Low Light/Darkness as Stressors of Multifactor-Induced Senescence in Rice Plants
Abstract
:1. Introduction
2. Results
2.1. Rice Growth and Low Light/Darkness
2.2. Mechanism of Low-Light Response in Rice
2.3. Mechanisms of Dark Response in Rice
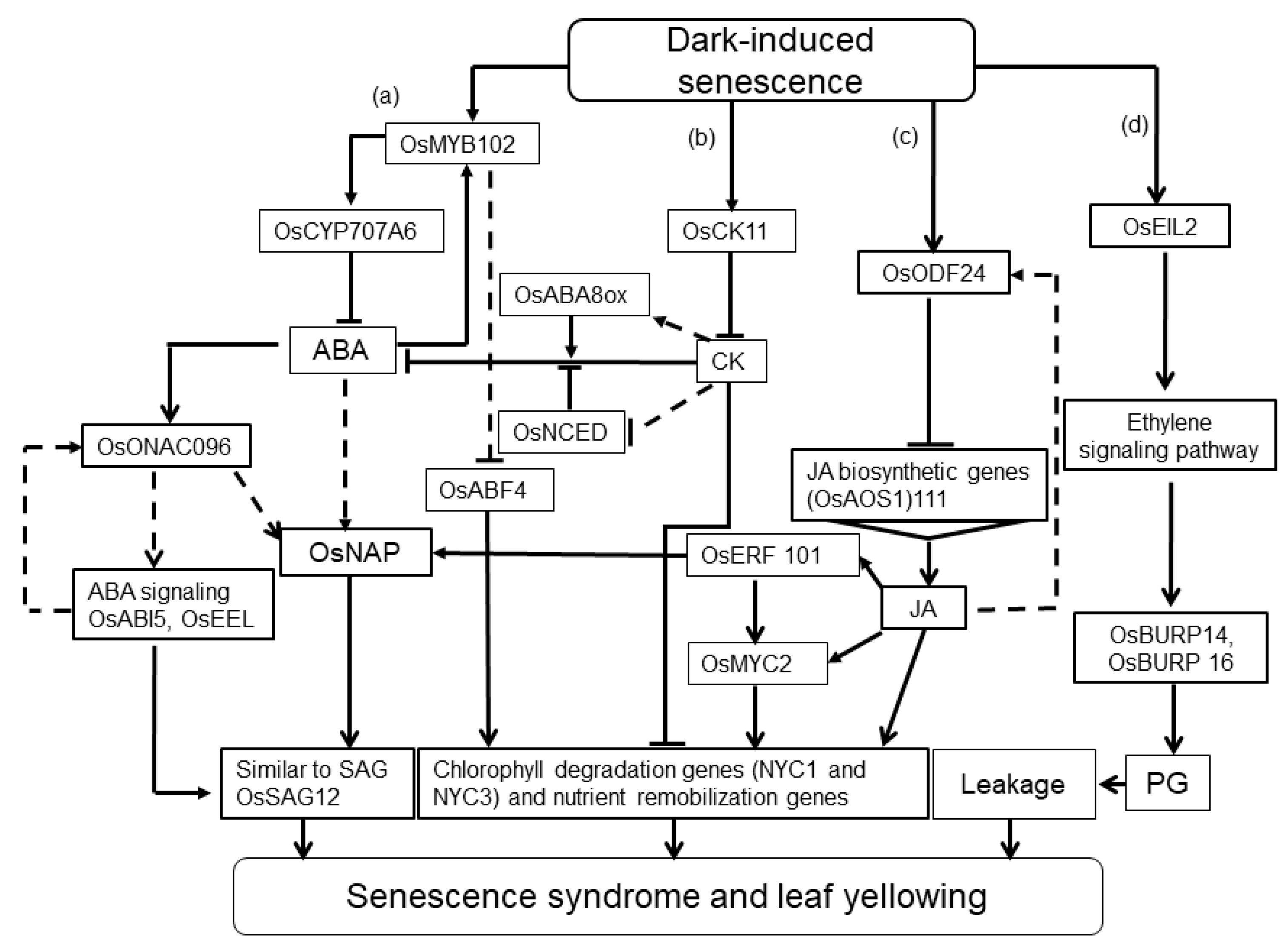
2.3.1. Hormonal Dynamics during Dark-Induced Senescence (DIS)
2.3.2. Signaling Molecule Regulation during DIS
2.3.3. Transcriptional Regulation during DIS
2.3.4. Protein Degradation during DIS
3. Conclusions and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Viji, M.; Thangaraj, M.; Jayapragasam, M. Low Irradiance Stress Tolerance in Rice (Oryza Sativa L.). Biol. Plant. 1997, 39, 251–256. [Google Scholar] [CrossRef]
- Panda, D.; Biswal, M.; Behera, L.; Baig, M.; Dey, P.; Nayak, L.; Sharma, S.; Samantaray, S.; Ngangkham, U.; Kumar, A. Impact of Low Light Stress on Physiological, Biochemical and Agronomic Attributes of Rice. J. Pharm. Phytochem. 2019, 8, 1814–1821. [Google Scholar]
- Kaiser, E.; Morales, A.; Harbinson, J.; Heuvelink, E.; Prinzenberg, A.E.; Marcelis, L.F. Metabolic and Diffusional Limitations of Photosynthesis in Fluctuating Irradiance in Arabidopsis Thaliana. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Morales, A.; Kaiser, E.; Yin, X.; Harbinson, J.; Molenaar, J.; Driever, S.M.; Struik, P.C. Dynamic Modelling of Limitations on Improving Leaf CO2 Assimilation under Fluctuating Irradiance. Plant Cell Environ. 2018, 41, 589–604. [Google Scholar] [CrossRef] [Green Version]
- Morales, A.; Kaiser, E. Photosynthetic Acclimation to Fluctuating Irradiance in Plants. Front. Plant Sci. 2020, 11, 268. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, S.; Panda, D.; Kumar, J.; Mohanty, N.; Biswal, M.; Baig, M.J.; Kumar, A.; Umakanta, N.; Samantaray, S.; Pradhan, S.K.; et al. Comparative Transcriptome Profiling of Low Light Tolerant and Sensitive Rice Varieties Induced by Low Light Stress at Active Tillering Stage. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Mendoza, M.; Velasco-Arroyo, B.; Santamaria, M.E.; González-Melendi, P.; Martinez, M.; Diaz, I. Plant Senescence and Proteolysis: Two Processes with One Destiny. Genet. Mol. Biol. 2016, 39, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.-H.; Liu, Y.-X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis Glucose Sensor HXK1 in Nutrient, Light, and Hormonal Signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusaba, M.; Tanaka, A.; Tanaka, R. Stay-Green Plants: What Do They Tell Us About the Molecular Mechanism of Leaf Senescence. Photosynth. Res. 2013, 117, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Law, S.R.; Chrobok, D.; Juvany, M.; Delhomme, N.; Lindén, P.; Brouwe, B.; Ahad, A.; Moritz, T.; Jansson, S.; Gardeström, P.; et al. Darkened Leaves Use Different Metabolic Strategies for Senescence and Survival. Plant Physiol. 2018, 177, 132–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to Environmental Stresses. Plant Cell 1995, 7, 1099. [Google Scholar] [CrossRef] [PubMed]
- Quirino, B.F.; Normanly, J.; Amasino, R.M. Diverse Range of Gene Activity during Arabidopsis Thaliana Leaf Senescence Includes Pathogen-Independent Induction of Defense-Related Genes. Plant Mol. Biol. 1999, 40, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Jeong, J.; Kang, M.-Y.; Kim, J.; Paek, N.-C.; Choi, G. Phytochrome-Interacting Transcription Factors PIF4 and PIF5 Induce Leaf Senescence in Arabidopsis. Nat. Commun. 2014, 5, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Kim, D.; Paek, N.-C. Salt Treatments and Induction of Senescence. In Plant Senescence; Springer: Berlin, Germany, 2018; pp. 141–149. [Google Scholar]
- Buchanan-Wollaston, V. The Molecular Biology of Leaf Senescence. J. Exp. Bot. 1997, 48, 181–199. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.H.; Wang, C.H.; Huang, L.T.; Chen, S.C.G. Leaf Senescence in Rice Plants: Cloning and Characterization of Senescence Up-Regulated Genes. J. Exp. Bot. 2001, 52, 1117–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kim, J.H.; Lyu, J.I.; Woo, H.R.; Lim, P.O. New Insights into the Regulation of Leaf Senescence in Arabidopsis. J. Exp. Bot. 2018, 69, 787–799. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Balazadeh, S.; Tanaka, R.; Mueller-Roeber, B.; Tanaka, A. Overproduction of Chl b Retards Senescence through Transcriptional Reprogramming in Arabidopsis. Plant Cell Physiol. 2012, 53, 505–517. [Google Scholar] [CrossRef]
- Neff, M.M.; Fankhauser, C.; Chory, J. Light: An Indicator of Time and Place. Genes Dev. 2000, 14, 257–271. [Google Scholar] [PubMed]
- Borthwick, H.A.; Hendricks, S.B.; Parker, M.W.; Toole, E.H.; Toole, V.K. A Reversible Photoreaction Controlling Seed Germination. Proc. Natl. Acad. Sci. USA 1952, 38, 662–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobieszczuk-Nowicka, E.; Wrzesiński, T.; Bagniewska-Zadworna, A.; Kubal, S.; Rucińska-Sobkowiak, R.; Polcyn, W.; Misztal, L.; Mattoo, A.K. Physio-Genetic Dissection of Dark-Induced Leaf Senescence and Timing Its Reversal in Barley. Plant Physiol. 2018, 178, 654–671. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Li, W.; Guo, Y.; Lu, Y.; Zhou, C.; Fan, D.; Weng, Q.; Zhu, C.; Huang, T.; Zhang, L.; et al. A Map of Rice Genome Variation Reveals the Origin of Cultivated Rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [Green Version]
- Pittendrigh, C.S. Temporal Organization: Reflections of a Darwinian Clock-Watcher. Annu. Rev. Physiol. 1993, 55, 17–54. [Google Scholar] [CrossRef]
- Bell, G.; Danneberger, T.; McMahon, M. Spectral Irradiance Available for Turfgrass Growth in Sun and Shade. Crop. Sci. 2000, 40, 189–195. [Google Scholar] [CrossRef]
- Liu, Q.-h.; Xiu, W.; CHEN, B.-c.; Jie, G. Effects of Low Light on Agronomic and Physiological Characteristics of Rice Including Grain Yield and Quality. Rice Sci. 2014, 21, 243–251. [Google Scholar] [CrossRef]
- Smith, H. Phytochromes and Light Signal Perception by Plants—An Emerging Synthesis. Nature 2000, 407, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.W. Agricultural Natural Resource; Science Press: Beijing, China, 1998; pp. 157–160. [Google Scholar]
- Barmudoi, B.; Bharali, B. Effects of Light Intensity and Quality On Physiological Changes in Winter Rice (Oryza Sativa L.). Int. J. Environ. Agric. Res. 2016, 2, 65–76. [Google Scholar]
- Chauhan, J.; Chaudhary, S.; Sodani, R.; Seema. High Light Stress Response and Tolerance Mechanism in Plant. Available online: https://www.biotecharticles.com/Agriculture-Article/High-Light-Stress-Response-and-Tolerance-Mechanism-in-Plants-3992.html (accessed on 10 April 2021).
- Nagatani, A.; Reed, J.W.; Chory, J. Isolation and Initial Characterization of Arabidopsis Mutants that are Deficient in Phytochrome A. Plant Physiol. 1993, 102, 269–277. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Li, Q.-P.; Zeng, Y.-L.; Deng, F.; Ren, W.-J. Effect of Different Shading Materials on Grain Yield and Quality of Rice. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Ren, W.; Yang, W.; Xu, J.; Fan, G.; Wang, L.; Guan, H. Impact of Low-Light Stress on Leaves Characteristics of Rice after Heading. J. Sichuan Agric. Univ. 2002, 20, 205–208. [Google Scholar]
- Gao, J.; Liu, Z.; Zhao, B.; Liu, P.; Zhang, J.-W. Physiological and Comparative Proteomic Analysis Provides New Insights into the Effects of Shade Stress in Maize (Zea Mays L.). BMC Plant Biol. 2020, 20, 60. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Cui, H.; Camberato, J.J.; Dong, S.; Liu, P.; Zhao, B.; Zhang, J. Effects of Shading on the Photosynthetic Characteristics and Mesophyll Cell Ultrastructure of Summer Maize. Sci. Nat. 2016, 103, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Ruiz-Sola, M.A.; Couso, A.; Veciana, N.; Monte, E. Red and Blue Light Differentially Impact Retrograde Signalling and Photoprotection in Rice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190402. [Google Scholar] [CrossRef]
- van Doorn, W.G. Is the Onset of Senescence in Leaf Cells of Intact Plants Due to Low or High Sugar Levels? J. Exp. Bot. 2008, 59, 1963–1972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Zhang, D.; Guo, J.; Wu, H.; Jin, M.; Lu, Q.; Lu, C.; Zhang, L. A Psb27 Homologue in Arabidopsis Thaliana is Required for Efficient Repair of Photo Damaged Photosystem II. Plant Mol. Biol. 2006, 61, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Roose, J.L.; Pakrasi, H.B. The Psb27 Protein Facilitates Manganese Cluster Assembly in Photosystem II. J. Biol. Chem. 2008, 283, 4044–4050. [Google Scholar] [CrossRef] [Green Version]
- Dutta, S.S.; Tyagi, W.; Pale, G.; Pohlong, J.; Aochen, C.; Pandey, A.; Pattanayak, A.; Rai, M. Marker–Trait Association for Low-Light Intensity Tolerance in Rice Genotypes from Eastern India. Mol. Genet. Genom. 2018, 293, 1493–1506. [Google Scholar] [CrossRef]
- Liu, Y.; Pan, T.; Tang, Y.; Zhuang, Y.; Liu, Z.; Li, P.; Li, H.; Huang, W.; Tu, S.; Ren, G.; et al. Proteomic Analysis of Rice Subjected to Low Light Stress and Overexpression of OsGAPB Increases the Stress Tolerance. Rice 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Ubierna, N.; Sun, W.; Kramer, D.M.; Cousins, A.B. The Efficiency of C4 Photosynthesis Under Low Light Conditions in Zea mays. Plant Cell Environ. 2013, 36, 365–381. [Google Scholar] [CrossRef]
- Bellasio, C.; Griffiths, H. Acclimation of C4 Metabolism to Low Light in Mature Maize Leaves could Limit Energetic Losses during Progressive Shading in a Crop Canopy. J. Exp. Bot. 2014, 65, 3725–3736. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Li, Y.; Li, Y.; Ma, L.; Ashraf, U.; Tang, X.; Pan, S.; Tian, H.; Mo, Z. Application of γ-Aminobutyric Acid under Low Light Conditions: Effects on Yield, Aroma, Element Status, and Physiological Attributes of Fragrant Rice. Ecotoxicol. Environ. Saf. 2021, 213, 111941. [Google Scholar] [CrossRef]
- Li, M.; Ashraf, U.; Tian, H.; Mo, Z.; Pan, S.; Anjum, S.A.; Duan, M.; Tang, X. Manganese-Induced Regulations in Growth, Yield Formation, Quality Characters, Rice Aroma and Enzyme Involved in 2-Acetyl-1-Pyrroline Biosynthesis in Fragrant Rice. Plant Physiol. Biochem. 2016, 103, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Kano, A.; Tamaoki, D.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Involvement of OsJAZ8 in Jasmonate-Induced Resistance to Bacterial Blight in Rice. Plant Cell Physiol. 2012, 53, 2060–2072. [Google Scholar] [CrossRef] [Green Version]
- Yoshitomi, K.; Taniguchi, S.; Tanaka, K.; Uji, Y.; Akimitsu, K.; Gomi, K. Rice Terpene Synthase 24 (OsTPS24) Encodes a Jasmonate-Responsive Monoterpene Synthase that Produces an Antibacterial γ-Terpinene Against Rice Pathogen. J. Plant Physiol. 2016, 191, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Miyamoto, K.; Nemoto, K.; Sawasaki, T.; Yamane, H.; Nojiri, H.; Okada, K. OsMYC2, An Essential Factor for JA-Inductive Sakuranetin Production In Rice, Interacts with MYC2-Like Proteins that Enhance Its Transactivation Ability. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Shan, X.; Wang, J.; Chua, L.; Jiang, D.; Peng, W.; Xie, D. The Role of Arabidopsis Rubisco Activase in Jasmonate-Induced Leaf Senescence. Plant Physiol. 2011, 155, 751–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Song, S.; Xie, D. Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between bHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Zhang, Y.; Di, C.; Zhang, Q.; Zhang, K.; Wang, C.; You, Q.; Yan, H.; Dai, S.Y.; Yuan, J.S.; et al. JAZ7 Negatively Regulates Dark-Induced Leaf Senescence in Arabidopsis. J. Exp. Bot. 2016, 67, 751–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.; Kang, K.; Shim, Y.; Sakuraba, Y.; An, G.; Paek, N.-C. Rice ETHYLENE RESPONSE FACTOR 101 Promotes Leaf Senescence Through Jasmonic Acid-Mediated Regulation of OsNAP and OsMYC2. Front. Plant Sci. 2020, 11, 1096. [Google Scholar] [CrossRef]
- Shim, Y.; Kang, K.; An, G.; Paek, N.-C. Rice DNA-Binding One Zinc Finger 24 (OsDOF24) Delays Leaf Senescence in a Jasmonate-Mediated Pathway. Plant Cell Physiol. 2019, 60, 2065–2076. [Google Scholar] [CrossRef]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant Senescence and Crop Productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef]
- Rashotte, A.M.; Carson, S.D.B.; To, J.P.C.; Kieber, J.J. Expression Profiling of Cytokinin Action in Arabidopsis. Plant Physiol. 2003, 132, 1998–2011. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Peng, K.; Cui, F.; Wang, D.; Zhao, J.; Zhang, Y.; Yu, N.; Wang, Y.; Zeng, D.; Wang, Y.; et al. Cytokinin Oxidase/Dehydrogenase OsCKX11 Coordinates Source and Sink Relationship in Rice by Simultaneous Regulation of Leaf Senescence and Grain Number. Plant Biotechnol. J. 2020, 19, 335–350. [Google Scholar] [CrossRef]
- Piao, W.; Kim, S.-H.; Lee, B.-D.; An, G.; Sakuraba, Y.; Paek, N.-C. Rice Transcription Factor OsMYB102 Delays Leaf Senescence by Down-Regulating Abscisic Acid Accumulation and Signaling. J. Exp. Bot. 2019, 70, 2699–2715. [Google Scholar] [CrossRef]
- Takahashi, A.; Kawasaki, T.; Wong, H.L.; Suharsono, U.; Hirano, H.; Shimamoto, K. Hyperphosphorylation of a Mitochondrial Protein, Prohibitin, is Induced by Calyculin A in a Rice Lesion-Mimic Mutant Cdr1. Plant Physiol. 2003, 132, 1861–1869. [Google Scholar] [CrossRef] [Green Version]
- Lim, P.O.; Kim, H.J.; Gil Nam, H. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeekens, S.; Ma, J.; Hanson, J.; Rolland, F. Sugar Signals and Molecular Networks Controlling Plant Growth. Curr. Opin. Plant Biol. 2010, 13, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Biswal, B.; Pandey, J. Loss of Photosynthesis Signals a Metabolic Reprogramming to Sustain Sugar Homeostasis during Senescence of Green Leaves: Role of cell wall hydrolases. Photosynthetica 2018, 56, 404–410. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, Q.; Cheng, F. Sugar Starvation Enhances Leaf Senescence and Genes Involved in Sugar Signaling Pathways Regulate Early Leaf Senescence in Mutant Rice. Rice Sci. 2020, 27, 201–214. [Google Scholar]
- Lee, R.H.; Hsu, J.H.; Huang, H.J.; Lo, S.F.; Grace Chen, S.C. Alkaline α-Galactosidase Degrades Thylakoid Membranes in the Chloroplast during Leaf Senescence in Rice. New Phytol. 2009, 184, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Jiao, B.-B.; Wang, J.-J.; Zhu, X.-D.; Zeng, L.-J.; Li, Q.; He, Z.-H. A Novel Protein RLS1 with NB–ARM Domains is Involved in Chloroplast Degradation during Leaf Senescence in Rice. Mol. Plant 2012, 5, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Tan, D.X.; Reiter, R.J.; Ye, T.; Yang, F.; Chan, Z. Melatonin Induces Class A1 Heat-Shock Factors (HSFA 1s) and Their Possible Involvement of Thermotolerance in Arabidopsis. J. Pineal Res. 2015, 58, 335–342. [Google Scholar] [CrossRef]
- Zhang, N.; Sun, Q.; Zhang, H.; Cao, Y.; Weeda, S.; Ren, S.; Guo, Y.D. Roles of Melatonin in Abiotic Stress Resistance in Plants. J. Exp. Bot. 2015, 66, 647–656. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Zhang, Y.; Sinumporn, S.; Yu, N.; Zhan, X.; Shen, X.; Chen, D.; Yu, P.; Wu, W.; Liu, Q.; et al. Premature Leaf Senescence 3, Encoding a Methyltransferase, is Required for Melatonin Biosynthesis in Rice. Plant J. 2018, 95, 877–891. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, H.; Xu, B.; Li, J.; Huang, B. Exogenous Melatonin Suppresses Dark-Induced Leaf Senescence by Activating the Superoxide Dismutase-Catalase Antioxidant Pathway and Down-Regulating Chlorophyll Degradation in Excised Leaves of Perennial Ryegrass (Lolium Perenne L.). Front. Plant Sci. 2016, 7, 1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.Y.; Cheng, S.H.; Kao, C.H. Senescence of Rice Leaves: VII. Proline Accumulation in Senescing Excised Leaves. Plant Physiol. 1982, 69, 1348–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondal, W.; Dey, B.; Choudhuri, M. Proline Accumulation as a Reliable Indicator of Monocarpic Senescence in Rice Cultivars. Experientia 1985, 41, 346–348. [Google Scholar] [CrossRef]
- Launay, A.; Cabassa-Hourton, C.; Eubel, H.; Maldiney, R.; Guivarc’h, A.; Crilat, E.; Planchais, S.; Lacoste, J.; Bordenave-Jacquemin, M.; Clément, G.; et al. Proline Oxidation Fuels Mitochondrial Respiration during Dark-Induced Leaf Senescence in Arabidopsis Thaliana. J. Exp. Bot. 2019, 70, 6203–6214. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Chen, Y.; Zhu, Z.; Liu, Y.; Yan, G.; Xu, S.; Xu, H.; Yang, M.; Dou, S.; Li, L.; et al. Integrated Analyses of Rice Dark Response and Leaf Color Regulation Reveal Links with Porphyrin and Chlorophyll Metabolism. Res. Square 2020. [Google Scholar] [CrossRef]
- Chao, Q.; Rothenberg, M.; Solano, R.; Roman, G.; Terzaghi, W.; Ecker, J.R. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 1997, 89, 1133–1144. [Google Scholar] [CrossRef] [Green Version]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear Events in Ethylene Signaling: A Transcriptional Cascade Mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef] [Green Version]
- An, F.; Zhao, Q.; Ji, Y.; Li, W.; Jiang, Z.; Yu, X.; Zhang, C.; Han, Y.; He, W.; Liu, Y.; et al. Ethylene-Induced Stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is Mediated by Proteasomal Degradation of EIN3 Binding F-box 1 and 2 that Requires EIN2 in Arabidopsis. Plant Cell 2010, 22, 2384–2401. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Teng, X.D.; Zheng, Q.Q.; Zhao, Y.Y.; Lu, J.Y.; Wang, Y.; Guo, H.; Yang, Z.N. Ethylene Signaling is Critical for Synergid Cell Functional Specification and Pollen Tube Attraction. Plant J. 2018, 96, 176–187. [Google Scholar] [CrossRef]
- Liu, H.; Ma, Y.; Chen, N.; Guo, S.; Liu, H.; Guo, X.; Chong, K.; Xu, Y. Overexpression of Stress-Inducible OsBURP16, the β Subunit of Polygalacturonase 1, Decreases Pectin Content and Cell Adhesion and Increases Abiotic Stress Sensitivity in Rice. Plant Cell Environ. 2014, 37, 1144–1158. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Duan, J.; Shan, C.; Mei, Z.; Chen, H.; Feng, H.; Zhu, J.; Cai, W. Ethylene Insensitive3-like2 (OsEIL2) Confers Stress Sensitivity by Regulating OsBURP16, the β Subunit of Polygalacturonase (PG1β-like) Subfamily Gene in Rice. Plant Sci. 2020, 292, 110353. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Shen, Y.-P.; Ma, L.-G.; Pan, Y.; Du, Y.-L.; Wang, D.-H.; Yang, J.Y.; Hu, L.D.; Liu, X.F.; Dong, C.X.; et al. Genome-Wide ORFeome Cloning and Analysis of Arabidopsis Transcription Factor Genes. Plant Physiol. 2004, 135, 773–782. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Liu, T.; Tian, C.; Sun, S.; Li, J.; Chen, M. Transcription Factors in Rice: A Genome-Wide Comparative Analysis between Monocots and Eudicots. Plant Mol. Biol. 2005, 59, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-Wide Analysis of NAC Transcription Factor Family in Rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, K.; Shim, Y.; Gi, E.; An, G.; Paek, N.-C. Mutation of ONAC096 Enhances Grain Yield by Increasing Panicle Number and Delaying Leaf Senescence during Grain Filling in Rice. Int. J. Mol. Sci. 2019, 20, 5241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Chen, W.; Foley, R.C.; Büttner, M.; Singh, K.B. Interactions between Distinct Types of DNA Binding Proteins Enhance Binding to Ocs Element Promoter Sequences. Plant Cell 1995, 7, 2241–2252. [Google Scholar]
- Yanagisawa, S.; Sheen, J. Involvement of Maize Dof Zinc Finger Proteins in Tissue-Specific and Light-Regulated Gene Expression. Plant Cell 1998, 10, 75–89. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, S. Dof1 and Dof2 Transcription Factors are Associated with Expression of Multiple Genes Involved in Carbon Metabolism in Maize. Plant J. 2000, 21, 281–288. [Google Scholar] [CrossRef]
- Uji, Y.; Akimitsu, K.; Gomi, K. Identification of OsMYC2-Regulated Senescence-Associated Genes in Rice. Planta 2017, 245, 1241–1246. [Google Scholar] [CrossRef]
- Cominelli, E.; Tonelli, C. A New Role for Plant R2R3-MYB Transcription Factors in Cell Cycle Regulation. Cell Res. 2009, 19, 1231–1232. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB Transcription Factors: Their Role in Drought Response Mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef] [Green Version]
- Jaradat, M.R.; Feurtado, J.A.; Huang, D.; Lu, Y.; Cutler, A. Multiple Roles of the Transcription Factor AtMYBR1/AtMYB44 in ABA Signaling, Stress Responses, and Leaf Senescence. BMC Plant Biol. 2013, 13, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer-Kilbienski, I.; Miao, Y.; Roitsch, T.; Zschiesche, W.; Humbeck, K.; Krupinska, K. Nuclear Targeted AtS40 Modulates Senescence Associated Gene Expression in Arabidopsis Thaliana during Natural Development and in Darkness. Plant Mol. Biol. 2010, 73, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Krupinska, K.; Dähnhardt, D.; Fischer-Kilbienski, I.; Kucharewicz, W.; Scharrenberg, C.; Trösch, M.; Buck, F. Identification of WHIRLY1 as a Factor Binding to the Promoter of the Stress-and Senescence-Associated Gene HvS40. J. Plant Growth Regul. 2014, 33, 91–105. [Google Scholar] [CrossRef]
- Jehanzeb, M.; Zheng, X.; Miao, Y. The Role of the S40 Gene Family in Leaf Senescence. Int. J. Mol. Sci. 2017, 18, 2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Jehanzeb, M.; Zhang, Y.; Li, L.; Miao, Y. Characterization of S40-Like Proteins and their Roles in Response to Environmental Cues and Leaf Senescence in Rice. BMC Plant Biol. 2019, 19, 174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, J.; Yoo, J.-H.; Yoo, S.-C.; Cho, S.-H.; Koh, H.-J.; Seo, H.-S.; Paek, N.-C. Rice Chlorina-1 and Chlorina-9 Encode ChlD and ChlI Subunits of Mg-Chelatase, a Key Enzyme for Chlorophyll Synthesis and Chloroplast Development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, N.; Kinoshita, K.; Kagawa, T.; Tanaka, A.; Ueno, O.; Shimada, H.; Takano, M.; Phytochrome, B. Phytochrome B Mediates the Regulation of Chlorophyll Biosynthesis through Transcriptional Regulation of ChlH and GUN4 in Rice Seedlings. PLoS ONE 2015, 10, e0135408. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, X.; He, B.; Diao, L.; Sheng, S.; Wang, J.; Guo, X.; Su, N.; Wang, L.; Jiang, L.; et al. A Chlorophyll-Deficient Rice Mutant with Impaired Chlorophyllide Esterification in Chlorophyll Biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, S.H.; Kao, C.H. The Role of Proteolytic Enzymes in Protein Degradation during Senescence of Rice Leaves. Physiol. Plant. 1984, 62, 231–237. [Google Scholar] [CrossRef]
- Roberts, I.N.; Caputo, C.; Criado, M.V.; Funk, C. Senescence Associated Proteases in Plants. Physiol. Plant. 2012, 145, 130–139. [Google Scholar] [CrossRef]
- Kinoshita, T.; Yamada, K.; Hiraiwa, N.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Vacuolar Processing Enzyme is Up-Regulated in the Lytic Vacuoles of Vegetative Tissues during Senescence and Under Various Stressed Conditions. Plant J. 1999, 19, 43–53. [Google Scholar] [CrossRef]
- Buet, A.; Costa, M.L.; Martínez, D.E.; Guiamet, J.J. Chloroplast protein degradation in senescing leaves: Proteases and lytic compartments. Front. Plant Sci. 2019, 10, 747. [Google Scholar] [CrossRef]
- Park, S.-Y.; Yu, J.-W.; Park, J.-S.; Li, J.; Yoo, S.-C.; Lee, N.-Y.; Lee, S.-K.; Jeong, S.-W.; Seo, H.-S.; Koh, H.-J.; et al. The Senescence-Induced Staygreen Protein Regulates Chlorophyll Degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two Short-Chain Dehydrogenase/Reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are Required for Chlorophyll b and Light-Harvesting Complex II Degradation during Senescence in Rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.; Sato, Y.; Masuda, Y.; Nishimura, M.; Kusaba, M. Defect in Non-Yellow Coloring 3, an α/β Hydrolase-Fold Family Protein, Causes a Stay-Green Phenotype during Leaf Senescence in Rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sullivan, R.W.; Kight, A.; Henry, R.L.; Huang, J.; Jones, A.M.; Korth, K.L. Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 2004, 136, 3594–3604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamatani, H.; Sato, Y.; Masuda, Y.; Kato, Y.; Morita, R.; Fukunaga, K.; Nagamura, Y.; Nishimura, M.; Sakamoto, W.; Tanaka, A.; et al. NYC4, the Rice Ortholog of Arabidopsis THF1, is Involved in the Degradation of Chlorophyll–Protein Complexes during Leaf Senescence. Plant J. 2013, 74, 652–662. [Google Scholar] [CrossRef]
- Krupinska, K. Fate and Activities of Plastids during Leaf Senescence. In The Structure and Function of Plastids; Springer: Berlin, Germany, 2007; pp. 433–449. [Google Scholar]
- Chiba, A.; Ishida, H.; Nishizawa, N.K.; Makino, A.; Mae, T. Exclusion of Ribulose-1, 5-Bisphosphate Carboxylase/Oxygenase from Chloroplasts by Specific Bodies in Naturally Senescing Leaves of Wheat. Plant Cell Physiol. 2003, 44, 914–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, H.; Yoshimoto, K.; Izumi, M.; Reisen, D.; Yano, Y.; Makino, A.; Ohsumi, Y.; Hanson, M.R.; Mae, T. Mobilization of Rubisco and Stroma-Localized Fluorescent Proteins of Chloroplasts to the Vacuole by An ATG Gene-Dependent Autophagic Process. Plant Physiol. 2008, 148, 142–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hensel, L.L.; Grbić, V.; Baumgarten, D.A.; Bleecker, A.B. Developmental and Age-Related Processes that Influence the Longevity and Senescence of Photosynthetic Tissues in Arabidopsis. Plant Cell 1993, 5, 553–564. [Google Scholar]
- Gregersen, P.L.; Holm, P.B. Transcriptome Analysis of Senescence in the Flag Leaf of Wheat (Triticum Aestivum L.). Plant Biotechnol. J. 2007, 5, 192–206. [Google Scholar] [CrossRef]
- Parrott, D.L.; Martin, J.M.; Fischer, A.M. Analysis of Barley (Hordeum Vulgare) Leaf Senescence and Protease Gene Expression: A Family C1A Cysteine Protease is Specifically Induced Under Conditions Characterized by High Carbohydrate, But Low to Moderate Nitrogen Levels. New Phytol. 2010, 187, 313–331. [Google Scholar] [CrossRef]
- Prins, A.; Van Heerden, P.D.; Olmos, E.; Kunert, K.J.; Foyer, C.H. Cysteine Proteinases Regulate Chloroplast Protein Content and Composition in Tobacco Leaves: A Model for Dynamic Interactions with Ribulose-1, 5-Bisphosphate Carboxylase/Oxygenase (Rubisco) Vesicular Bodies. J. Exp. Bot. 2008, 59, 1935–1950. [Google Scholar] [CrossRef] [Green Version]
- Kato, Y.; Murakami, S.; Yamamoto, Y.; Chatani, H.; Kondo, Y.; Nakano, T.; Yokota, A.; Sato, F. The DNA-Binding Protease, CND41, and the Degradation of Ribulose-1, 5-Bisphosphate Carboxylase/Oxygenase in Senescent Leaves of Tobacco. Planta 2004, 220, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, M.; Wang, G.; Galili, G. Autophagy: An Important Biological Process that Protects Plants from Stressful Environments. Front. Plant Sci. 2017, 7, 2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Wang, W.; Bittner, F.; Schmidt, N.; Berkey, R.; Zhang, L.; King, H.; Zhang, Y.; Feng, J.; Wen, Y.; et al. Dual and Opposing Roles of Xanthine Dehydrogenase in Defense-Associated Reactive Oxygen Species Metabolism in Arabidopsis. Plant Cell 2016, 28, 1108–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, R.; He, X.; Pan, X.; Shi, Q.; Wu, Z. Enhancing xanthine dehydrogenase activity is an effective way to delay leaf senescence and increase rice yield. Rice 2020, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Rasheed, A.; Wang, Y.; Wu, Z.; Tang, S.; Shi, Q.; Wu, Z.M. Silencing of OsXDH Reveals the Role of Purine Metabolism in Dark Tolerance in Rice Seedlings. J. Integr. Agric. 2018, 17, 1736–1744. [Google Scholar] [CrossRef]
- Zhu, P.; Yang, S.-M.; Ma, J.; Li, S.-X.; Chen, Y. Effect of Shading on the Photosynthetic Characteristics and Yield at Later Growth Stage of Hybrid Rice Combination. Acta Agron. Sin. 2008, 34, 2003–2009. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gad, A.G.; Habiba; Zheng, X.; Miao, Y. Low Light/Darkness as Stressors of Multifactor-Induced Senescence in Rice Plants. Int. J. Mol. Sci. 2021, 22, 3936. https://doi.org/10.3390/ijms22083936
Gad AG, Habiba, Zheng X, Miao Y. Low Light/Darkness as Stressors of Multifactor-Induced Senescence in Rice Plants. International Journal of Molecular Sciences. 2021; 22(8):3936. https://doi.org/10.3390/ijms22083936
Chicago/Turabian StyleGad, Ahmed G., Habiba, Xiangzi Zheng, and Ying Miao. 2021. "Low Light/Darkness as Stressors of Multifactor-Induced Senescence in Rice Plants" International Journal of Molecular Sciences 22, no. 8: 3936. https://doi.org/10.3390/ijms22083936





