Bile Acids and Microbiota: Multifaceted and Versatile Regulators of the Liver–Gut Axis
Abstract
1. Introduction
2. Bile Acids (BAs) and Enterohepatic Circulation
3. Local Site Microbiota in the Gut and Bile
4. Crosstalk between BAs and Microbiota
5. Functional Consequences
5.1. Infectious Disease
5.2. Inflammatory Bowel Disease (IBD)
5.3. Immune-Mediated Disease of the Liver
5.4. Hepato-Intestinal Carcinogenesis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AAD | antibiotic-associated diarrhea |
| ASBT | apical sodium-dependent bile acid transporter |
| BA | bile acid |
| BFT | Bacteroides fragilis toxin |
| BSH | bile salt hydrolases |
| CA | cholic acid |
| CD | Crohn’s disease |
| CDCA | chenodeoxycholic acid |
| CRC | colorectal cancer |
| DCA | deoxycholic acid |
| EHC | enterohepatic circulation |
| ETBF | enterotoxigenic Bacteroides fragilis |
| FXR | nuclear farnesoid X receptor |
| GI | gastrointestinal tract |
| Gly | glycine |
| HCC | hepatocellular carcinoma |
| IBD | inflammatory bowel disease |
| IFNg | interferon gamma |
| IgA | immunoglobulin A |
| IL1-b | interleukin-1 beta |
| IL-6 | interleukin-6 |
| IL-10 | interleukin-10 |
| LCA | lithocholic acid |
| LPS | lipopolysaccharide |
| NASH | nonalcoholic steatohepatitis |
| NKT | natural killer T cells |
| PBC | primary biliary cirrhosis |
| PSC | primary sclerosing cholangitis |
| Tau | taurine |
| TDG | thymine DNA glycosylase |
| TGF-b | transforming growth factor beta |
| TNF-a | tumor necrosis factor alpha |
| T3SS | type-3-secretion system |
| UC | ulcerative colitis |
| UCDA | ursodeoxycholic acid |
References
- Schroeder, B.O.; Backhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef]
- Belkaid, Y.; Harrison, O.J. Homeostatic Immunity and the Microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Nunez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Xavier, J.B.; Young, V.B.; Skufca, J.; Ginty, F.; Testerman, T.; Pearson, A.T.; Macklin, P.; Mitchell, A.; Shmulevich, I.; Xie, L.; et al. The Cancer Microbiome: Distinguishing Direct and Indirect Effects Requires a Systemic View. Trends Cancer 2020, 6, 192–204. [Google Scholar] [CrossRef]
- Sharon, G.; Sampson, T.R.; Geschwind, D.H.; Mazmanian, S.K. The Central Nervous System and the Gut Microbiome. Cell 2016, 167, 915–932. [Google Scholar] [CrossRef]
- de Souza, H.S.P.; Fiocchi, C.; Iliopoulos, D. The IBD interactome: An integrated view of aetiology, pathogenesis and therapy. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Baier, J.; Gansbauer, M.; Giessler, C.; Arnold, H.; Muske, M.; Schleicher, U.; Lukassen, S.; Ekici, A.; Rauh, M.; Daniel, C.; et al. Arginase impedes the resolution of colitis by altering the microbiome and metabolome. J. Clin. Investig. 2020, 130, 5703–5720. [Google Scholar] [CrossRef] [PubMed]
- Ferdinandusse, S.; Houten, S.M. Peroxisomes and bile acid biosynthesis. Biochim. Biophys. Acta 2006, 1763, 1427–1440. [Google Scholar] [CrossRef]
- Hofmann, A.F.; Borgstroem, B. The Intraluminal Phase of Fat Digestion in Man: The Lipid Content of the Micellar and Oil Phases of Intestinal Content Obtained during Fat Digestion and Absorption. J. Clin. Investig. 1964, 43, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Metz, P.; Tjan, M.J.H.; Wu, S.; Pervaiz, M.; Hermans, S.; Shettigar, A.; Sears, C.L.; Ritschel, T.; Dutilh, B.E.; Boleij, A. Drug Discovery and Repurposing Inhibits a Major Gut Pathogen-Derived Oncogenic Toxin. Front. Cell Infect. Microbiol. 2019, 9, 364. [Google Scholar] [CrossRef]
- Wotzka, S.Y.; Kreuzer, M.; Maier, L.; Arnoldini, M.; Nguyen, B.D.; Brachmann, A.O.; Berthold, D.L.; Zund, M.; Hausmann, A.; Bakkeren, E.; et al. Escherichia coli limits Salmonella Typhimurium infections after diet shifts and fat-mediated microbiota perturbation in mice. Nat. Microbiol. 2019, 4, 2164–2174. [Google Scholar] [CrossRef]
- Hernandez, S.B.; Cota, I.; Ducret, A.; Aussel, L.; Casadesus, J. Adaptation and preadaptation of Salmonella enterica to Bile. PLoS Genet. 2012, 8, e1002459. [Google Scholar] [CrossRef]
- Johnson, R.; Ravenhall, M.; Pickard, D.; Dougan, G.; Byrne, A.; Frankel, G. Comparison of Salmonella enterica Serovars Typhi and Typhimurium Reveals Typhoidal Serovar-Specific Responses to Bile. Infect. Immun. 2018, 86, e00490-17. [Google Scholar] [CrossRef]
- Rosenberg, E.Y.; Bertenthal, D.; Nilles, M.L.; Bertrand, K.P.; Nikaido, H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 2003, 48, 1609–1619. [Google Scholar] [CrossRef]
- Sistrunk, J.R.; Nickerson, K.P.; Chanin, R.B.; Rasko, D.A.; Faherty, C.S. Survival of the Fittest: How Bacterial Pathogens Utilize Bile To Enhance Infection. Clin. Microbiol. Rev. 2016, 29, 819–836. [Google Scholar] [CrossRef]
- Kramer, V.C.; Nickerson, K.W.; Hamlett, N.V.; O’Hara, C. Prevalence of extreme detergent resistance among the Enterobacteriaceae. Can. J. Microbiol. 1984, 30, 711–713. [Google Scholar] [CrossRef] [PubMed]
- Kwan, B.W.; Lord, D.M.; Peti, W.; Page, R.; Benedik, M.J.; Wood, T.K. The MqsR/MqsA toxin/antitoxin system protects Escherichia coli during bile acid stress. Environ. Microbiol. 2015, 17, 3168–3181. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, C.; Ray, S.; Chowdhury, R. Fine tuning of virulence regulatory pathways in enteric bacteria in response to varying bile and oxygen concentrations in the gastrointestinal tract. Gut. Pathog. 2014, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Giel, J.L.; Sorg, J.A.; Sonenshein, A.L.; Zhu, J. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS ONE 2010, 5, e8740. [Google Scholar] [CrossRef]
- Sorg, J.A.; Sonenshein, A.L. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 2010, 192, 4983–4990. [Google Scholar] [CrossRef]
- Weingarden, A.R.; Chen, C.; Bobr, A.; Yao, D.; Lu, Y.; Nelson, V.M.; Sadowsky, M.J.; Khoruts, A. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G310–G319. [Google Scholar] [CrossRef]
- Fitzpatrick, L.R.; Jenabzadeh, P. IBD and Bile Acid Absorption: Focus on Pre-clinical and Clinical Observations. Front. Physiol. 2020, 11, 564. [Google Scholar] [CrossRef]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360. [Google Scholar] [CrossRef]
- Hofmann, A.F. The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med. 1999, 159, 2647–2658. [Google Scholar] [CrossRef]
- Chen, I.; Cassaro, S. Physiology, Bile Acids; StatPearls: Treasure Island, FL, USA, 2020. [Google Scholar]
- Di Ciaula, A.; Garruti, G.; Lunardi Baccetto, R.; Molina-Molina, E.; Bonfrate, L.; Wang, D.Q.; Portincasa, P. Bile Acid Physiology. Ann. Hepatol. 2017, 16, s4–s14. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef] [PubMed]
- Gerard, P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013, 3, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.P.; Xie, G.; Raufman, J.P.; Hogan, S.; Griffin, T.L.; Packard, C.A.; Chatfield, D.A.; Hagey, L.R.; Steinbach, J.H.; Hofmann, A.F. Human cecal bile acids: Concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G256–G263. [Google Scholar] [CrossRef]
- Hofmann, A.F.; Hagey, L.R.; Krasowski, M.D. Bile salts of vertebrates: Structural variation and possible evolutionary significance. J. Lipid Res. 2010, 51, 226–246. [Google Scholar] [CrossRef]
- Fiorucci, S.; Mencarelli, A.; Palladino, G.; Cipriani, S. Bile-acid-activated receptors: Targeting TGR5 and farnesoid-X-receptor in lipid and glucose disorders. Trends Pharm. Sci. 2009, 30, 570–580. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Hollister, K.; Sowers, L.C.; Forman, B.M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 1999, 3, 543–553. [Google Scholar] [CrossRef]
- Maruyama, T.; Miyamoto, Y.; Nakamura, T.; Tamai, Y.; Okada, H.; Sugiyama, E.; Nakamura, T.; Itadani, H.; Tanaka, K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 2002, 298, 714–719. [Google Scholar] [CrossRef]
- Kawamata, Y.; Fujii, R.; Hosoya, M.; Harada, M.; Yoshida, H.; Miwa, M.; Fukusumi, S.; Habata, Y.; Itoh, T.; Shintani, Y.; et al. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003, 278, 9435–9440. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Wolf, P.G.; Gaskins, H.R. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 2016, 7, 201–215. [Google Scholar] [CrossRef]
- Song, M.; Yang, Q.; Zhang, F.L.; Chen, L.; Su, H.; Yang, X.H.; He, H.W.; Liu, F.F.; Zheng, J.S.; Ling, M.F.; et al. Hyodeoxycholic acid (HDCA) suppresses intestinal epithelial cell proliferation through FXR-PI3K/AKT pathway, accompanied by alteration of bile acids metabolism profiles induced by gut bacteria. Faseb. J. 2020, 34, 7103–7117. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.Y.; Yun, C.Y.; Sun, L.L.; Xia, J.L.; Wu, Q.; Wang, Y.; Wang, L.N.; Zhang, Y.M.; Liang, X.Y.; Wang, L.Y.; et al. Gut microbiota-bile acid-interleukin-22 axis orchestrates polycystic ovary syndrome. Nat. Med. 2019, 25, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Marksteiner, J.; Blasko, I.; Kemmler, G.; Koal, T.; Humpel, C. Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer’s disease. Metabolomics 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Zhao, J.F.; Gui, W.F.; Sun, D.; Dai, H.J.; Xiao, L.; Chu, H.K.; Du, F.; Zhu, Q.J.; Schnabl, B.; et al. Tauroursodeoxycholic acid inhibits intestinal inflammation and barrier disruption in mice with non-alcoholic fatty liver disease. Br. J. Pharm. 2018, 175, 469–484. [Google Scholar] [CrossRef]
- Vandewynckel, Y.P.; Laukens, D.; Devisscher, L.; Paridaens, A.; Bogaerts, E.; Verhelst, X.; Van den Bussche, A.; Raevens, S.; Van Steenkiste, C.; Van Troys, M.; et al. Tauroursodeoxycholic Acid Dampens Oncogenic Apoptosis Induced by Endoplasmic Reticulum Stress during Hepatocarcinogen Exposure. J. Hepatol. 2015, 62, S402–S403. [Google Scholar] [CrossRef]
- He, J.; Liang, J.R.; Zhu, S.; Zhao, W.N.; Zhang, Y.M.; Sun, W.J. Protective effect of taurohyodeoxycholic acid from Pulvis Fellis Suis on trinitrobenzene sulfonic acid induced ulcerative colitis in mice. Eur. J. Pharm. 2011, 670, 229–235. [Google Scholar] [CrossRef]
- Li, F.; Jiang, C.T.; Krausz, K.W.; Li, Y.F.; Albert, I.; Hao, H.P.; Fabre, K.M.; Mitchell, J.B.; Patterson, A.D.; Gonzalez, F.J. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 2013, 4, 2384. [Google Scholar] [CrossRef]
- Pan, X.B.; Elliott, C.T.; McGuinness, B.; Passmore, P.; Kehoe, P.G.; Holscher, C.; McClean, P.L.; Graham, S.F.; Green, B.D. Metabolomic Profiling of Bile Acids in Clinical and Experimental Samples of Alzheimer’s Disease. Metabolites 2017, 7, 28. [Google Scholar] [CrossRef]
- McGlone, E.R.; Bloom, S.R. Bile acids and the metabolic syndrome. Ann. Clin. Biochem. 2019, 56, 326–337. [Google Scholar] [CrossRef]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile Acids Activated Receptors Regulate innate immunity. Front. Immunol. 2018, 9, 1853. [Google Scholar] [CrossRef]
- Wammers, M.; Schupp, A.K.; Bode, J.G.; Ehlting, C.; Wolf, S.; Deenen, R.; Kohrer, K.; Haussinger, D.; Graf, D. Reprogramming of pro-inflammatory human macrophages to an anti-inflammatory phenotype by bile acids. Sci. Rep. 2018, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Biagioli, M.; Carino, A.; Cipriani, S.; Francisci, D.; Marchiano, S.; Scarpelli, P.; Sorcini, D.; Zampella, A.; Fiorucci, S. The Bile Acid Receptor GPBAR1 Regulates the M1/M2 Phenotype of Intestinal Macrophages and Activation of GPBAR1 Rescues Mice from Murine Colitis. J. Immunol. 2017, 199, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Gahan, C.G.; Hill, C. The interaction between bacteria and bile. Fems. Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Brestoff, J.R.; Artis, D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013, 14, 676–684. [Google Scholar] [CrossRef]
- Zeng, M.Y.; Inohara, N.; Nunez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef]
- Molinero, N.; Ruiz, L.; Milani, C.; Gutierrez-Diaz, I.; Sanchez, B.; Mangifesta, M.; Segura, J.; Cambero, I.; Campelo, A.B.; Garcia-Bernardo, C.M.; et al. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome 2019, 7, 100. [Google Scholar] [CrossRef]
- Folseraas, T.; Melum, E.; Rausch, P.; Juran, B.D.; Ellinghaus, E.; Shiryaev, A.; Laerdahl, J.K.; Ellinghaus, D.; Schramm, C.; Weismuller, T.J.; et al. Extended analysis of a genome-wide association study in primary sclerosing cholangitis detects multiple novel risk loci. J. Hepatol. 2012, 57, 366–375. [Google Scholar] [CrossRef]
- Verdier, J.; Luedde, T.; Sellge, G. Biliary Mucosal Barrier and Microbiome. Viszeralmedizin 2015, 31, 156–161. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Grill, J.P.; Perrin, S.; Schneider, F. Bile salt toxicity to some bifidobacteria strains: Role of conjugated bile salt hydrolase and pH. Can. J. Microbiol. 2000, 46, 878–884. [Google Scholar] [CrossRef] [PubMed]
- Dussurget, O.; Cabanes, D.; Dehoux, P.; Lecuit, M.; Buchrieser, C.; Glaser, P.; Cossart, P. European Listeria Genome Consortium. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol 2002, 45, 1095–1106. [Google Scholar] [CrossRef]
- Carbonero, F.; Benefiel, A.C.; Alizadeh-Ghamsari, A.H.; Gaskins, H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012, 3, 448. [Google Scholar] [CrossRef] [PubMed]
- Out, C.; Patankar, J.V.; Doktorova, M.; Boesjes, M.; Bos, T.; de Boer, S.; Havinga, R.; Wolters, H.; Boverhof, R.; van Dijk, T.H.; et al. Gut microbiota inhibit Asbt-dependent intestinal bile acid reabsorption via Gata4. J. Hepatol. 2015, 63, 697–704. [Google Scholar] [CrossRef]
- Sayin, S.I.; Wahlstrom, A.; Felin, J.; Jantti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyotylainen, T.; Oresic, M.; Backhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef]
- Wahlstrom, A.; Sayin, S.I.; Marschall, H.U.; Backhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Chaix, A.; Xu, Z.Z.; Chang, M.W.; Marotz, C.A.; Saghatelian, A.; Knight, R.; Panda, S. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 2018, 9, 2872. [Google Scholar] [CrossRef]
- Long, S.L.; Gahan, C.G.M.; Joyce, S.A. Interactions between gut bacteria and bile in health and disease. Mol. Asp. Med. 2017, 56, 54–65. [Google Scholar] [CrossRef]
- Urdaneta, V.; Casadesus, J. Interactions between Bacteria and Bile Salts in the Gastrointestinal and Hepatobiliary Tracts. Front. Med. (Lausanne) 2017, 4, 163. [Google Scholar] [CrossRef]
- Bernstein, C.; Bernstein, H.; Payne, C.M.; Beard, S.E.; Schneider, J. Bile salt activation of stress response promoters in Escherichia coli. Curr. Microbiol. 1999, 39, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Islam, K.B.; Fukiya, S.; Hagio, M.; Fujii, N.; Ishizuka, S.; Ooka, T.; Ogura, Y.; Hayashi, T.; Yokota, A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011, 141, 1773–1781. [Google Scholar] [CrossRef] [PubMed]
- Kakiyama, G.; Pandak, W.M.; Gillevet, P.M.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; Takei, H.; Muto, A.; Nittono, H.; Ridlon, J.M.; et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 2013, 58, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, T.; Moschetta, A.; Lee, Y.K.; Peng, L.; Zhao, G.; Downes, M.; Yu, R.T.; Shelton, J.M.; Richardson, J.A.; Repa, J.J.; et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 3920–3925. [Google Scholar] [CrossRef]
- Vavassori, P.; Mencarelli, A.; Renga, B.; Distrutti, E.; Fiorucci, S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol. 2009, 183, 6251–6261. [Google Scholar] [CrossRef]
- Cipriani, S.; Mencarelli, A.; Chini, M.G.; Distrutti, E.; Renga, B.; Bifulco, G.; Baldelli, F.; Donini, A.; Fiorucci, S. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS ONE 2011, 6, e25637. [Google Scholar] [CrossRef]
- David, L.A.; Materna, A.C.; Friedman, J.; Campos-Baptista, M.I.; Blackburn, M.C.; Perrotta, A.; Erdman, S.E.; Alm, E.J. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014, 15, R89. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353.e1321. [Google Scholar] [CrossRef]
- Alvarez-Ordonez, A.; Begley, M.; Prieto, M.; Messens, W.; Lopez, M.; Bernardo, A.; Hill, C. Salmonella spp. survival strategies within the host gastrointestinal tract. Microbiology 2011, 157, 3268–3281. [Google Scholar] [CrossRef]
- Devendran, S.; Shrestha, R.; Alves, J.M.P.; Wolf, P.G.; Ly, L.; Hernandez, A.G.; Mendez-Garcia, C.; Inboden, A.; Wiley, J.; Paul, O.; et al. Clostridium scindens ATCC 35704: Integration of Nutritional Requirements, the Complete Genome Sequence, and Global Transcriptional Responses to Bile Acids. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Grau, K.R.; Zhu, S.; Peterson, S.T.; Helm, E.W.; Philip, D.; Phillips, M.; Hernandez, A.; Turula, H.; Frasse, P.; Graziano, V.R.; et al. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat. Microbiol. 2020, 5, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Abdulrab, S.; Al-Maweri, S.; Halboub, E. Ursodeoxycholic acid as a candidate therapeutic to alleviate and/or prevent COVID-19-associated cytokine storm. Med. Hypotheses 2020, 143, 109897. [Google Scholar] [CrossRef]
- Carino, A.; Moraca, F.; Fiorillo, B.; Marchiano, S.; Sepe, V.; Biagioli, M.; Finamore, C.; Bozza, S.; Francisci, D.; Distrutti, E.; et al. Hijacking SARS-CoV-2/ACE2 Receptor Interaction by Natural and Semi-synthetic Steroidal Agents Acting on Functional Pockets on the Receptor Binding Domain. Front. Chem. 2020, 8, 572885. [Google Scholar] [CrossRef]
- Abraham, C.; Cho, J.H. Inflammatory bowel disease. New. Engl. J. Med. 2009, 361, 2066–2078. [Google Scholar] [CrossRef]
- Baumgart, D.C.; Carding, S.R. Inflammatory bowel disease: Cause and immunobiology. Lancet 2007, 369, 1627–1640. [Google Scholar] [CrossRef]
- Engel, M.A.; Neurath, M.F. New pathophysiological insights and modern treatment of IBD. J. Gastroenterol. 2010, 45, 571–583. [Google Scholar] [CrossRef]
- Braun, J.; Wei, B. Body traffic: Ecology, genetics, and immunity in inflammatory bowel disease. Annu. Rev. Pathol. 2007, 2, 401–429. [Google Scholar] [CrossRef][Green Version]
- Connors, J.; Dunn, K.A.; Allott, J.; Bandsma, R.; Rashid, M.; Otley, A.R.; Bielawski, J.P.; Van Limbergen, J. The relationship between fecal bile acids and microbiome community structure in pediatric Crohn’s disease. ISME J. 2020, 14, 702–713. [Google Scholar] [CrossRef]
- Dawson, P.A.; Karpen, S.J. Intestinal transport and metabolism of bile acids. J. Lipid Res. 2015, 56, 1085–1099. [Google Scholar] [CrossRef] [PubMed]
- Pavlidis, P.; Powell, N.; Vincent, R.P.; Ehrlich, D.; Bjarnason, I.; Hayee, B. Systematic review: Bile acids and intestinal inflammation-luminal aggressors or regulators of mucosal defence? Aliment. Pharm. 2015, 42, 802–817. [Google Scholar] [CrossRef] [PubMed]
- Rau, M.; Stieger, B.; Monte, M.J.; Schmitt, J.; Jahn, D.; Frey-Wagner, I.; Raselli, T.; Marin, J.J.; Mullhaupt, B.; Rogler, G.; et al. Alterations in Enterohepatic Fgf15 Signaling and Changes in Bile Acid Composition Depend on Localization of Murine Intestinal Inflammation. Inflamm. Bowel Dis. 2016, 22, 2382–2389. [Google Scholar] [CrossRef] [PubMed]
- Guard, B.C.; Honneffer, J.B.; Jergens, A.E.; Jonika, M.M.; Toresson, L.; Lawrence, Y.A.; Webb, C.B.; Hill, S.; Lidbury, J.A.; Steiner, J.M.; et al. Longitudinal assessment of microbial dysbiosis, fecal unconjugated bile acid concentrations, and disease activity in dogs with steroid-responsive chronic inflammatory enteropathy. J. Vet. Intern. Med. 2019, 33, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Jahnel, J.; Fickert, P.; Hauer, A.C.; Hogenauer, C.; Avian, A.; Trauner, M. Inflammatory bowel disease alters intestinal bile acid transporter expression. Drug Metab. Dispos. 2014, 42, 1423–1431. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.; Fantin, A.C.; Scheurer, U.; Fried, M.; Kullak-Ublick, G.A. Human ileal bile acid transporter gene ASBT (SLC10A2) is transactivated by the glucocorticoid receptor. Gut 2004, 53, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Sun, X.; Oh, S.F.; Wu, M.; Zhang, Y.; Zheng, W.; Geva-Zatorsky, N.; Jupp, R.; Mathis, D.; Benoist, C.; et al. Microbial bile acid metabolites modulate gut RORgamma+ regulatory T cell homeostasis. Nature 2020, 577, 410–415. [Google Scholar] [CrossRef]
- Paramsothy, S.; Nielsen, S.; Kamm, M.A.; Deshpande, N.P.; Faith, J.J.; Clemente, J.C.; Paramsothy, R.; Walsh, A.J.; van den Bogaerde, J.; Samuel, D.; et al. Specific Bacteria and Metabolites Associated With Response to Fecal Microbiota Transplantation in Patients With Ulcerative Colitis. Gastroenterology 2019, 156, 1440–1454 e1442. [Google Scholar] [CrossRef]
- Mattner, J. Impact of Microbes on the Pathogenesis of Primary Biliary Cirrhosis (PBC) and Primary Sclerosing Cholangitis (PSC). Int. J. Mol. Sci. 2016, 17, 1864. [Google Scholar] [CrossRef]
- Liwinski, T.; Zenouzi, R.; John, C.; Ehlken, H.; Ruhlemann, M.C.; Bang, C.; Groth, S.; Lieb, W.; Kantowski, M.; Andersen, N.; et al. Alterations of the bile microbiome in primary sclerosing cholangitis. Gut 2020, 69, 665–672. [Google Scholar] [CrossRef]
- Ruhlemann, M.C.; Solovjeva, M.E.L.; Zenouzi, R.; Liwinski, T.; Kummen, M.; Lieb, W.; Hov, J.R.; Schramm, C.; Franke, A.; Bang, C. Gut mycobiome of primary sclerosing cholangitis patients is characterised by an increase of Trichocladium griseum and Candida species. Gut 2020, 69, 1890–1892. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Yamamoto, K. Role of gut microbiota in liver diseases. Hepatol. Res. 2013, 43, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Pflughoeft, K.J.; Versalovic, J. Human microbiome in health and disease. Annu. Rev. Pathol. 2012, 7, 99–122. [Google Scholar] [CrossRef] [PubMed]
- Mattner, J.; Savage, P.B.; Leung, P.; Oertelt, S.S.; Wang, V.; Trivedi, O.; Scanlon, S.T.; Pendem, K.; Teyton, L.; Hart, J.; et al. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe 2008, 3, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, J.P.; Fusakio, M.E.; Rainbow, D.B.; Moule, C.; Fraser, H.I.; Clark, J.; Todd, J.A.; Peterson, L.B.; Savage, P.B.; Wills-Karp, M.; et al. Identification of Cd101 as a susceptibility gene for Novosphingobium aromaticivorans-induced liver autoimmunity. J. Immunol. 2011, 187, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Eaton, J.E.; Talwalkar, J.A.; Lazaridis, K.N.; Gores, G.J.; Lindor, K.D. Pathogenesis of primary sclerosing cholangitis and advances in diagnosis and management. Gastroenterology 2013, 145, 521–536. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.; Palmela, C.; Brito, H.; Bao, X.; Ruiqi, H.; Moura-Santos, P.; da Silva, J.P.; Oliveira, A.; Vieira, C.; Perez, K.; et al. The gut microbiota, bile acids and their correlation in primary sclerosing cholangitis associated with inflammatory bowel disease. United Eur. Gastroenterol. J. 2018, 6, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Rossen, N.G.; Fuentes, S.; Boonstra, K.; D’Haens, G.R.; Heilig, H.G.; Zoetendal, E.G.; de Vos, W.M.; Ponsioen, C.Y. The mucosa-associated microbiota of PSC patients is characterized by low diversity and low abundance of uncultured Clostridiales II. J. Crohns Colitis 2015, 9, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Lichtman, S.N.; Okoruwa, E.E.; Keku, J.; Schwab, J.H.; Sartor, R.B. Degradation of endogenous bacterial cell wall polymers by the muralytic enzyme mutanolysin prevents hepatobiliary injury in genetically susceptible rats with experimental intestinal bacterial overgrowth. J. Clin. Investig. 1992, 90, 1313–1322. [Google Scholar] [CrossRef]
- Tabibian, J.H.; Talwalkar, J.A.; Lindor, K.D. Role of the microbiota and antibiotics in primary sclerosing cholangitis. Biomed. Res. Int. 2013, 2013, 389537. [Google Scholar] [CrossRef]
- Weingarden, A.R.; Chen, C.; Zhang, N.; Graiziger, C.T.; Dosa, P.I.; Steer, C.J.; Shaughnessy, M.K.; Johnson, J.R.; Sadowsky, M.J.; Khoruts, A. Ursodeoxycholic Acid Inhibits Clostridium difficile Spore Germination and Vegetative Growth, and Prevents the Recurrence of Ileal Pouchitis Associated With the Infection. J. Clin. Gastroenterol. 2016, 50, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Finlay, B.B.; Goldszmid, R.; Honda, K.; Trinchieri, G.; Wargo, J.; Zitvogel, L. Can we harness the microbiota to enhance the efficacy of cancer immunotherapy? Nat. Rev. Immunol. 2020, 20, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Vetizou, M.; Pitt, J.M.; Daillere, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillere, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Anker, J.F.; Naseem, A.F.; Mok, H.; Schaeffer, A.J.; Abdulkadir, S.A.; Thumbikat, P. Multi-faceted immunomodulatory and tissue-tropic clinical bacterial isolate potentiates prostate cancer immunotherapy. Nat. Commun. 2018, 9, 1591. [Google Scholar] [CrossRef]
- Wang, S.; Dong, W.; Liu, L.; Xu, M.; Wang, Y.; Liu, T.; Zhang, Y.; Wang, B.; Cao, H. Interplay between bile acids and the gut microbiota promotes intestinal carcinogenesis. Mol. Carcinog. 2019, 58, 1155–1167. [Google Scholar] [CrossRef]
- Hassan, H.M.; Isovic, M.; Kolendowski, B.; Bauer-Maison, N.; Onabote, O.; Cecchini, M.; Haig, A.; Maleki Vareki, S.; Underhill, T.M.; Torchia, J. Loss of Thymine DNA Glycosylase Causes Dysregulation of Bile Acid Homeostasis and Hepatocellular Carcinoma. Cell Rep. 2020, 31, 107475. [Google Scholar] [CrossRef]
- Phelan, J.P.; Reen, F.J.; Caparros-Martin, J.A.; O’Connor, R.; O’Gara, F. Rethinking the bile acid/gut microbiome axis in cancer. Oncotarget 2017, 8, 115736–115747. [Google Scholar] [CrossRef]
- Ren, X.H.; Xu, J.; Zhang, Y.Y.; Chen, G.D.; Zhang, Y.W.; Huang, Q.; Liu, Y.L. Bacterial Alterations in Post-Cholecystectomy Patients Are Associated With Colorectal Cancer. Front. Oncol. 2020, 10, 1418. [Google Scholar] [CrossRef]
- Yoon, W.J.; Kim, H.N.; Park, E.; Ryu, S.; Chang, Y.; Shin, H.; Kim, H.L.; Yi, S.Y. The Impact of Cholecystectomy on the Gut Microbiota: A Case-Control Study. J. Clin. Med. 2019, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Wang, J.F.; Li, J.L.; Yan, P.P.; Jin, Y.; Zhang, R.Y.; Yue, W.; Guo, Q.; Geng, J.W. Cholecystectomy Damages Aging-Associated Intestinal Microbiota Construction. Front. Microbiol. 2018, 9, 1402. [Google Scholar] [CrossRef] [PubMed]
- Sarashina-Kida, H.; Negishi, H.; Nishio, J.; Suda, W.; Nakajima, Y.; Yasui-Kato, M.; Iwaisako, K.; Kang, S.J.; Endo, N.; Yanai, H.; et al. Gallbladder-derived surfactant protein D regulates gut commensal bacteria for maintaining intestinal homeostasis. Proc. Natl. Acad. Sci. USA 2017, 114, 10178–10183. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Xu, M.; Dong, W.; Deng, B.; Wang, S.; Zhang, Y.; Wang, S.; Luo, S.; Wang, W.; Qi, Y.; et al. Secondary bile acid-induced dysbiosis promotes intestinal carcinogenesis. Int. J. Cancer 2017, 140, 2545–2556. [Google Scholar] [CrossRef]
- Hale, V.L.; Chen, J.; Johnson, S.; Harrington, S.C.; Yab, T.C.; Smyrk, T.C.; Nelson, H.; Boardman, L.A.; Druliner, B.R.; Levin, T.R.; et al. Shifts in the Fecal Microbiota Associated with Adenomatous Polyps. Cancer Epidemiol. Biomark. Prev. 2017, 26, 85–94. [Google Scholar] [CrossRef]
- Chen, D.; Jin, D.; Huang, S.; Wu, J.; Xu, M.; Liu, T.; Dong, W.; Liu, X.; Wang, S.; Zhong, W.; et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020, 469, 456–467. [Google Scholar] [CrossRef]
- Zeng, H.; Safratowich, B.D.; Wang, T.T.Y.; Hamlin, S.K.; Johnson, L.K. Butyrate Inhibits Deoxycholic-Acid-Resistant Colonic Cell Proliferation via Cell Cycle Arrest and Apoptosis: A Potential Pathway Linking Dietary Fiber to Cancer Prevention. Mol. Nutr. Food Res. 2020, 64, e1901014. [Google Scholar] [CrossRef]
- Mao, J.; Chen, X.; Wang, C.; Li, W.; Li, J. Effects and mechanism of the bile acid (farnesoid X) receptor on the Wnt/beta-catenin signaling pathway in colon cancer. Oncol. Lett. 2020, 20, 337–345. [Google Scholar] [CrossRef]
- Coutzac, C.; Jouniaux, J.M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef]
- Mattner, J. Natural killer T (NKT) cells in autoimmune hepatitis. Curr. Opin. Immunol. 2013, 25, 697–703. [Google Scholar] [CrossRef]
- Sydor, S.; Best, J.; Messerschmidt, I.; Manka, P.; Vilchez-Vargas, R.; Brodesser, S.; Lucas, C.; Wegehaupt, A.; Wenning, C.; Assmuth, S.; et al. Altered Microbiota Diversity and Bile Acid Signaling in Cirrhotic and Noncirrhotic NASH-HCC. Clin. Transl. Gastroenterol. 2020, 11, e00131. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Takashina, Y.; Watanabe, M.; Nagamine, R.; Saito, Y.; Kamada, N.; Saito, H. Bile acid metabolism regulated by the gut microbiota promotes non-alcoholic steatohepatitis-associated hepatocellular carcinoma in mice. Oncotarget 2018, 9, 9925–9939. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Wang, X.; Zhao, A.; Yan, J.; Chen, W.; Jiang, R.; Ji, J.; Huang, F.; Zhang, Y.; Lei, S.; et al. Sex-dependent effects on gut microbiota regulate hepatic carcinogenic outcomes. Sci. Rep. 2017, 7, 45232. [Google Scholar] [CrossRef] [PubMed]
- Cassano, M.; Offner, S.; Planet, E.; Piersigilli, A.; Jang, S.M.; Henry, H.; Geuking, M.B.; Mooser, C.; McCoy, K.D.; Macpherson, A.J.; et al. Polyphenic trait promotes liver cancer in a model of epigenetic instability in mice. Hepatology 2017, 66, 235–251. [Google Scholar] [CrossRef]
- Kohli, R.M.; Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Cho, I.; Yamanishi, S.; Cox, L.; Methe, B.A.; Zavadil, J.; Li, K.; Gao, Z.; Mahana, D.; Raju, K.; Teitler, I.; et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 2012, 488, 621–626. [Google Scholar] [CrossRef]
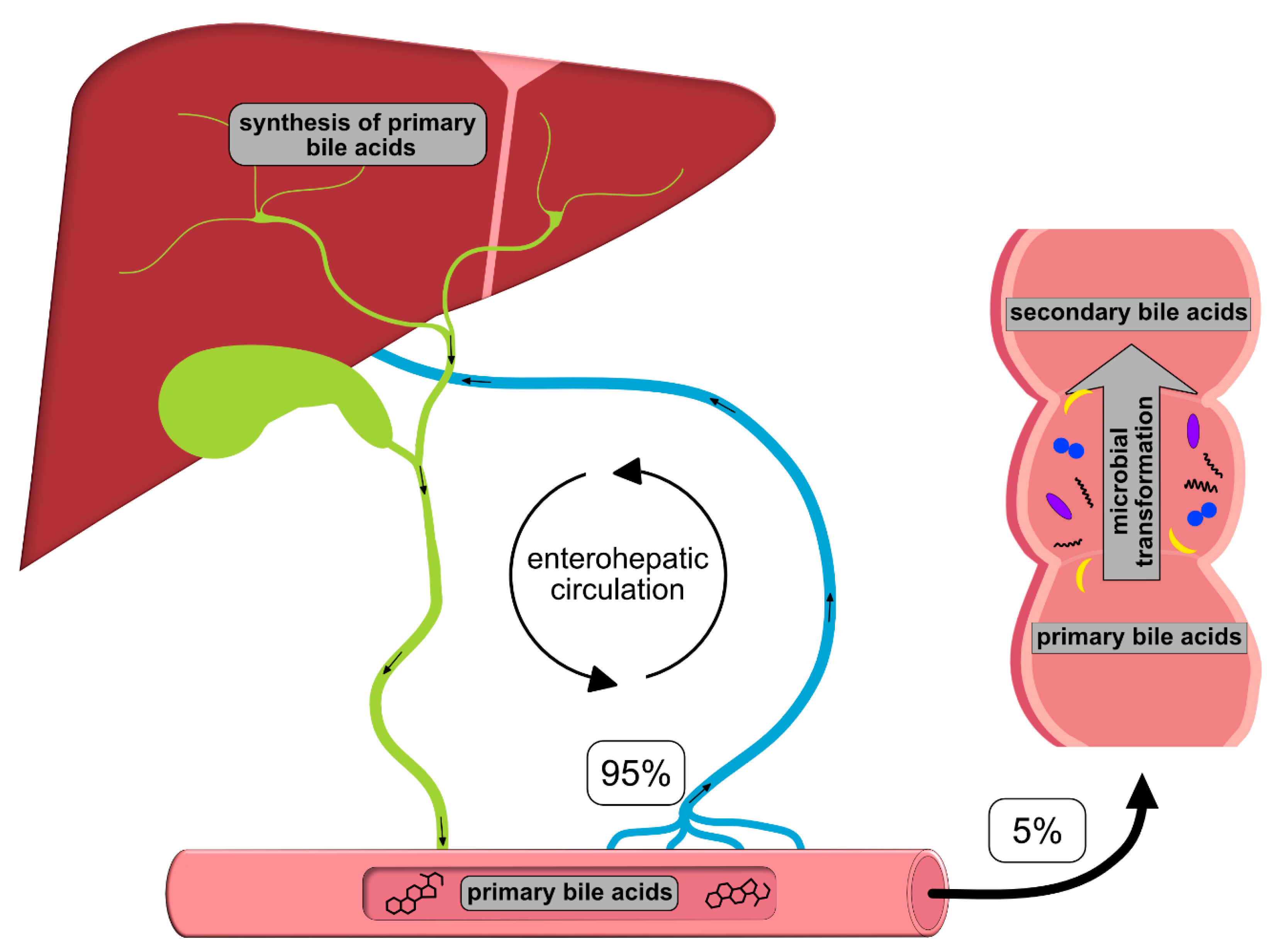

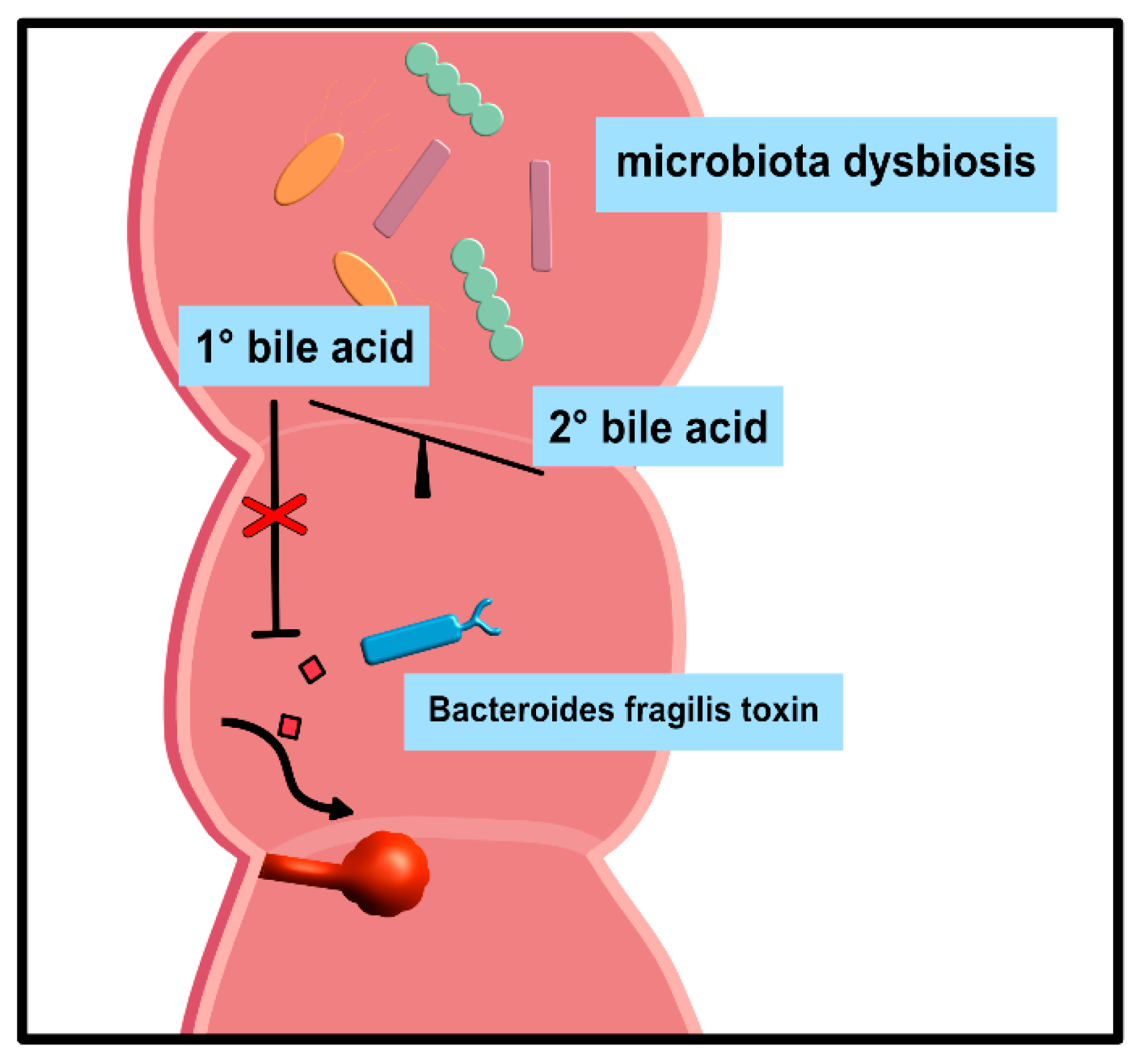
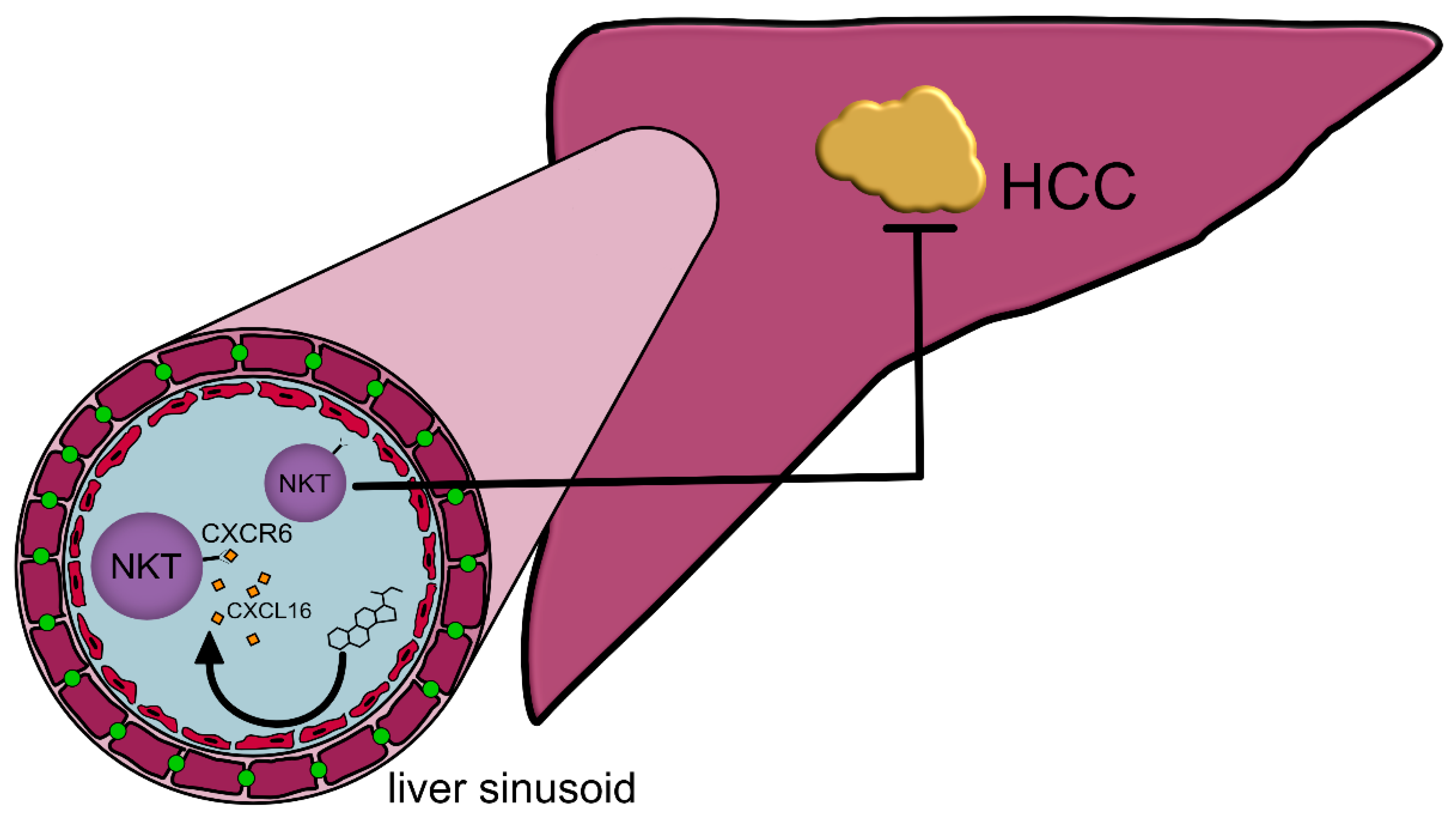
| Abbrevation | Bile Acid Name | Physiologic Functions | Pathophysiologic Functions |
|---|---|---|---|
| CA | Cholic acid | facilitate digestion [14]; emulsify hydrophobic food components such as fats into micelles for intestinal absorption [14]; protect from CRC induced by Bacteroides fragilis toxin [15] | favor infections with pathogenic bacteria (Salmonella spp., E. coli, Shigella dysenteriae) [16,17,18,19,20,21,22,23] and the germination of C. difficile spores [24,25,26]; can cause colitis and diarrhea in UC patients [27]; promote the accumu-lation of CXCR6+-NKT-cells in the liver and protects from HCC [28] |
| CDCA | Chenodeoxycholic acid | ||
| GCA | Glycocholic acid | ||
| GCDCA | Glycochenodeoxycholic acid | ||
| TCA | Taurocholic acid | ||
| TCDCA | Taurochenodeoxycholic acid |
| Abbrevation | Bile Acid Name | Physiologic Functions | Pathophysiologic Functions |
|---|---|---|---|
| DCA | Deoxycholic acid | exhibit antimicrobial activity [32]; maintain colonic microbiota [32]; perpetuate endocrine functions via binding to nuclear factor X receptor (FXR) and G-protein-coupled bile acid receptor (TGR5) [37,38,39,40] | enhanced levels are associated with CRC development [41] |
| UDCA | Ursodeoxycholic acid | drug for the treatment of primary biliary cirrhosis (PBC) | |
| HDCA | Hyodeoxycholic acid | suppresses intestinal cell proliferation and enhances abundance of microbiota [42] | |
| GDCA | Glycodeoxycholic acid | induces interleukin-22 production by ILC3s and improves ovulatory dysfunction and insulin resistance in patients suffering from polycystic ovary syndrome (POCS) [43] | |
| GUDCA | Glycoursodeoxycholic acid | neuroprotective agent (https://pubchem.ncbi.nlm.nih.gov/compound/Glycoursodeoxycholic-acid) | |
| LCA | Lithocholic acid | enhanced levels in patients suffering from Alzheimer´s disease [44] | |
| GLCA | Glycolithocholic acid | ||
| TLCA | Taurolithocholic acid | induces biliary dysbiosis in PSC patients | |
| TDCA | Taurodeoxycholic acid | ||
| TUDCA | Tauroursodeoxycholic acid | protective effects in NASH [45]; associated with attenuated hepatocarcinogenesis [46] | |
| THDCA | Taurohyodeoxycholic acid | protective effects in experimentally induced colitis [47] | |
| TMCA (a+b) | Tauromuricholic acid (alpha + beta) | maintains lipid and glucose metabolism [48]; reduced levels in the plasma of APP/PS1 mice, which are a mouse model for Alzheimer’s disease [49] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grüner, N.; Mattner, J. Bile Acids and Microbiota: Multifaceted and Versatile Regulators of the Liver–Gut Axis. Int. J. Mol. Sci. 2021, 22, 1397. https://doi.org/10.3390/ijms22031397
Grüner N, Mattner J. Bile Acids and Microbiota: Multifaceted and Versatile Regulators of the Liver–Gut Axis. International Journal of Molecular Sciences. 2021; 22(3):1397. https://doi.org/10.3390/ijms22031397
Chicago/Turabian StyleGrüner, Niklas, and Jochen Mattner. 2021. "Bile Acids and Microbiota: Multifaceted and Versatile Regulators of the Liver–Gut Axis" International Journal of Molecular Sciences 22, no. 3: 1397. https://doi.org/10.3390/ijms22031397
APA StyleGrüner, N., & Mattner, J. (2021). Bile Acids and Microbiota: Multifaceted and Versatile Regulators of the Liver–Gut Axis. International Journal of Molecular Sciences, 22(3), 1397. https://doi.org/10.3390/ijms22031397





