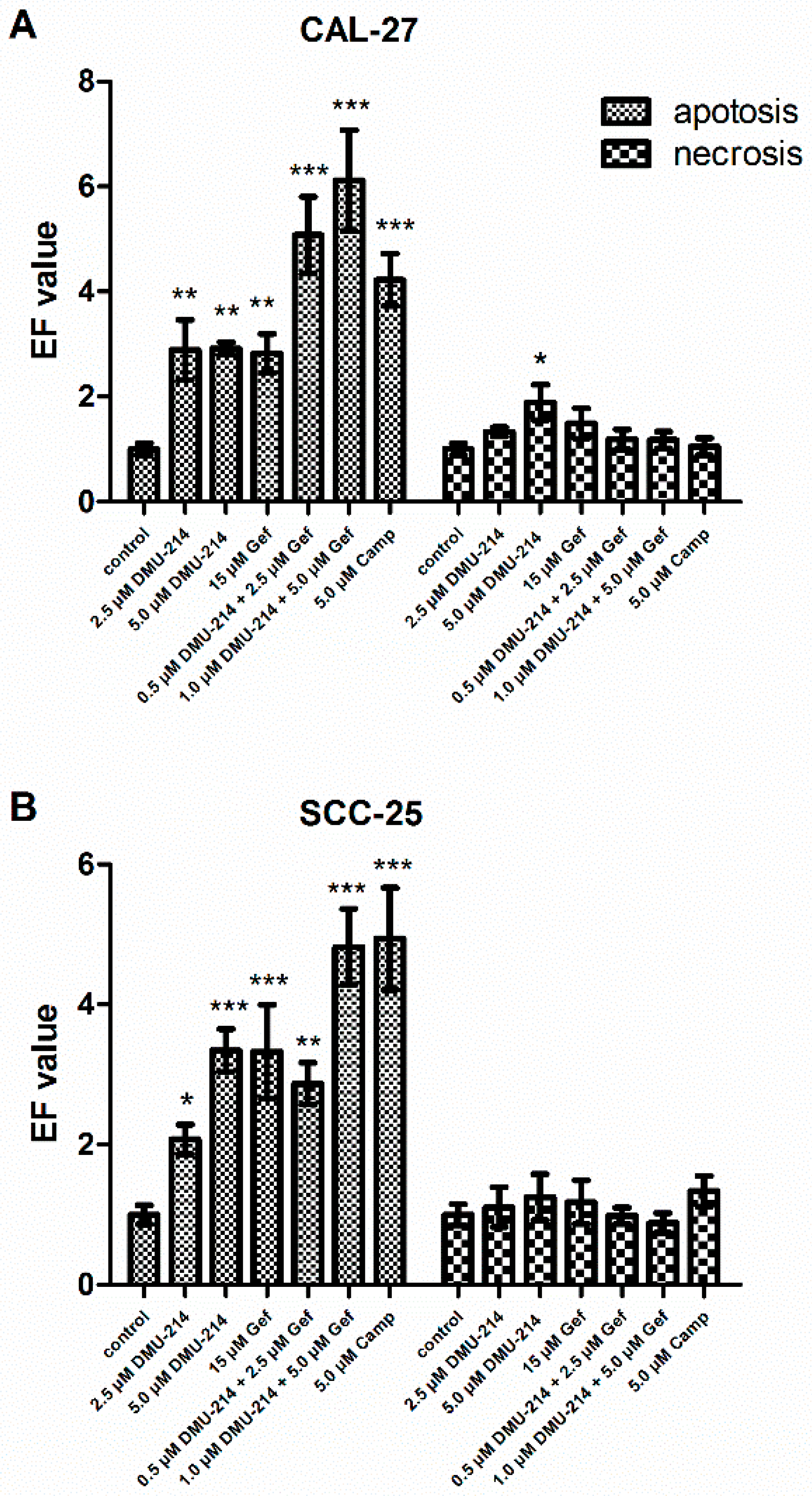
2.2. Effect of DMU-214, Gef, and the Combination of DMU-214 and Gef on Apoptosis and Necrosis Induction
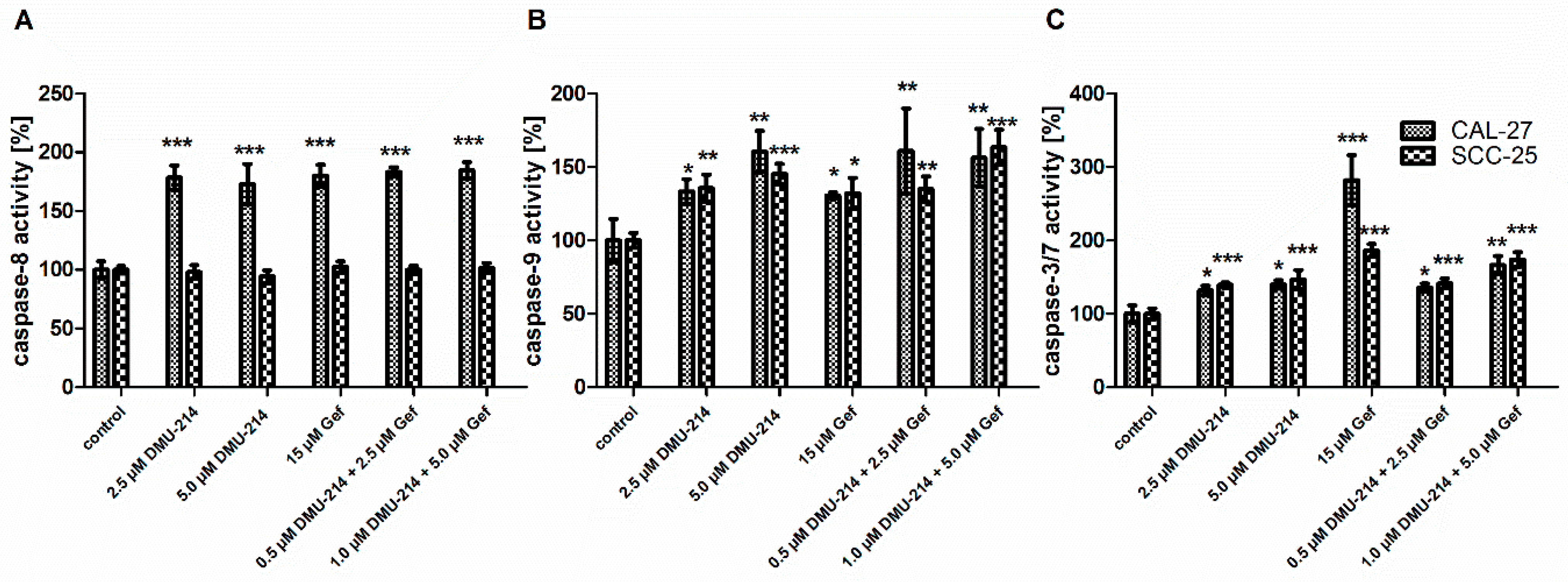
2.3. Effect of DMU-214, Gef, and the Combination of DMU-214 and Gef on Caspase-8, -9, -3/7 Activation in CAL-27 and SCC-25 Cell Lines
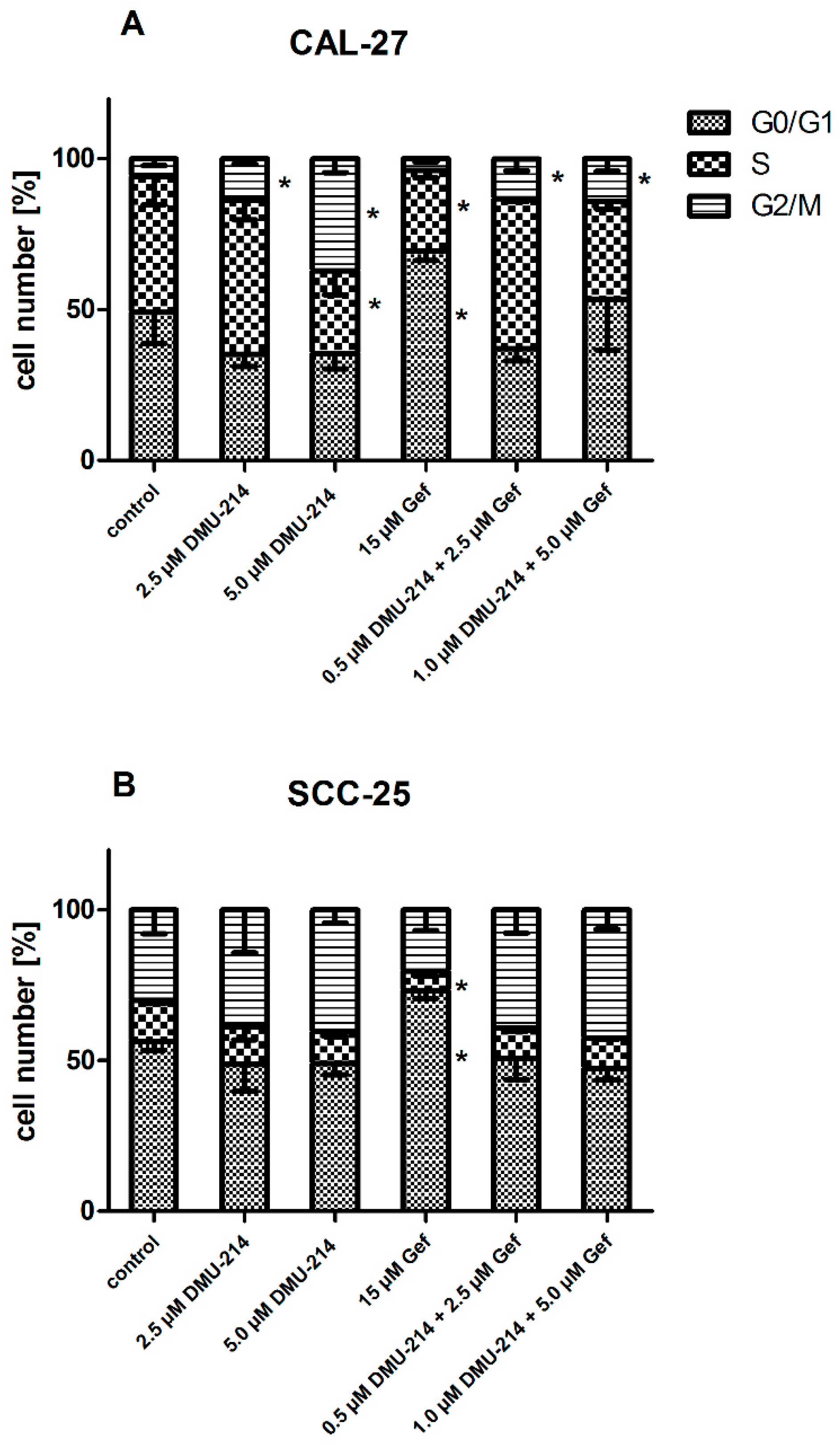
2.4. Cell Cycle Analysis
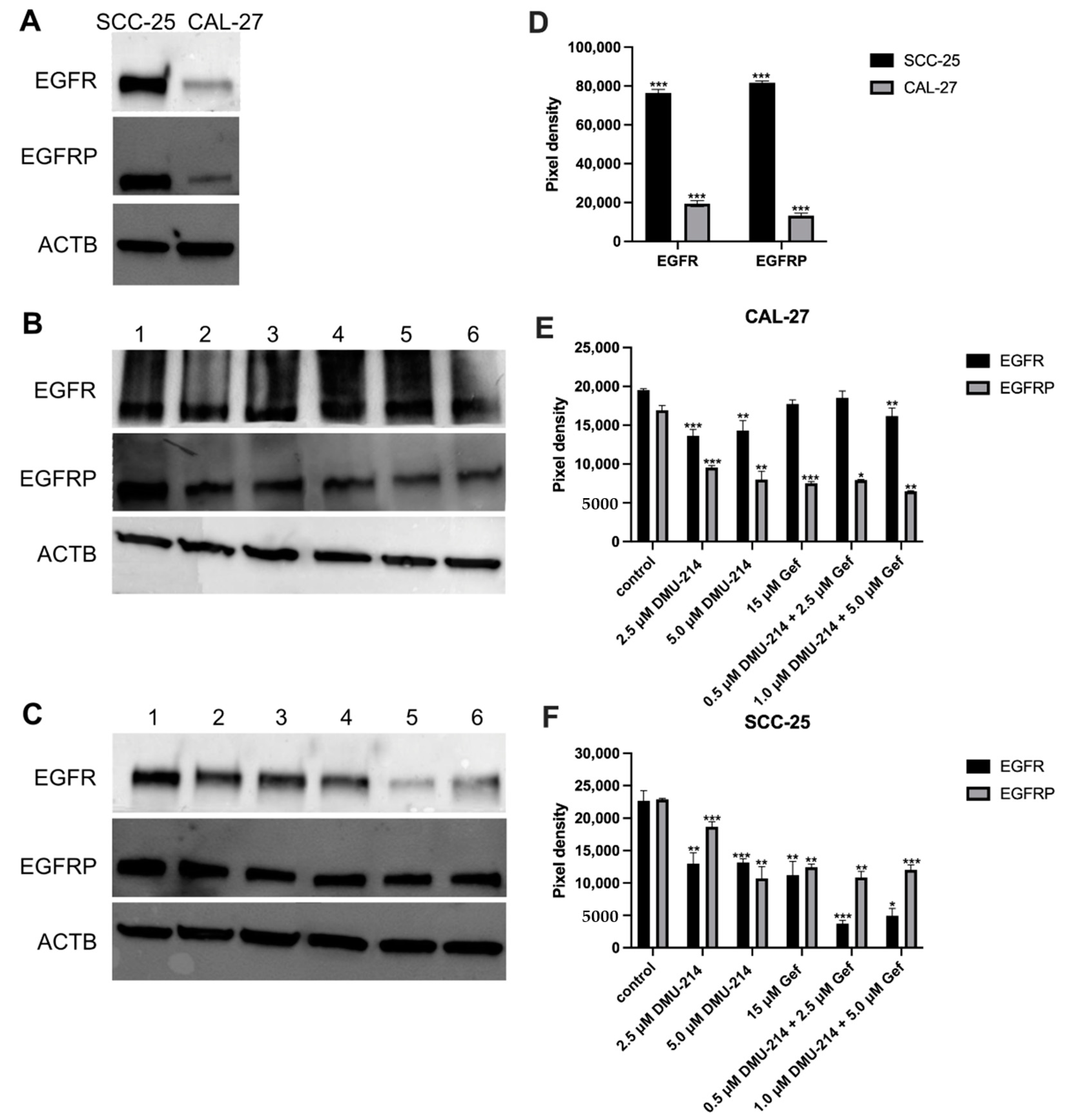
2.5. Protein Expression Analyses
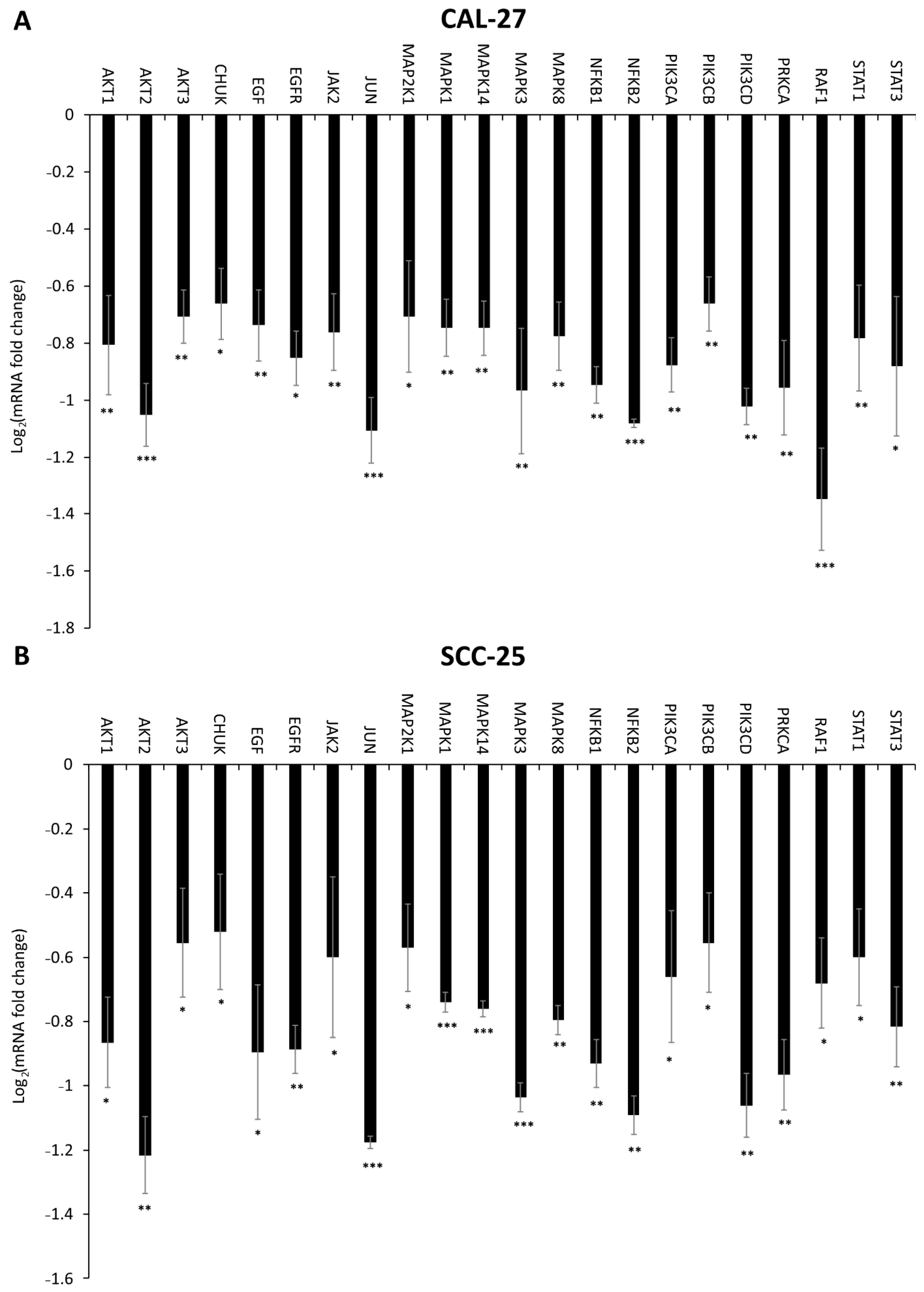
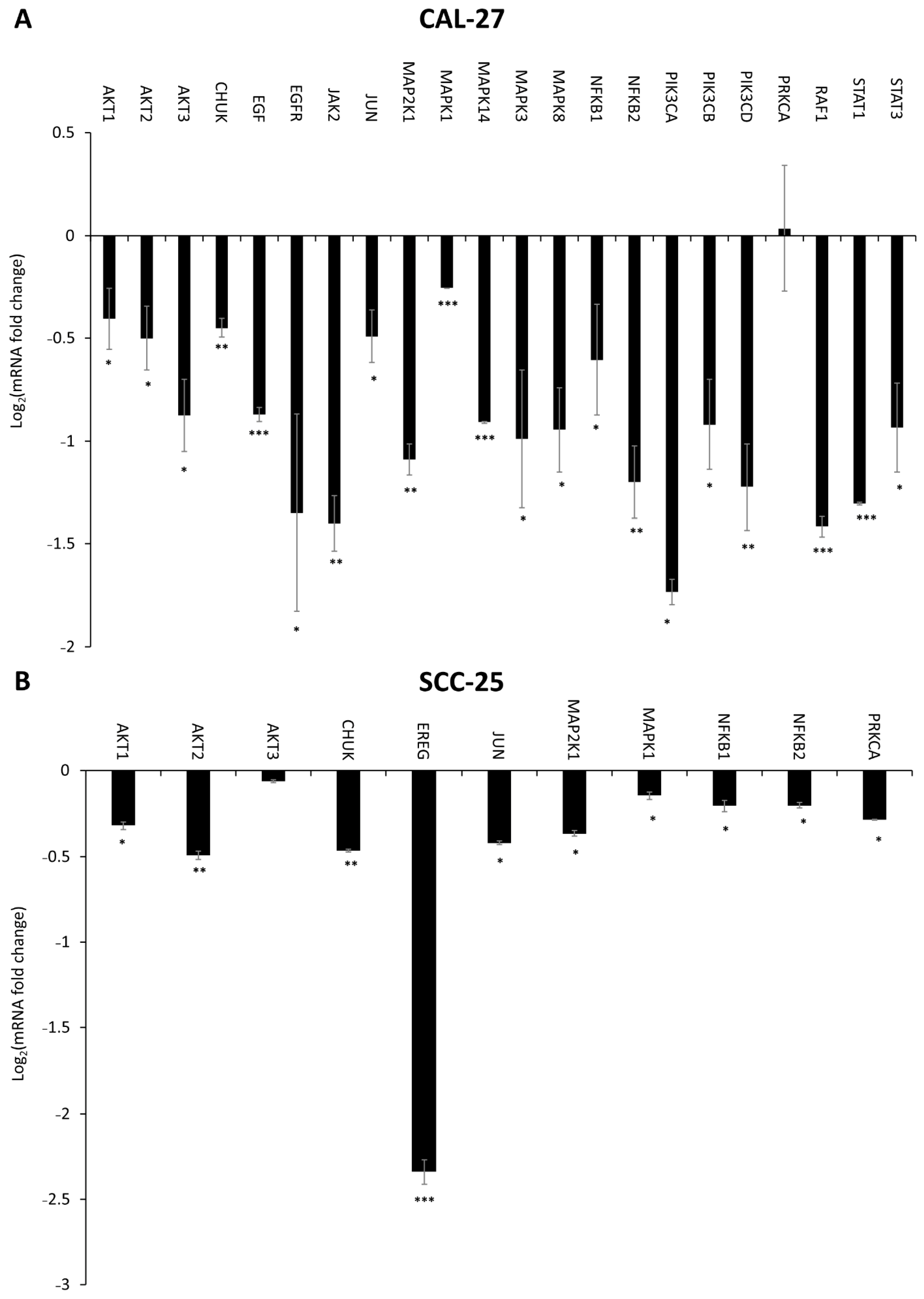
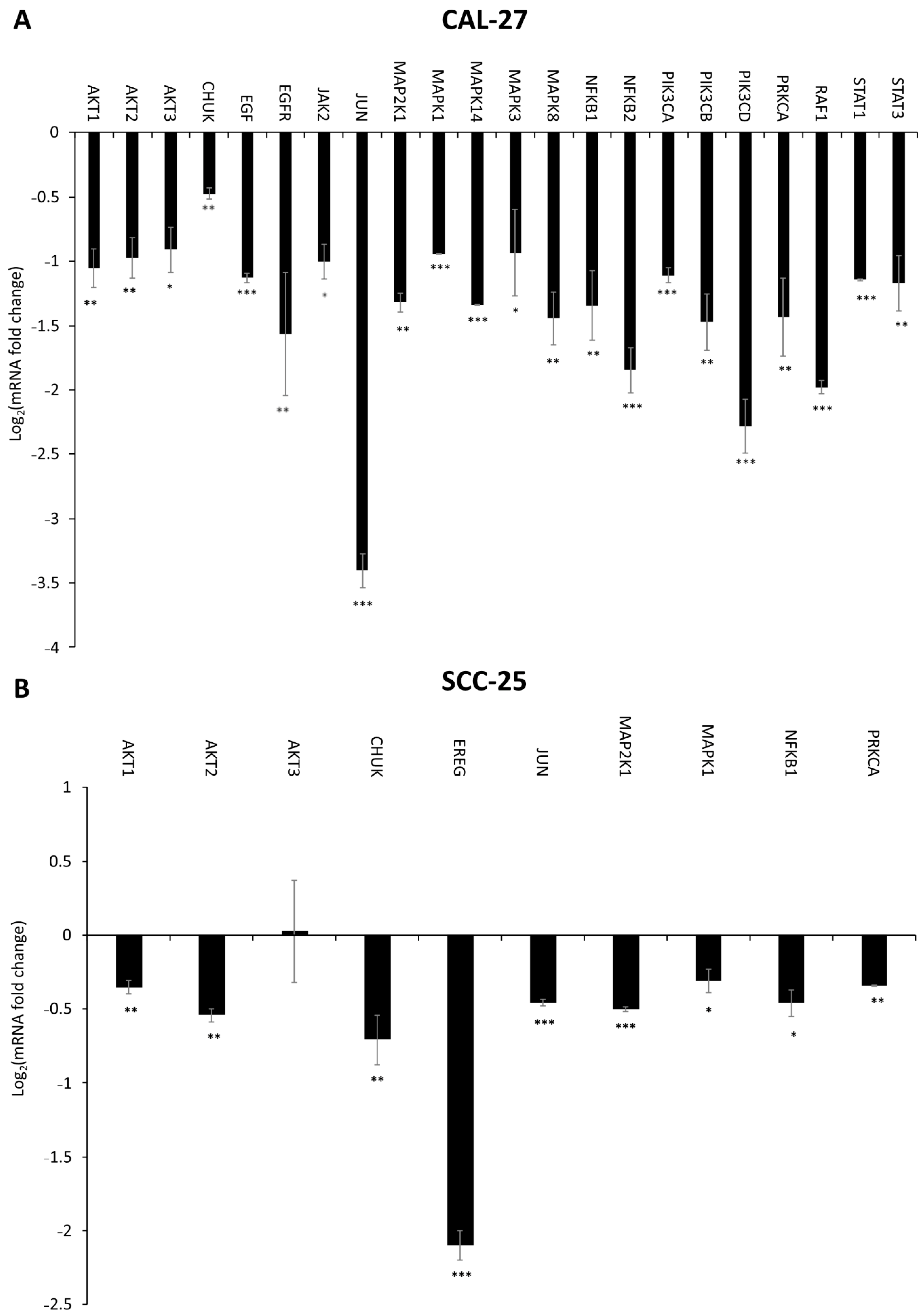
2.6. Effect of DMU-214, Gef, and the Combination of DMU-214 and Gef on the Expression of EGFR Signaling Pathway-Related Genes
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture and Cell Viability
4.3. Assessment of Apoptosis and Necrosis Induction
4.4. Determination of Caspase-8, -9 and Caspase-3/7 Activity
4.5. Cell Cycle Analysis
4.6. The RNA Isolation and Real-Time Quantitative PCR-Array (qPCR-array) Analysis
4.7. Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Western Blotting Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Agency for Research on Cancer. Available online: https://gco.iarc.fr/today/online-analysis-table?v=2018&mode=cancer&mode_population=continents&population=900&populations=900&key=asr&sex=0&cancer=39&type=0&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&group_cancer=1&include_nmsc=1&include_nmsc_other=1 (accessed on 18 April 2021).
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Saiko, P.; Szakmary, A.; Jaeger, W.; Szekeres, T. Resveratrol and its analogs: Defense against cancer, coronary disease and neurodegenerative maladies or just a fad? Mutat. Res. 2008, 658, 68–94. [Google Scholar] [CrossRef]
- Whitlock, N.C.; Baek, S.J. The anticancer effects of resveratrol: Modulation of transcription factors. Nutr. Cancer 2012, 64, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Walle, T.; Hsieh, F.; DeLegge, M.H.; Oatis, J.E., Jr.; Walle, U.K. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab. Dispos. 2004, 32, 1377–1382. [Google Scholar] [CrossRef]
- Androutsopoulos, V.P.; Ruparelia, K.C.; Papakyriakou, A.; Filippakis, H.; Tsatsakis, A.M.; Spandidos, D.A. Anticancer effects of the metabolic products of the resveratrol analogue, DMU-212: Structural requirements for potency. Eur. J. Med. Chem. 2011, 46, 2586–2595. [Google Scholar] [CrossRef]
- Piotrowska-Kempisty, H.; Ruciński, M.; Borys, S.; Kucińska, M.; Kaczmarek, M.; Zawierucha, P.; Wierzchowski, M.; Łażewski, D.; Murias, M.; Jodynis-Liebert, J. 3′-hydroxy-3,4,5,4′-tetramethoxystilbene, the metabolite of resveratrol analogue DMU-212, inhibits ovarian cancer cell growth in vitro and in a mice xenograft model. Sci. Rep. 2016, 6, 32627. [Google Scholar] [CrossRef]
- Seshacharyulu, P.; Ponnusamy, M.P.; Haridas, D.; Jain, M.; Ganti, A.K.; Batra, S.K. Targeting the EGFR signaling pathway in cancer therapy. Expert Opin. Ther. Targets 2012, 16, 15–31. [Google Scholar] [CrossRef]
- Ryott, M.; Wangsa, D.; Heselmeyer-Haddad, K.; Lindholm, J.; Elmberger, G.; Auer, G.; Avall Lundqvist, E.; Ried, T.; Munck-Wikland, E. EGFR protein overexpression and gene copy number increases in oral tongue squamous cell carcinoma. Eur. J. Cancer. 2009, 45, 1700–1708. [Google Scholar] [CrossRef]
- Kriegs, M.; Clauditz, T.S.; Hoffer, K.; Bartels, J.; Buhs, S.; Gerull, H.; Zech, H.B.; Bußmann, L.; Struve, N.; Rieckmann, T.; et al. Analyzing expression and phosphorylation of the EGF receptor in HNSCC. Sci. Rep. 2019, 9, 13564. [Google Scholar] [CrossRef]
- Mendelsohn, J.; Baselga, J. Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J. Clin. Oncol. 2003, 21, 2787–2799. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Zouhair, A.; Azria, D.; Ozsahin, M. The epidermal growth factor receptor (EGFR) in head and neck cancer: Its role and treatment implications. Radiat. Oncol. 2006, 1, 11. [Google Scholar] [CrossRef]
- Bruzzese, F.; Leone, A.; Rocco, M.; Carbone, C.; Piro, G.; Caraglia, M.; Di Gennaro, E.; Budillon, A. HDAC inhibitor vorinostat enhances the antitumor effect of gefitinib in squamous cell carcinoma of head and neck by modulating ErbB receptor expression and reverting EMT. J. Cell. Physiol. 2011, 226, 2378–2390. [Google Scholar] [CrossRef] [PubMed]
- Helal, H.H.; Qi, C.E.; Zhao, Y.Y.; Yao, C.S.; Li, D.C. The effects of combination of gefitinib and cisplatin on tongue squamous cell carcinoma cell lines. J. Can. Res. Ther. 2015, 11, 37–40. [Google Scholar] [CrossRef]
- Sekiguchi, Y.; Yamada, M.; Noguchi, T.; Noomote, C.; Tsuchida, M.; Kudoh, Y.; Matsuzawa, A. The anti-cancer drug gefitinib accelerates Fas-mediated apoptosis by enhancing caspase-8 activation in cancer cells. J. Toxicol. Sci. 2019, 44, 435–440. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, M.; Chen, F.; Zhu, Y.; Yong, W.; Lin, H.; Sun, Y.; Han, X. Gefitinib inhibits the proliferation of pancreatic cancer cells via cell cycle arrest. Anat. Rec. 2009, 292, 1122–1127. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, H.; Myszkowski, K.; Ziolkowska, A.; Kulcenty, K.; Wierzchowski, M.; Kaczmarek, M.; Murias, M.; Kwiatkowska-Borowczyk, E.; Jodynis-Liebert, J. Resveratrol analogue 3,4,4’,5-tetramethoxystilbene inhibits growth, arrests cell cycle and induces apoptosis in ovarian SKOV-3 and A-2780 cancer cells. Toxicol. Appl. Pharmacol. 2012, 263, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Purba, E.R.; Saita, E.I.; Maruyama, I.N. Activation of the EGF Receptor by Ligand Binding and Oncogenic Mutations: The “Rotation Model”. Cells 2017, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.K.; Velmurugan, K.; Xiong, K.N.; Alexander, C.M.; Xiong, J.; Brooks, R. Epiregulin is required for lung tumor promotion in a murine two-stage carcinogenesis model. Mol. Carcinog. 2017, 56, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Johnston, P.A.; Grandis, J.R. STAT3 signaling: Anticancer strategies and challenges. Mol. Interv. 2011, 11, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, G.D.; Shi, Z.; Qi, L.L.; Zhou, L.Y.; Fu, Z.X. The Ras/Raf/MEK/ERK signaling pathway and its role in the occurrence and development of HCC. Oncol. Lett. 2016, 12, 3045–3050. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 2007, 1773, 1263–1284. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Lu, Y.; Chen, Y.; Cheng, J.; Zhang, W. Dual Inhibition of MAPK and JAK2/STAT3 Pathways Is Critical for the Treatment of BRAF Mutant Melanoma. Mol. Ther. Oncolytics 2020, 18, 100–108. [Google Scholar] [CrossRef]
- Perner, F.; Perner, C.; Ernst, T.; Heidel, F.H. Roles of JAK2 in Aging, Inflammation, Hematopoiesis and Malignant Transformation. Cells 2019, 8, 854. [Google Scholar] [CrossRef] [PubMed]
- Freudlsperger, C.; Burnett, J.R.; Friedman, J.A.; Kannabiran, V.R.; Chen, Z.; Van Waes, C. EGFR–PI3K–AKT–mTOR signaling in head and neck squamous cell carcinomas: Attractive targets for molecular-oriented therapy. Expert Opin. Ther. Targets 2011, 15, 63–74. [Google Scholar] [CrossRef]
- Xie, Y.; Shi, X.; Sheng, K.; Han, G.; Li, W.; Zhao, Q.; Jiang, B.; Feng, J.; Li, J.; Gu, Y. PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review). Mol. Med. Rep. 2019, 19, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Rotman, T.; Etkovitz, N.; Spiegel, A.; Rubinstein, S.; Breitbart, H. Protein kinase A and protein kinase C(alpha)/PPP1CC2 play opposing roles in the regulation of phosphatidylinositol 3-kinase activation in bovine sperm. Reproduction 2010, 140, 43–56. [Google Scholar] [CrossRef]
- Kumar, A.; Rajendran, V.; Sethumadhavan, R.; Purohit, R. AKT kinase pathway: A leading target in cancer research. Sci. World J. 2013, 2013, 756134. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Tan, S.; Zhou, Y.; Lin, J.; Wang, H.; Oyang, L.; Tian, Y.; Liu, L.; Su, M.; Wang, H.; et al. Role of the NFκB-signaling pathway in cancer. Onco Targets Ther. 2018, 11, 2063–2073. [Google Scholar] [CrossRef]
- Häcker, H.; Karin, M. Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006, 2006, re13. [Google Scholar] [CrossRef]
- Stewart, J.S.; Cohen, E.E.; Licitra, L.; Van Herpen, C.M.; Khorprasert, C.; Soulieres, D.; Vodvarka, P.; Rischin, D.; Garin, A.M.; Hirsch, F.R.; et al. Phase III study of gefitinib compared with intravenous methotrexate for recurrent squamous cell carcinoma of the head and neck. J. Clin. Oncol. 2009, 27, 1864–1871. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
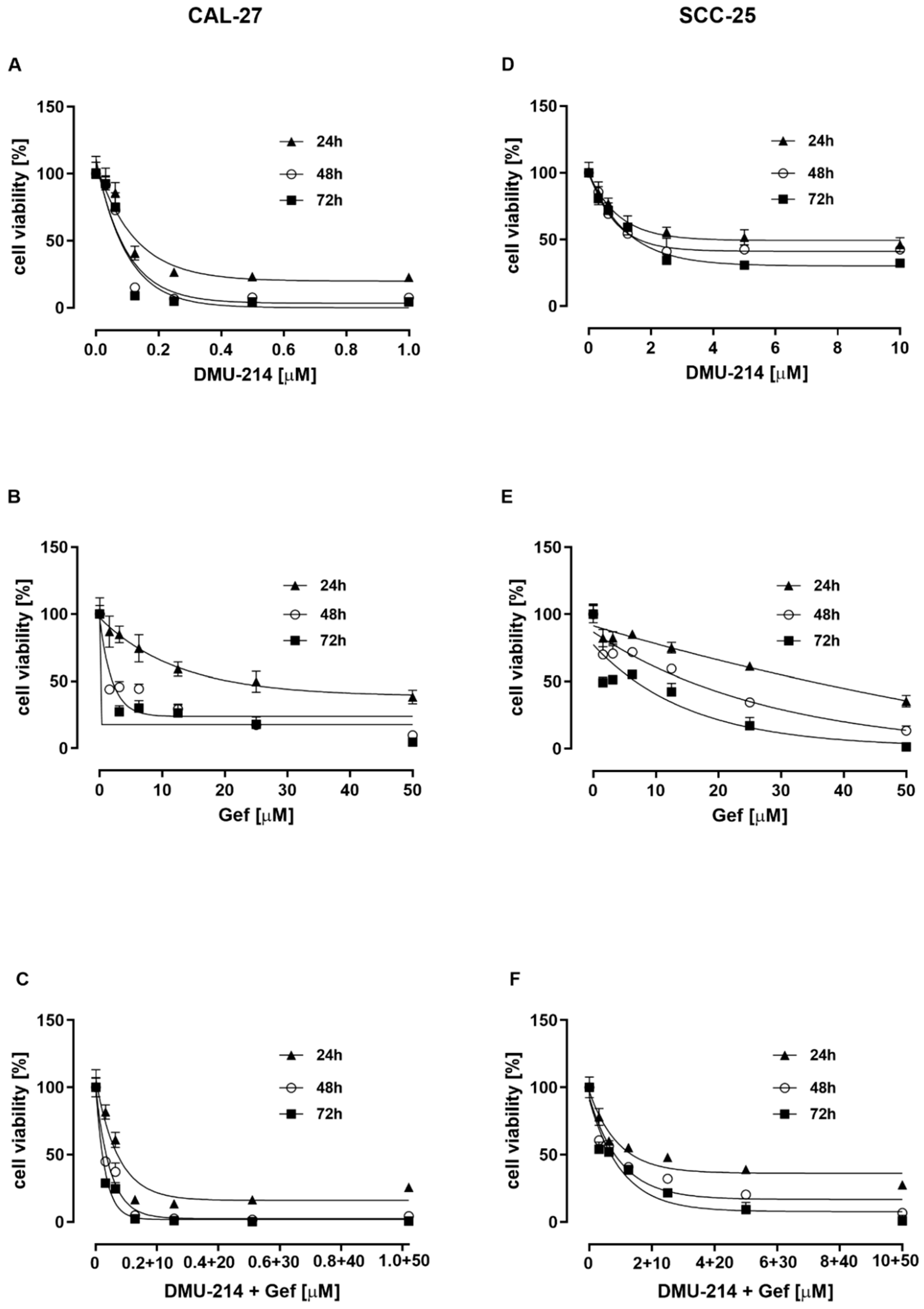
| Compounds Tested | Incubation Time (h) | IC50 Value (μM) | |
|---|---|---|---|
| CAL-27 | SCC-25 | ||
| DMU-214 | 24 | 0.13 ± 0.01 | 5.31 ± 1.70 |
| 48 | 0.08 ± 0.01 | 1.75 ± 0.19 | |
| 72 | 0.07 ± 0.01 | 1.63 ± 0.17 | |
| Gef | 24 | 17.14 ± 5.00 | 36.15 ± 1.09 |
| 48 | 2.00 ± 0.03 | 17.66 ± 3.33 | |
| 72 | 1.03 ± 0.01 | 8.64 ± 2.67 | |
| Cell Line | Compound Tested | Concentration Range [μM] |
|---|---|---|
| CAL-27 | DMU-214 | 1; 0.5; 0.25; 0.125; 0.0625; 0.03125 |
| GEFITINIB | 50; 25; 12.5; 6.25; 3.125; 1.5625 | |
| DMU-214 + GEFITINIB | (1 DMU-214 + 50 Gef) (0.5 DMU-214 + 25 Gef) (0.25 DMU-214 + 12.5 Gef) (0.125 DMU-214 + 6.25 Gef) (0.0625 DMU-214 + 3.125 Gef) (0.03125 DMU-214 + 1.5625 Gef) | |
| SCC-25 | DMU-214 | 10; 5; 2.5; 1.25; 0.625; 0.3125 |
| GEFITINIB | 50; 25; 12.5; 6.2 5; 3.125; 1.5625 | |
| DMU-214 + GEFITINIB | (10 DMU-214 + 50 Gef) (5 DMU-214 + 25 Gef) (2.5 DMU-214 + 12.5 Gef) (1.25 DMU-214 + 6.25 Gef) (0.625 DMU-214 + 3.125 Gef) (0.3125 DMU-214 + 1.5625 Gef) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jozkowiak, M.; Dyszkiewicz-Konwinska, M.; Ramlau, P.; Kranc, W.; Spaczynska, J.; Wierzchowski, M.; Kaczmarek, M.; Jodynis-Liebert, J.; Piotrowska-Kempisty, H. Individual and Combined Treatments with Methylated Resveratrol Analogue DMU-214 and Gefitinib Inhibit Tongue Cancer Cells Growth via Apoptosis Induction and EGFR Inhibition. Int. J. Mol. Sci. 2021, 22, 6180. https://doi.org/10.3390/ijms22126180
Jozkowiak M, Dyszkiewicz-Konwinska M, Ramlau P, Kranc W, Spaczynska J, Wierzchowski M, Kaczmarek M, Jodynis-Liebert J, Piotrowska-Kempisty H. Individual and Combined Treatments with Methylated Resveratrol Analogue DMU-214 and Gefitinib Inhibit Tongue Cancer Cells Growth via Apoptosis Induction and EGFR Inhibition. International Journal of Molecular Sciences. 2021; 22(12):6180. https://doi.org/10.3390/ijms22126180
Chicago/Turabian StyleJozkowiak, Malgorzata, Marta Dyszkiewicz-Konwinska, Piotr Ramlau, Wieslawa Kranc, Julia Spaczynska, Marcin Wierzchowski, Mariusz Kaczmarek, Jadwiga Jodynis-Liebert, and Hanna Piotrowska-Kempisty. 2021. "Individual and Combined Treatments with Methylated Resveratrol Analogue DMU-214 and Gefitinib Inhibit Tongue Cancer Cells Growth via Apoptosis Induction and EGFR Inhibition" International Journal of Molecular Sciences 22, no. 12: 6180. https://doi.org/10.3390/ijms22126180
APA StyleJozkowiak, M., Dyszkiewicz-Konwinska, M., Ramlau, P., Kranc, W., Spaczynska, J., Wierzchowski, M., Kaczmarek, M., Jodynis-Liebert, J., & Piotrowska-Kempisty, H. (2021). Individual and Combined Treatments with Methylated Resveratrol Analogue DMU-214 and Gefitinib Inhibit Tongue Cancer Cells Growth via Apoptosis Induction and EGFR Inhibition. International Journal of Molecular Sciences, 22(12), 6180. https://doi.org/10.3390/ijms22126180










