Effect of Cultivation Parameters on Fermentation and Hydrogen Production in the Phylum Thermotogae
Abstract
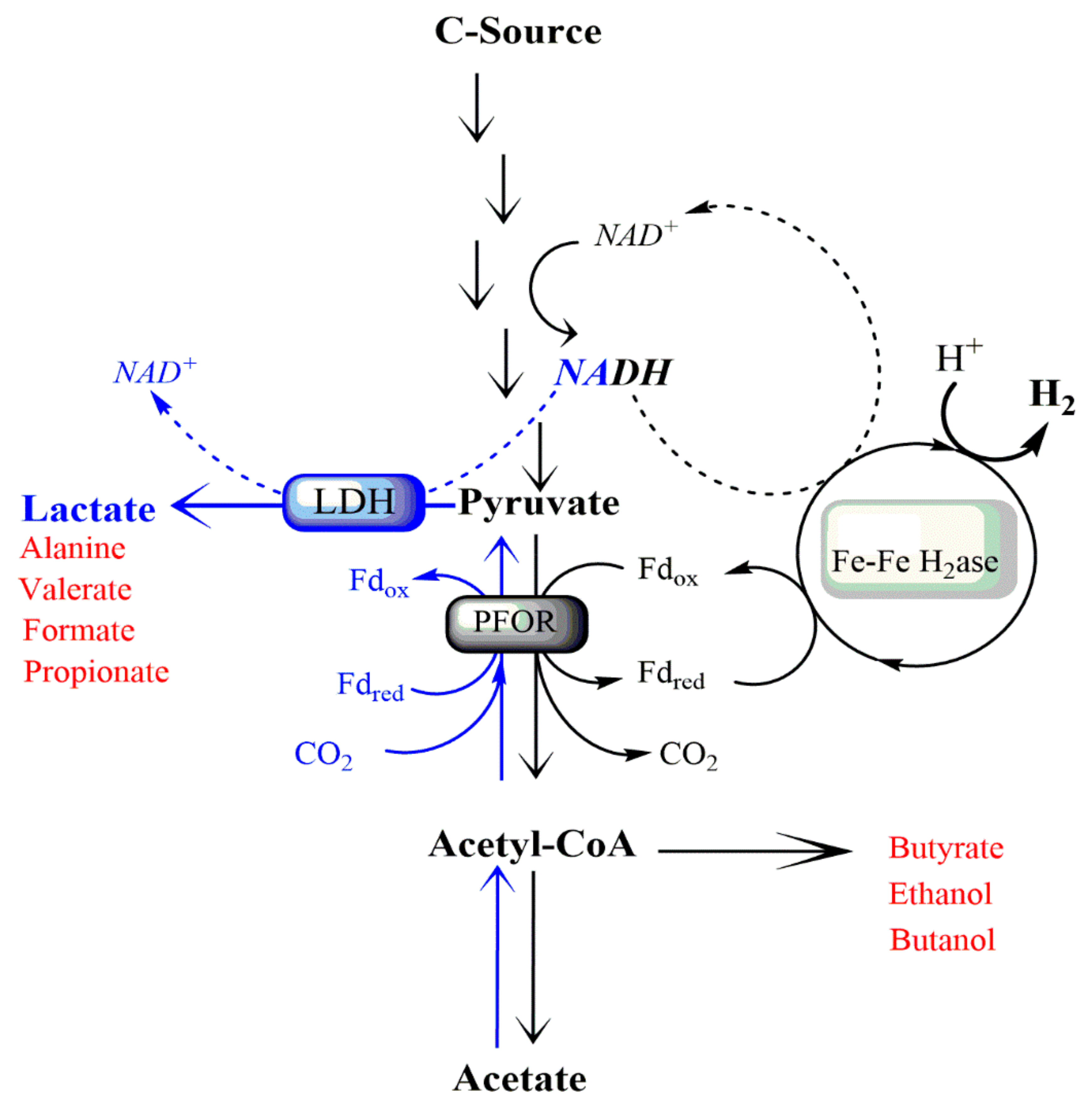
1. Introduction
2. Operating Conditions
2.1. H2 Partial Pressure (PH2)
2.2. Shaking Speed, Culture/Headspace Volume Ratio, Gas Sparging, and Inoculum
2.3. pH
2.4. Temperature
2.5. Oxygen (O2)
3. Nitrogen Containing-Compounds
4. Sodium Chloride and Phosphate
5. Sulfur-Containing Compounds
6. Metal Ions
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Huber, R.H.M. Thermotogales; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 899–922. [Google Scholar]
- Bhandari, V.; Gupta, R.S. The Phylum Thermotogae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Huber, R.; Langworthy, T.A.; König, H.; Thomm, M.; Woese, C.R.; Sleytr, U.B.; Stetter, K.O. Thermotoga maritima Sp. Nov. Represents a New Genus of Unique Extremely Thermophilic Eubacteria Growing up to 90 °C. Arch. Microbiol. 1986, 144, 324–333. [Google Scholar] [CrossRef]
- Belahbib, H.; Summers, Z.M.; Fardeau, M.; Joseph, M.; Tamburini, C.; Dolla, A.; Ollivier, B.; Armougom, F. Towards a Congruent Reclassification and Nomenclature of the Thermophilic Species of the Genus Pseudothermotoga within the Order Thermotogales. Syst. Appl. Microbiol. 2018, 41, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.K.C.; Morgan, H.W.; Daniel, R.M. Fervidobacterium nodosum Gen. Nov. and Spec. Nov., a New Chemoorganotrophic, Caldoactive, Anaerobic Bacterium. Arch. Microbiol. 1985, 141, 63–69. [Google Scholar] [CrossRef]
- Windberger, E.; Huber, R.; Trincone, A.; Fricke, H.; Stetter, K.O. Thermotoga thermarum Sp. Nov. and Thermotoga Neapolitana Occurring in African Continental Solfataric Springs. Arch. Microbiol. 1989, 151, 506–512. [Google Scholar] [CrossRef]
- DiPippo, J.L.; Nesbø, C.L.; Dahle, H.; Doolittle, W.F.; Birkland, N.K.; Noll, K.M. Kosmotoga olearia Gen. Nov., Sp. Nov., a Thermophilic, Anaerobic Heterotroph Isolated from an Oil Production Fluid. Int. J. Syst. Evol. Microbiol. 2009, 59, 2991–3000. [Google Scholar] [CrossRef] [PubMed]
- Nesbø, C.L.; Bradnan, D.M.; Adebusuyi, A.; Dlutek, M.; Petrus, A.K.; Foght, J.; Doolittle, W.F.; Noll, K.M. Mesotoga prima Gen. Nov., Sp. Nov., the First Described Mesophilic Species of the Thermotogales. Extremophile 2012, 16, 387–393. [Google Scholar] [CrossRef]
- Ben Hania, W.; Godbane, R.; Postec, A.; Hamdi, M.; Ollivier, B.; Fardeau, M.L. Defluviitoga tunisiensis Gen. Nov., Sp. Nov., a Thermophilic Bacterium Isolated from a Mesothermic and Anaerobic Whey Digester. Int. J. Syst. Evol. Microbiol. 2012, 62, 1377–1382. [Google Scholar] [CrossRef]
- Davey, M.E.; Wood, W.A.; Key, R.; Nakamura, K.; Stahl, D.A. Isolation of Three Species of Geotoga and Petrotoga: Two New Genera, Representing a New Lineage in the Bacterial Line of Descent Distantly Related to the “Thermotogales”. Syst. Appl. Microbiol. 1993, 16, 191–200. [Google Scholar] [CrossRef]
- Wery, N.; Lesongeur, F.; Pignet, P.; Derennes, V.; Cambon-Bonavita, M.A.; Godfroy, A.; Barbier, G. Marinitoga camini Gen. Nov., Sp. Nov., a Rod-Shaped Bacterium Belonging to the Order Thermotogales, Isolated from a Deep-Sea Hydrothermal Vent. Int. J. Syst. Evol. Microbiol. 2001, 51, 495–504. [Google Scholar] [CrossRef]
- Jayasinghearachchi, H.S.; Lal, B. Oceanotoga Teriensis Gen. Nov., Sp. Nov., a Thermophilic Bacterium Isolated from Offshore Oil-Producing Wells. Int. J. Syst. Evol. Microbiol. 2011, 61, 554–560. [Google Scholar] [CrossRef]
- Reysenbach, A.L.; Liu, Y.; Lindgren, A.R.; Wagner, I.D.; Sislak, C.D.; Mets, A.; Schouten, S. Mesoaciditoga lauensis Gen. Nov., Sp. Nov., a Moderately Thermoacidophilic Member of the Order Thermotogales from a Deep-Sea Hydrothermal Vent. Int. J. Syst. Evol. Microbiol. 2013, 63, 4724–4729. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Onishi, M.; Kato, S.; Iino, T.; Sakamoto, M.; Kudo, T.; Takashina, T.; Ohkuma, M. Athalassotoga saccharophila Gen. Nov., Sp. Nov., Isolated from an Acidic Terrestrial Hot Spring, and Proposal of Mesoaciditogales Ord. Nov. and Mesoaciditogaceae Fam. Nov. in the Phylum Thermotogae. Int. J. Syst. Evol. Microbiol. 2016, 66, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Cheng, L.; Zhang, X.; Li, X.; Deng, Y.; Zhang, H. Thermococcoides shengliensis Gen. Nov., Sp. Nov., a New Member of the Order Thermotogales Isolated from Oil-Production Fluid. Int. J. Syst. Evol. Microbiol. 2010, 60, 932–937. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.; Clayton, R.; Gill, S.; Gwinn, M.; Dodson, R.; Haft, D.; Hickey, E.; Peterson, J.; Nelson, W.; Ketchum, K.; et al. Evidence for Lateral Gene Transfer between Archae and Bacteria from Genome Sequence of Thermotoga maritima. Nature 1999, 399, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Conners, S.B.; Mongodin, E.F.; Johnson, M.R.; Montero, C.I.; Nelson, K.E.; Kelly, R.M. Microbial Biochemistry, Physiology, and Biotechnology of Hyperthermophilic Thermotoga Species. FEMS Microbiol. Rev. 2006, 30, 872–905. [Google Scholar] [CrossRef]
- Rodionov, D.A.; Rodionova, I.A.; Li, X.; Ravcheev, I.; Tarasova, Y.; Portnoy, V.A.; Zengler, K.; Osterman, A.L. Transcriptional Regulation of the Carbohydrate Utilization Network in Thermotoga maritima. Front. Microbiol. 2013, 4, 244. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Noll, K.M.; Romano, A.H. The Glucose Transport System of the Hyperthermophilic Anaerobic Bacterium Thermotoga neapolitana. Appl. Environ. Microbiol. 1996, 62, 2915–2918. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, I.T.; Nguyen, L.; Sliwinski, M.K.; Rabus, R.; Jr, M.H.S. Microbial Genome Analyses: Comparative Transport Capabilities in Eighteen Prokaryotes. J. Mol. Biol. 2000, 301, 75–100. [Google Scholar] [CrossRef]
- Nanavati, D.; Thirangoon, K.; Noll, K.M. Several Archaeal Homologs of Putative Oligopeptide-Binding Proteins Encoded by Thermotoga maritima Bind Sugars. Appl. Environ. Microbiol. 2006, 72, 1336–1345. [Google Scholar] [CrossRef]
- Latif, H.; Sahin, M.; Tarasova, J.; Tarasova, Y.; Portnoy, V.A.; Nogales, J.; Zengler, K. Adaptive Evolution of Thermotoga maritima Reveals Plasticity of the ABC Transporter Network. Appl. Environ. Microbiol. 2015, 81, 5477–5485. [Google Scholar] [CrossRef]
- Boucher, N.; Noll, K.M. Substrate Adaptabilities of Thermotogae Mannan Binding Proteins as a Function of Their Evolutionary Histories. Extremophiles 2016, 20, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Thauer, R.K.; Jungermann, K.; Decker, K. Energy Conservation in Chemotrophic Anaerobic Bacteria. Bacteriol. Rev. 1977, 41, 100–180. [Google Scholar] [CrossRef] [PubMed]
- Ben Hania, W.; Ghodbane, R.; Postec, A.; Brochier-Armanet, C.; Hamdi, M.; Fardeau, M.L.; Ollivier, B. Cultivation of the First Mesophilic Representative (“Mesotoga”) within the Order Thermotogales. Syst. Appl. Microbiol. 2011, 34, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Hania, W.B.; Postec, A.; Aüllo, T.; Ranchou-Peyruse, A.; Erauso, G.; Brochier-Armanet, C.; Hamdi, M.; Ollivier, B.; Saint-Laurent, S.; Magot, M.; et al. Mesotoga infera Sp. Nov., a Mesophilic Member of the Order Thermotogales, Isolated from an Underground Gas Storage Aquifer. Int. J. Syst. Evol. Microbiol. 2013, 63, 3003–3008. [Google Scholar] [CrossRef] [PubMed]
- Fadhlaoui, K.; Hania, W.B.; Armougom, F.; Bartoli, M.; Fardeau, M.L.; Erauso, G.; Brasseur, G.; Aubert, C.; Hamdi, M.; Brochier-Armanet, C.; et al. Obligate Sugar Oxidation in Mesotoga Spp., Phylum Thermotogae, in the Presence of Either Elemental Sulfur or Hydrogenotrophic Sulfate-Reducers as Electron Acceptor. Environ. Microbiol. 2017, 20, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.H.; Morgan, H.W. Heterotrophic Sulfur Reduction by Thermotoga Sp. Strain FjSS3.B1. FEMS Microbiol. Lett. 1992, 96, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Schröder, C.; Selig, M.; Schönheit, P. Glucose Fermentation to Acetate, CO2 and H2 in the Anaerobic Hyperthermophilic Eubacterium Thermotoga maritima: Involvement of the Embden-Meyerhof Pathway. Arch. Microbiol. 1994, 161, 460–470. [Google Scholar] [CrossRef]
- D’Ippolito, G.; Dipasquale, L.; Vella, F.M.; Romano, I.; Gambacorta, A.; Cutignano, A.; Fontana, A. Hydrogen Metabolism in the Extreme Thermophile Thermotoga neapolitana. Int. J. Hydrogen Energy 2010, 35, 2290–2295. [Google Scholar] [CrossRef]
- Dipasquale, L.; d’Ippolito, G.; Fontana, A. Capnophilic Lactic Fermentation and Hydrogen Synthesis by Thermotoga neapolitana: An Unexpected Deviation from the Dark Fermentation Model. Int. J. Hydrogen Energy 2014, 39, 4857–4862. [Google Scholar] [CrossRef]
- Wushke, S.; Fristensky, B.; Zhang, X.L.; Spicer, V.; Krokhin, O.V.; Levin, D.B.; Stott, M.B.; Sparling, R. A Metabolic and Genomic Assessment of Sugar Fermentation Profiles of the Thermophilic Thermotogales, Fervidobacterium pennivorans. Extremophiles 2018, 22, 965–974. [Google Scholar] [CrossRef]
- Vijayakumar, J.; Aravindan, R.; Viruthagiri, T. Lactic Acid and Its Potential Applications in Industries. Bioprod. Biosyst. Eng. 2007, 42, 101–103. [Google Scholar]
- Roehr, M.; Kosaric, N.; Vardar-Sukan, F.; Pieper, H.J.; Senn, T. The Biotechnology of Ethanol. Classical and Future Applications; Wiley: Hoboken, NJ, USA, 2005; p. 245. ISBN 978-3-527-60234-6. [Google Scholar]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic Enzymes: Sources, Uses, and Molecular Mechanisms for Thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Ul Haq, I.; Hussain, Z.; Khan, M.; Muneer, B.; Afzal, S.; Majeed, S.; Akram, F. Kinetic and Thermodynamic Study of Cloned Thermostable Endo-1, 4- b -Xylanase from Thermotoga petrophila in Mesophilic Host. Mol. Biol. Rep. 2012, 39, 7251–7261. [Google Scholar] [CrossRef] [PubMed]
- Ul Haq, I.; Tahir, S.F.; Aftab, M.N.; Akram, F.; ur Rehman, A.; Nawaz, A.; Mukhtar, H. Purification and Characterization of a Thermostable Cellobiohydrolase from Thermotoga petrophila. Protein Pept. Lett. 2018, 25, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Colussi, F.; Viviam, M.; Ian, S.; Junio, M. Oligomeric State and Structural Stability of Two Hyperthermophilic β—Glucosidases from Thermotoga petrophila. Amino Acids 2015, 47, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Pollo, S.M.J.; Zhaxybayeva, O.; Nesbø, C.L. Insights into Thermoadaptation and the Evolution of Mesophily from the Bacterial Phylum Thermotogae. Can. J. Microbiol. 2015, 61, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Fatima, B.; Aftab, M.; Ul Haq, I. Cloning, Purifiication, and Characterization of Xylose Isomerase from Thermotoga naphthophila RKU-10. J. Basic Microbiol. 2016, 56, 949–962. [Google Scholar] [CrossRef]
- Lopes, J.L.S.; Yoneda, J.S.; Martins, J.M.; Demarco, R. Environmental Factors Modulating the Stability and Enzymatic Activity of the Petrotoga mobilis Esterase (PmEst). PLoS ONE 2016, 11, e0158146. [Google Scholar] [CrossRef]
- Intagun, W.; Kanoksilapatham, W. A Review: Biodegradation and Applications of Keratin Degrading Microorganisms and Keratinolytic Enzymes, Focusing on Thermophiles and Thermostable Serine Proteases. Am. J. Appl. Sci. Rev. 2017, 14, 1016–1023. [Google Scholar] [CrossRef]
- Hamid, A.; Aftab, M.N. Cloning, Purification, and Characterization of Recombinant Thermostable β -Xylanase Tnap_0700 from Thermotoga naphthophila. Appl. Biochem. Biotechnol. 2019, 189, 1274–1290. [Google Scholar] [CrossRef]
- Kang, E.; Jin, H.; La, J.W.; Park, S.; Kim, W.; Lee, W. Identification of Keratinases from Fervidobacterium islandicum AW-1 Using Dynamic Gene Expression pro Fi Ling. Microb. Biotechnol. 2019, 13, 442–457. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Communication from the Commission to the European Parliament, the Council, the European Economic and Social Committee and the Committee of the Regions; A Hydrogen Strategy for a Climate-Neutral Europe; 8.7.2020 COM 301 Final; European Commission: Brussels, Belgium, 2020. [Google Scholar]
- Dipasquale, L.; d’Ippolito, G.; Gallo, C.; Vella, F.M.; Gambacorta, A.; Picariello, G.; Fontana, A. Hydrogen Production by the Thermophilic Eubacterium Thermotoga neapolitana from Storage Polysaccharides of the CO2-Fixing Diatom Thalassiosira weissflogii. Int. J. Hydrogen Energy 2012, 37, 12250–12257. [Google Scholar] [CrossRef]
- Angenent, L.; Karim, K.; Al-Dahhan, M.; Wrenn, B.; Domiguez-Espinosa, R. Production of Bioenergy and Biochemicals from Industrial and Agricultural Wastewater. Trends Biotechnol. 2004, 22, 477–485. [Google Scholar] [CrossRef] [PubMed]
- De Vrije, T.; de Haas, G.G.; Tan, G.B.; Keijsers, E.R.P.; Claassen, P.A.M. Pretreatment of Miscanthus for Hydrogen Production by Thermotoga elfii. Hydrog. Energy 2002, 27, 1381–1390. [Google Scholar] [CrossRef]
- De Vrije, T.; Budde, M.A.W.; Lips, S.J.; Bakker, R.R.; Mars, A.E.; Claassen, P.A.M. Hydrogen Production from Carrot Pulp by the Extreme Thermophiles Caldicellulosiruptor saccharolyticus and Thermotoga neapolitana. Int. J. Hydrogen Energy 2010, 35, 13206–13213. [Google Scholar] [CrossRef]
- Mars, A.E.; Veuskens, T.; Budde, M.A.W.; Van Doeveren, P.F.; Lips, S.J.; Bakker, R.R.; De Vrije, T.; Claassen, P.A.M. Biohydrogen Production from Untreated and Hydrolyzed Potato Steam Peels by the Extreme Thermophiles Caldicellulosiruptor saccharolyticus and Thermotoga neapolitana. Int. J. Hydrogen Energy 2010, 35, 7730–7737. [Google Scholar] [CrossRef]
- Cappelletti, M.; Bucchi, G.; De Sousa Mendes, J.; Alberini, A.; Fedi, S.; Bertin, L.; Frascari, D. Biohydrogen Production from Glucose, Molasses and Cheese Whey by Suspended and Attached Cells of Four Hyperthermophilic Thermotoga Strains. J. Chem. Technol. Biotechnol. 2012, 87, 1291–1301. [Google Scholar] [CrossRef]
- Saidi, R.; Liebgott, P.P.; Gannoun, H.; Ben Gaida, L.; Miladi, B.; Hamdi, M.; Bouallagui, H.; Auria, R. Biohydrogen Production from Hyperthermophilic Anaerobic Digestion of Fruit and Vegetable Wastes in Seawater: Simplification of the Culture Medium of Thermotoga maritima. Waste Manag. 2018, 71, 474–484. [Google Scholar] [CrossRef]
- Saidi, R.; Liebgott, P.P.; Hamdi, M.; Auria, R.; Bouallagui, H. Enhancement of Fermentative Hydrogen Production by Thermotoga maritima through Hyperthermophilic Anaerobic Co-Digestion of Fruit-Vegetable and Fish Wastes. Int. J. Hydrogen Energy 2018, 43, 23168–23177. [Google Scholar] [CrossRef]
- Saidi, R.; Hamdi, M.; Bouallagui, H. Hyperthermophilic Hydrogen Production in a Simplified Reaction Medium Containing Onion Wastes as a Source of Carbon and Sulfur. Environ. Sci. Pollut. Res. 2020, 27, 17382–17392. [Google Scholar] [CrossRef]
- Amend, J.P.; Shock, E.L. Energetics of Overall Metabolic Reactions of Thermophilic and Hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 2001, 25, 175–243. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, N.; Dipasquale, L.; d’Ippolito, G.; Panico, A.; Lens, P.N.L.; Esposito, G.; Fontana, A. Hydrogen Production by the Thermophilic Bacterium Thermotoga neapolitana. Int. J. Mol. Sci. 2015, 16, 12578–12600. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.B.; Pitt, L.; Love, M. Biohydrogen Production: Prospects and Limitations to Practical Application. Int. J. Hydrogen Energy 2004, 29, 173–185. [Google Scholar] [CrossRef]
- Schut, G.J.; Adams, M.W.W. The Iron-Hydrogenase of Thermotoga maritima Utilizes Ferredoxin and NADH Synergistically: A New Perspective on Anaerobic Hydrogen Production. J. Bacteriol. 2009, 191, 4451–4457. [Google Scholar] [CrossRef]
- Yu, J.; Varga, M.; Mityas, C.; Noll, K.M. Liposome-Mediated DNA Uptake and Transient Expression in Thermotoga. Extremophiles 2001, 5, 53–60. [Google Scholar] [CrossRef]
- Xu, H.; Han, D.; Xu, Z. Expression of Heterologous Cellulases in Thermotoga Sp. Strain RQ2. BioMed Res. Int. 2015, 2015, 304523. [Google Scholar] [CrossRef]
- Han, D.; Xu, Z. Development of a PyrE—Based Selective System for Thermotoga sp. Extremophiles 2017, 21, 297–306. [Google Scholar] [CrossRef]
- Pradhan, N.; Dipasquale, L.; Panico, A.; Lens, P.N.L.; Esposito, G.; d’Ippolito, G.; Fontana, A. Hydrogen and Lactic Acid Synthesis by the Wild-Type and a Laboratory Strain of the Hyperthermophilic Bacterium Thermotoga neapolitana DSMZ 4359 under Capnophilic Lactic Fermentation Conditions. Int. J. Hydrogen Energy 2017, 42, 16023–16030. [Google Scholar] [CrossRef]
- Grogan, D.W.; Carver, G.T.; Drake, J.W. Genetic fidelity under harsh conditions: Analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA 2001, 98, 7928–7933. [Google Scholar] [CrossRef]
- Nguyen, T.A.D.; Pyo Kim, J.; Sun Kim, M.; Kwan Oh, Y.; Sim, S.J. Optimization of Hydrogen Production by Hyperthermophilic Eubacteria, Thermotoga maritima and Thermotoga neapolitana in Batch Fermentation. Int. J. Hydrogen Energy 2008, 33, 1483–1488. [Google Scholar] [CrossRef]
- Munro, S.A.; Zinder, S.H.; Walker, L.P. The Fermentation Stoichiometry of Thermotoga neapolitana and Influence of Temperature, Oxygen, and PH on Hydrogen Production. Biotechnol. Prog. 2009, 25, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, M.; Zannoni, D.; Postec, A.; Ollivier, B. Members of the Order Thermotogales: From Microbiology to Hydrogen Production. In Microbial BioEnergy: Hydrogen Production, Advances in Photosynthesis and Respiration; Springer: Dordrecht, The Netherlands, 2014; Volume 38, pp. 197–224. [Google Scholar]
- Pradhan, N.; d’Ippolito, G.; Dipasquale, L.; Esposito, G.; Panico, A.; Lens, P.N.L.; Fontana, A. Simultaneous Synthesis of Lactic Acid and Hydrogen from Sugars via Capnophilic Lactic Fermentation by Thermotoga neapolitana Cf Capnolactica. Biomass Bioenergy 2019, 125, 17–22. [Google Scholar] [CrossRef]
- Dreschke, G.; d’Ippolito, G.; Panico, A.; Lens, P.N.L.; Esposito, G.; Fontana, A. Enhancement of Hydrogen Production Rate by High Biomass Concentrations of Thermotoga Neapolitana. Int. J. Hydrogen Energy 2018, 43, 13072–13080. [Google Scholar] [CrossRef]
- Dreschke, G.; Papirio, S.; Panico, A.; Lens, P.N.L.; Esposito, G.; d’Ippolito, G.; Fontana, A. H2-Rich Biogas Recirculation Prevents Hydrogen Supersaturation and Enhances Hydrogen Production by Thermotoga neapolitana Cf. Capnolactica. Int. J. Hydrogen Energy 2019, 44, 19698–19708. [Google Scholar] [CrossRef]
- Dipasquale, L.; Pradhan, N.; d’Ippolito, G.; Fontana, A. Potential of Hydrogen Fermentative Pathways in Marine Thermophilic Bacteria: Dark Fermentation and Capnophilic Lactic Fermentation in Thermotoga and Pseudothermotoga Species. In Grand Challenges in Marine Biotechnology; Springer: Dordrecht, The Netherlands, 2018; pp. 217–235. [Google Scholar]
- Nuzzo, G.; Landi, S.; Esercizio, N.; Manzo, E.; Fontana, A.; d’Ippolito, G. Capnophilic Lactic Fermentation from Thermotoga neapolitana: A Resourceful Pathway to Obtain Almost Enantiopure L-Lactic Acid. Fermentation 2019, 5, 34. [Google Scholar] [CrossRef]
- Takahata, Y.; Nishijima, M.; Hoaki, T.; Maruyama, T. Thermotoga petrophila sp. Nov. and Thermotoga naphthophila Sp. Nov., Two Hyperthermophilic Bacteria from the Kubiki Oil Reservoir in Niigata, Japan. Int. J. Syst. Evol. Microbiol. 2001, 51, 1901–1909. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Yamazoe, A.; Hosoyama, A.; Ohji, S.; Fujita, N.; Ishibashi, J.I.; Kimura, H.; Suzuki, K.I. Thermotoga profunda Sp. Nov. and Thermotoga caldifontis Sp. Nov., Anaerobic Thermophilic Bacteria Isolated from Terrestrial Hot Springs. Int. J. Syst. Evol. Microbiol. 2014, 64, 2128–2136. [Google Scholar] [CrossRef] [PubMed]
- Jannasch, H.W.; Huber, R.; Belkin, S.; Stetter, K.O. Thermotoga neapolitana Sp. Nov. of the Extremely Thermophilic, Eubacterial Genus Thermotoga. Arch. Microbiol. 1988, 150, 103–104. [Google Scholar] [CrossRef]
- Balk, M.; Weijma, J.; Stams, A.J.M. Thermotoga Lettingae Sp. Nov., a Novel Thermophilic, Methanol-Degrading Bacterium Isolated from a Thermophilic Anaerobic Reactor. Int. J. Syst. Evol. Microbiol. 2002, 52, 1361–1368. [Google Scholar] [CrossRef] [PubMed]
- Ravot, G.; Magot, M.; Fardeau, M.L.; Patel, B.K.C.; Prensier, G.; Egan, A.; Garcia, J.L.; Ollivier, B. Thermotoga elfii Sp. Nov., a Novel Thermophilic Bacterium from an African Oil-Producing Well. Int. J. Syst. Bacteriol. 1995, 45, 308–314. [Google Scholar] [CrossRef]
- Fardeau, M.L.; Ollivier, B.C.; Patel, B.K.; Magot, M.; Thomas, P.; Rimbault, A.; Rocchiccioli, F.; Garcia, J.L. Thermotoga hypogea Sp. Nov., a Xylanolytic, Thermophilic Bacterium from an Oil-Producing Well. Int. J. Syst. Bacteriol. 1997, 147, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Jeanthon, C.; Reysenbach, A.L.; L’Haridon, S.; Gambacorta, A.; Pace, N.R.; Glénat, P.; Prieur, D. Thermotoga subterranea Sp. Nov., a New Thermophilic Bacterium Isolated from a Continental Oil Reservoir. Arch. Microbiol. 1995, 164, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Friedricht, A.B.; Antranikian, G. Keratin Degradation by Fervidobacterium pennavorans, a Novel Thermophilic Anaerobic Species of the Order Thermotogales. Appl. Environ. Microbiol. 1996, 62, 2875–2882. [Google Scholar] [CrossRef] [PubMed]
- Huber, R.; Woese, C.R.; Langworthy, T.A.; Kristjansson, J.K.; Stetter, K.O. Fervidobacterium islandicum Sp. Nov., a New Extremely Thermophilic Eubacterium Belonging to the “Thermotogales”. Arch. Microbiol. 1990, 154, 105–111. [Google Scholar] [CrossRef]
- Podosokorskaya, O.A.; Merkel, Y.A.; Kolganova, T.V.; Chernyh, N.A.; Miroshnichenko, M.L.; Bonch-Osmolovskaya, E.A.; Kublanov, I.V. Fervidobacterium riparium Sp. Nov., a Thermophilic Anaerobic Cellulolytic Bacterium Isolated from a Hot Spring. Int. J. Syst. Evol. Microbiol. 2011, 61, 2697–2701. [Google Scholar] [CrossRef]
- Andrews, K.T.; Patel, B.K.C. Fervidobacterium gondwanense Sp. Nov., a New Thermophilic Anaerobic Bacterium Isolated from Nonvolcanically Heated Geothermal Waters of the Great Artesian Basin of Australia. Int. J. Syst. Bacteriol. 1996, 46, 265–269. [Google Scholar] [CrossRef]
- Kanoksilapatham, W.; Pasomsup, P.; Keawram, P.; Cuecas, A.; Portillo, M.C.; Gonzalez, J.M. Fervidobacterium thailandense Sp. Nov., an Extremely Thermophilic Bacterium Isolated from a Hot Spring. Int. J. Syst. Evol. Microbiol. 2016, 66, 5023–5027. [Google Scholar] [CrossRef]
- Cai, J.; Wang, Y.; Liu, D.; Zeng, Y.; Xue, Y.; Ma, Y.; Feng, Y. Fervidobacterium changbaicum Sp. Nov., a Novel Thermophilic Anaerobic Bacterium Isolated from a Hot Spring of the Changbai Mountains, China. Int. J. Syst. Evol. Microbiol. 2007, 57, 2333–2336. [Google Scholar] [CrossRef]
- Huber, R.; Woese, C.R.; Langworthy, T.A.; Fricke, H.; Stetter, K.O. Thermosipho Africanus Gen. Nov., Represents a New Genus of Thermophilic Eubacteria within the “Thermotogales”. Syst. Appl. Microbiol. 1989, 12, 32–37. [Google Scholar] [CrossRef]
- Takai, K.; Horikoshi, K. Thermosipho japonicus Sp. Nov., an Extremely Thermophilic Bacterium Isolated from a Deep-Sea Hydrothermal Vent in Japan. Extremophiles 2000, 4, 9–17. [Google Scholar] [CrossRef]
- L’Haridon, S.; Miroshnichenko, M.L.; Hippe, H.; Fardeau, M.L.; Bonch-Osmolovskaya, E.; Stackebrandt, E.; Jeanthon, C. Thermosipho geolei Sp. Nov., a Thermophilic Bacterium Isolated from a Continental Petroleum Reservoir in Western Siberia. Int. J. Syst. Evol. Microbiol. 2001, 51, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Podosokorskaya, O.A.; Kublanov, I.V.; Reysenbach, A.L.; Kolganova, T.V.; Bonch-Osmolovskaya, E.A. Thermosipho affectus Sp. Nov., a Thermophilic, Anaerobic, Cellulolytic Bacterium Isolated from a Mid-Atlantic Ridge Hydrothermal Vent. Int. J. Syst. Evol. Microbiol. 2011, 61, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, T.; Kawasaki, A.; Uda, I.; Sugai, A. Thermosipho globiformans Sp. Nov., an Anaerobic Thermophilic Bacterium That Transforms into Multicellular Spheroids with a Defect in Peptidoglycan Formation. Int. J. Syst. Evol. Microbiol. 2011, 61, 1622–1627. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Antoine, E.; Cilia, V.; Meunier, J.R.; Guezennec, J.; Lesongeur, F.; Barbier, G. Thermosipho melanesiensis Sp. Nov., a New Thermophilic Anaerobic Bacterium Belonging to the Order Thermotogales, Isolated from Deep-Sea Hydrothermal Vents in the Southwestern Pacific Ocean. Int. J. Syst. Bacteriol. 1997, 47, 1118–1123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Podosokorskaya, O.A.; Bonch-Osmolovskaya, E.A.; Godfroy, A.; Gavrilov, S.N.; Beskorovaynaya, D.A.; Sokolova, T.G.; Kolganova, T.V.; Toshchakov, S.V.; Kublanov, I.V. Thermosipho activus Sp. Nov., a Thermophilic, Anaerobic, Hydrolytic Bacterium Isolated from a Deep-Sea Sample. Int. J. Syst. Evol. Microbiol. 2014, 64, 3307–3313. [Google Scholar] [CrossRef] [PubMed]
- Urios, L.; Cueff-Gauchard, V.; Pignet, P.; Postec, A.; Fardeau, M.L.; Ollivier, B.; Barbier, G. Thermosipho atlanticus Sp. Nov., a Novel Member of the Thermotogales Isolated from a Mid-Atlantic Ridge Hydrothermal Vent. Int. J. Syst. Evol. Microbiol. 2004, 54, 1953–1957. [Google Scholar] [CrossRef]
- L’Haridon, S.; Miroshnichenko, M.L.; Hippe, H.; Fardeau, M.L.; Stackebrandt, E.; Jeanthon, C. Petrotoga olearia Sp. Nov. and Petrotoga sibirica Sp. Nov., Two Thermophilic Bacteria Isolated from a Continental Petroleum Reservoir in Western Siberia. Int. J. Syst. Evol. Microbiol. 2002, 52, 1715–1722. [Google Scholar] [CrossRef]
- Lien, T.; Madsen, M.; Rainey, F.A.; Birkeland, N.K. Petrotoga mobilis Sp. Nov., from a North Sea Oil-Production Well. Int. J. Syst. Bacteriol. 1998, 48, 1007–1013. [Google Scholar] [CrossRef]
- Miranda-Tello, E.; Fardeau, M.L.; Joulian, C.; Magot, M.; Thomas, P.; Tholozan, J.L.; Olivier, B. Petrotoga halophila Sp. Nov., a Thermophilic, Moderately Halophilic, Fermentative Bacterium Isolated from an Offshore Oil Well in Congo. Int. J. Syst. Evol. Microbiol. 2007, 57, 40–44. [Google Scholar] [CrossRef]
- Miranda-Tello, E.; Fardeau, M.L.; Thomas, P.; Ramirez, F.; Casalot, L.; Cayol, J.L.; Garcia, J.L.; Ollivier, B. Petrotoga mexicana Sp. Nov., a Novel Thermophilic, Anaerobic and Xylanolytic Bacterium Isolated from an Oil-Producing Well in the Gulf of Mexico. Int. J. Syst. Evol. Microbiol. 2004, 54, 169–174. [Google Scholar] [CrossRef]
- Purwasena, I.A.; Sugai, Y.; Sasaki, K. Petrotoga japonica Sp. Nov., a Thermophilic, Fermentative Bacterium Isolated from Yabase Oilfield in Japan. Arch Microbiol. 2014, 196, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Alain, K.; Marteinsson, V.T.; Miroshnichenko, M.L.; Bonch-Osmolovskaya, E.A.; Prieur, D.; Birrien, J.-L. Marinitoga piezophila Sp. Nov., a Rod-Shaped, Thermo-Piezophilic Bacterium Isolated under High Hydrostatic Pressure from a Deep-Sea Hydrothermal Vent. Int. J. Syst. Evol. Microbiol. 2002, 52, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Postec, A.; Ciobanu, M.; Birrien, J.L.; Bienvenu, N.; Prieur, D.; Le Romancer, M. Marinitoga litoralis Sp. Nov., a Thermophilic, Heterotrophic Bacterium Isolated from a Coastal Thermal Spring on Île Saint-Paul, Southern Indian Ocean. Int. J. Syst. Evol. Microbiol. 2010, 60, 1778–1782. [Google Scholar] [CrossRef] [PubMed]
- Nunoura, T.; Oida, H.; Miyazaki, M.; Suzuki, Y.; Takai, K.; Horikoshi, K. Marinitoga okinawensis Sp. Nov., a Novel Thermophilic and Anaerobic Heterotroph Isolated from a Deep-Sea Hydrothermal Field, Southern Okinawa Trough. Int. J. Syst. Evol. Microbiol. 2007, 57, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Postec, A.; Le Breton, C.; Fardeau, M.L.; Lesongeur, F.; Pignet, P.; Querellou, J.; Ollivier, B.; Godfroy, A. Marinitoga hydrogenitolerans Sp. Nov., a Novel Member of the Order Thermotogales Isolated from a Black Smoker Chimney on the Mid-Atlantic Ridge. Int. J. Syst. Evol. Microbiol. 2005, 55, 1217–1221. [Google Scholar] [CrossRef] [PubMed]
- Steinsbu, B.O.; Røyseth, V.; Thorseth, I.H.; Steen, I.H. Marinitoga arctica Sp. Nov., a Thermophilic, Anaerobic Heterotroph Isolated from a Mid-Ocean Ridge Vent Field. Int. J. Syst. Evol. Microbiol. 2016, 66, 5070–5076. [Google Scholar] [CrossRef]
- Nunoura, T.; Hirai, M.; Imachi, H.; Miyazaki, M.; Makita, H.; Hirayama, H.; Furushima, Y.; Yamamoto, H.; Takai, K. Kosmotoga arenicorallina Sp. Nov. a Thermophilic and Obligately Anaerobic Heterotroph Isolated from a Shallow Hydrothermal System Occurring within a Coral Reef, Southern Part of the Yaeyama Archipelago, Japan, Reclassification of Thermococcoides shengliensis. Arch. Microbiol. 2010, 192, 811–819. [Google Scholar] [CrossRef] [PubMed]
- L’Haridon, S.; Jiang, L.; Alain, K.; Chalopin, M.; Rouxel, O.; Beauverger, M.; Xu, H.; Shao, Z.; Jebbar, M. Kosmotoga pacifica Sp. Nov., a Thermophilic Chemoorganoheterotrophic Bacterium Isolated from an East Pacific Hydrothermal Sediment. Extremophiles 2014, 18, 81–88. [Google Scholar] [CrossRef]
- Van Ooteghem, S.A.; Beer, S.K.; Yue, P.C. Hydrogen Production by the Thermophilic Bacterium Thermotoga neapolitana. Appl. Biochem. Biotechnol. 2002, 98–100, 177–189. [Google Scholar] [CrossRef]
- Eriksen, N.T.; Nielsen, T.M.; Iversen, N. Hydrogen Production in Anaerobic and Microaerobic Thermotoga neapolitana. Biotechnol. Lett. 2008, 30, 103–109. [Google Scholar] [CrossRef]
- Boileau, C.; Auria, R.; Davidson, S.; Casalot, L.; Christen, P.; Liebgott, P.P.; Combet-Blanc, Y. Hydrogen Production by the Hyperthermophilic Bacterium Thermotoga maritima Part I: Effects of Sulfured Nutriments, with Thiosulfate as Model, on Hydrogen Production and Growth. Biotechnol. Biofuels 2016, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, E.W.J.; Budde, M.A.W.; De Haas, G.; van der Wal, F.J.; Claassen, P.A.M.; Stams, A.J.M. Distinctive Properties of High Hydrogen Producing Extreme Thermophiles, Caldicellulosiruptor saccharolyticus and Thermotoga elfii. Int. J. Hydrogen Energy 2002, 27, 1391–1398. [Google Scholar] [CrossRef]
- Hawkes, F.; Hussy, I.; Kyazze, G.; Dinsdale, R.; Hawkes, D. Continuous Dark Fermentative Hydrogen Production by Mesophilic Microflora: Principles and Progress. Int. J. Hydrogen Energy 2007, 32, 172–184. [Google Scholar] [CrossRef]
- Nguyen, T.A.D.; Han, S.J.; Kim, J.P.; Kim, M.S.; Sim, S.J. Hydrogen Production of the Hyperthermophilic Eubacterium, Thermotoga neapolitana under N2 Sparging Condition. Bioresour. Technol. 2010, 101, S38–S41. [Google Scholar] [CrossRef]
- Karadagli, F.; Marcus, A.K.; Rittmann, B.E. Role of Hydrogen (H2) Mass Transfer in Microbiological H2-Threshold Studies. Biodegradation 2019, 30, 113–125. [Google Scholar] [CrossRef]
- Nath, K.; Das, D. Improvement of Fermentative Hydrogen Production: Various Approaches. Appl. Microbiol. Biotechnol. 2004, 65, 520–529. [Google Scholar] [CrossRef]
- Ngo, T.A.; Mi-sun, K.; Sim, S.J. Thermophilic Hydrogen Fermentation Using Thermotoga neapolitana DSM 4359 by Fed-Batch Culture. Int. J. Hydrogen Energy 2011, 36, 14014–14023. [Google Scholar] [CrossRef]
- Pradhan, N.; Dipasquale, L.; d’Ippolito, G.; Fontana, A.; Panico, A.; Lens, P.N.L.; Pirozzi, F.; Esposito, G. Kinetic modeling of fermentative hydrogen production by Thermotoga neapolitana. Int. J. Hydrogen Energy 2016, 41, 1–10. [Google Scholar] [CrossRef]
- Mizuno, O.; Dinsdale, R.; Hawkes, F.R.; Hawkes, D.L. Enhancement of Hydrogen Production from Glucose by Nitrogen Gas Sparging. Bioresour. Technol. 2000, 73, 59–65. [Google Scholar] [CrossRef]
- Ngo, T.A.; Kim, M.S.; Sim, S.J. High-Yield Biohydrogen Production from Biodiesel Manufacturing Waste by Thermotoga neapolitana. Int. J. Hydrogen Energy 2011, 36, 5836–5842. [Google Scholar] [CrossRef]
- D’Ippolito, G.; Dipasquale, L.; Fontana, A. Recycling of Carbon Dioxide and Acetate as Lactic Acid by the Hydrogen-Producing Bacterium Thermotoga neapolitana. ChemSuschem 2014, 7, 2678–2683. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, N.; Dipasquale, L.; d’Ippolito, G.; Fontana, A.; Panico, A.; Pirozzi, F.; Lens, P.N.L.; Esposito, G. Model Development and Experimental Validation of Capnophilic Lactic Fermentation and Hydrogen Synthesis by Thermotoga neapolitana. Water Res. 2016, 99, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Dipasquale, L.; Adessi, A.; d’Ippolito, G.; Rossi, F.; Fontana, A.; De Philippis, R. Introducing Capnophilic Lactic Fermentation in a Combined Dark-Photo Fermentation Process: A Route to Unparalleled H2 Yields. Appl. Microbiol. Biotechnol. 2015, 99, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- D’Ippolito, G.; Landi, S.; Esercizio, N.; Lanzilli, M.; Vastano, M.; Dipasquale, L.; Pradhan, N.; Fontana, A. CO2-Induced Transcriptional Reorganization: Molecular Basis of Capnophillic Lactic Fermentation in Thermotoga neapolitana. Front. Microbiol. 2020, 11, 171. [Google Scholar] [CrossRef] [PubMed]
- Van Ooteghem, S.A.; Jones, A.; Van Der Lelie, D.; Dong, B.; Mahajan, D. H2 Production and Carbon Utilization by Thermotoga neapolitana under Anaerobic and Microaerobic Growth Conditions. Biotechnol. Lett. 2004, 26, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Ngo, T.A.; Sim, S.J. Dark Fermentation of Hydrogen from Waste Glycerol Using Hyperthermophilic Eubacterium Thermotoga neapolitana. Environ. Prog. Sustain. Energy 2011, 31, 466–473. [Google Scholar] [CrossRef]
- Juszczak, A.; Aono, S.; Adams, M.W.W. The Extremely Thermophilic Eubacterium, Thermotoga maritima Contains a Novel Iron-Hydrogenase Whose Cellular Activity Is Dependent Upon Tungsten. J. Bacteriol. Chem. 1991, 226, 13834–13841. [Google Scholar] [PubMed]
- Rusch, A.; Walpersdorf, E.; DeBeer, D.; Gurrier, S.; Amend, P.J. Microbial Communities near the Oxic/Anoxic Interface in the Hydrothermal System of Vulcano Island, Italy. Chem. Geol. 2005, 224, 169–182. [Google Scholar] [CrossRef]
- Belkin, S.; Wirsen, C.O.; Jannasch, H.W. A New Sulfur-Reducing, Extremely Thermophilic Eubacterium from a Submarine Thermal Vent. Appl. Environ. Microbiol. 1986, 51, 1180–1185. [Google Scholar] [CrossRef]
- Childers, S.E.; Vargas, M.; Noll, K.M. Improved Methods for Cultivation of the Extremely Thermophilic Bacterium Thermotoga neapolitana. Appl. Environ. Microbiol. 1992, 58, 3949–3953. [Google Scholar] [CrossRef]
- Le Fourn, C.; Fardeau, M.L.; Ollivier, B.; Lojou, E.; Dolla, A. The Hyperthermophilic Anaerobe Thermotoga maritima Is Able to Cope with Limited Amount of Oxygen: Insights into Its Defence Strategies. Environ. Microbiol. 2008, 10, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Tosatto, S.C.E.; Toppo, S.; Carbonera, D.; Giacometti, G.M.; Costantini, P. Comparative Analysis of [FeFe] Hydrogenase from Thermotogales Indicates the Molecular Basis of Resistance to Oxygen Inactivation. Int. J. Hydrogen Energy 2008, 33, 570–578. [Google Scholar] [CrossRef]
- Lakhal, R.; Auria, R.; Davidson, S.; Ollivier, B.; Dolla, A.; Hamdi, M.; Combet-Blanc, Y. Effect of Oxygen and Redox Potential on Glucose Fermentation in Thermotoga maritima under Controlled Physicochemical Conditions. Int. J. Microbiol. 2010, 2010, 896510. [Google Scholar] [CrossRef] [PubMed]
- Kaslin, S.A.; Childers, S.E.; Noll, K.M. Membrane-Associated Redox Activities in Thermotoga neapolitana. Arch. Microbiol. 1998, 170, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ma, K. Purification and Characterization of an NADH Oxidase from Extremely Thermophilic Anaerobic Bacterium Thermotoga hypogea. Arch. Microbiol. 2005, 183, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ma, K. Characterization of an Exceedingly Active NADH Oxidase from the Anaerobic Hyperthermophilic Bacterium Thermotoga maritima. J. Bacteriol. 2007, 189, 3312–3317. [Google Scholar] [CrossRef]
- Mangayil, R.; Aho, T.; Karp, M.; Santala, V. Improved Bioconversion of Crude Glycerol to Hydrogen by Statistical Optimization of Media Components. Renew. Energy 2015, 75, 583–589. [Google Scholar] [CrossRef]
- Abdullah, M.F.; Md Jahim, J.; Abdul, P.M.; Mahmod, S.S. Effect of Carbon/Nitrogen Ratio and Ferric Ion on the Production of Biohydrogen from Palm Oil Mill Effluent (POME). Biocatal. Agric. Biotechnol. 2020, 23, 101445. [Google Scholar] [CrossRef]
- Munro, S.A.; Choe, L.; Zinder, S.H.; Lee, K.H.; Walker, L.P. Proteomic and Physiological Experiments to Test Thermotoga neapolitana Constraint-Based Model Hypotheses of Carbon Source Utilization. Biotechnol. Prog. 2012, 28, 312–318. [Google Scholar] [CrossRef]
- Rinker, K.D.; Kelly, R.M. Effect of Carbon and Nitrogen Sources on Growth Dynamics and Exopolysaccharide Production for the Hyperthermophilic Archaeon Thermococcus Litoralis and Bacterium Thermotoga maritima. Biotechnol. Bioeng. 2000, 69, 537–547. [Google Scholar] [CrossRef]
- Van Niel, E.W.J.; Hahn-Hägerdal, B. Nutrient Requirements of Lactococci in Defined Growth Media. Appl. Microbiol. Biotechnol. 1999, 52, 617–627. [Google Scholar] [CrossRef]
- Maru, B.T.; Bielen, A.A.M.; Kengen, S.W.M.; Constantí, M.; Medinaa, F. Biohydrogen Production from Glycerol Using Thermotoga Spp. Energy Procedia 2012, 29, 300–307. [Google Scholar] [CrossRef]
- Kengen, S.W.M.; Stams, A.J.M. Formation of L-Alanine as a Reduced End Product in Carbohydrate Fermentation by the Hyperthermophilic Archaeon Pyrococcus Furiosus. Arch. Microbiol. 1994, 161, 168–175. [Google Scholar] [CrossRef]
- Weiner, R.; Langille, S.; Quintero, E. Structure, Function, and Immu- Nochemistry of Bacterial Exopolysaccharides. J. Ind. Microbiol. 1995, 15, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Pierra, M.; Trably, E.; Godon, J.J.; Bernet, N. Fermentative Hydrogen Production under Moderate Halophilic Conditions. Int. J. Hydrogen Energy 2014, 39, 7508–7517. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, W.; Zhang, X.; Yu, L.; Zhou, J.; Xu, Y.; Qian, G. Phosphate Enhancing Fermentative Hydrogen Production from Substrate with Municipal Solid Waste Composting Leachate as a Nutrient. Bioresour. Technol. 2015, 190, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.; Lin, C. Calcium Effect on Fermentative Hydrogen Production in an Anaerobic Up-Flow Sludge Blanket System. Water Sci. Technol. 2006, 54, 105–112. [Google Scholar] [CrossRef]
- Ravot, G.; Ollivier, B.; Magot, M.; Patel, B.K.C.; Crolet, J.L.; Fardeau, M.L.; Garcia, J.L. Thiosulfate Reduction, an Important Physiological Feature Shared by Members of the Order Thermotogales. Appl. Environ. Microbiol. 1995, 61, 2053–2055. [Google Scholar] [CrossRef]
- Ravot, G.; Ollivier, B.; Fardeau, M.L.; Patel, B.K.C.; Andrews, K.T.; Magot, M.; Garcia, J.L. L-Alanine Production from Glucose Fermentation by Hyperthermophilic Members of the Domains Bacteria and Archaea: A Remnant of an Ancestral Metabolism? Appl. Environ. Microbiol. 1996, 62, 2657–2659. [Google Scholar] [CrossRef]
- Ainala, S.K.; Seol, E.; Kim, J.R.; Park, S. Effect of Culture Medium on Fermentative and CO-Dependent H2 Production Activity in Citrobacter Amalonaticus Y19. Int. J. Hydrogen Energy 2016, 41, 6734–6742. [Google Scholar] [CrossRef]
- Huber, R.; Stetter, K.O. The Order Thermotogales. In The Prokaryotes; Springer: New York, NY, USA, 1992; pp. 93809–93815. [Google Scholar]
- Hania, W.B.; Fadhlaoui, K.; Brochier-Armanet, C.; Persillon, C.; Postec, A.; Hamdi, M.; Dolla, A.; Ollivier, B.; Fardeau, M.L.; Le Mer, J.; et al. Draft Genome Sequence of Mesotoga Strain PhosAC3, a Mesophilic Member of the Bacterial Order Thermotogales, Isolated from a Digestor Treating Phosphogypsum in Tunisia. Stand. Genom. Sci. 2015, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nesbø, C.L.; Charchuk, R.; Pollo, S.M.J.; Budwill, K.; Kublanov, I.V.; Haverkamp, T.H.A.; Foght, J. Genomic Analysis of the Mesophilic Thermotogae Genus Mesotoga Reveals Phylogeographic Structure and Genomic Determinants of Its Distinct Metabolism. Environ. Microbiol. 2019, 21, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Yang, Q.; Yao, F.; Huang, X.; Wu, Y.; Du, M.; Chen, S.; Liu, X.; Li, X.; Wang, D. The Inhibitory Effect of Thiosulfinate on Volatile Fatty Acid and Hydrogen Production from Anaerobic Co-Fermentation of Food Waste and Waste Activated Sludge. Bioresour. Technol. 2020, 297, 122428. [Google Scholar] [CrossRef] [PubMed]
- Slobodkin, A.I. Thermophilic Microbial Metal Reduction. Microbiologia 2005, 74, 581–595. [Google Scholar] [CrossRef]
- Gomez-Romero, J.; Gonzalez-Garcia, A.; Chairez, I.; Torres, L.; García-Peña, E.I. Selective Adaptation of an Anaerobic Microbial Community: Biohydrogen Production by Co-Digestion of Cheese Whey and Vegetables Fruit Waste. Int. J. Hydrogen Energy 2014, 39, 12541–12550. [Google Scholar] [CrossRef]
- Llanos, J.; Capasso, C.; Parisi, E.; Prieur, D.; Jeanthon, C. Susceptibility to Heavy Metals and Cadmium Accumulation in Aerobic and Anaerobic Thermophilic Microorganisms Isolated from Deep-Sea Hydrothermal Vents. Curr. Microbiol. 2000, 41, 0201–0205. [Google Scholar] [CrossRef]
- Vargas, M.; Kashefi, K.; Blunt-harris, E.L.; Lovley, D.R. Fe (III) Reduction on Early Earth. Nature 1998, 395, 65–67. [Google Scholar] [CrossRef]
- Balch, W.E.; Fox, G.E.; Magrum, L.J.; Woese, C.R.; Wolfe, R.S. Methanogens: Reevaluation of a Unique Biological Group. Microbiol. Rev. 1979, 43, 260–296. [Google Scholar] [CrossRef]
- Lee, Y.J.; Miyahara, T.; Noike, T. Effect of Iron Concentration on Hydrogen Fermentation. Bioresour. Technol. 2001, 80, 227–231. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, J. Effect of Temperature and Iron Concentration on the Growth and Hydrogen Production of Mixed Bacteria. Int. J. Hydrogen Energy 2006, 31, 441–446. [Google Scholar] [CrossRef]
- Ying, X.; Wang, Y.; Badiei, H.R.; Karanassios, V.; Ma, K. Purification and Characterization of an Iron-Containing Alcohol Dehydrogenase in Extremely Thermophilic Bacterium Thermotoga hypogea. Arch. Microbiol. 2007, 187, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, S.; Rao, S.R.; Gehlot, H.S. Nucleic Acid Stability in Thermophilic Prokaryotes: A Review Nucleic Acid Stability in Thermophilic Prokaryotes: A Review. J. Cell Mol. Biol. 2005, 4, 61–69. [Google Scholar]
| Genus | Species | Isolation | Temp. Range/ Optimal (°C) | pH Range/ Optimal | Cell Dimension (Long by Wide) (µm) | Growth Substrates | NaCl Range/ Optimal (%) | Electron Acceptor | End Products | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Thermotoga | Thermotoga petrophila | Oil reservoir, Japan | 47–88/ 80 | 5.2–9.0/ 7.0 | 2.0–7.0 by 0.7–1.0 | YE, peptone, glucose, fructose, ribose, arabinose, sucrose, lactose, maltose, starch, cellulose | 0.1–5.5/ 1.0 | S0; Thio | AA, LA, CO2, H2 | [72] |
| Thermotoga naphthophila | Oil reservoir, Japan | 48–86/ 80 | 5.4–9.0/ 7.0 | 2.0–7.0 by 0.8–1.2 | YE, peptone, glucose, galactose, fructose, mannitol, ribose, arabinose, sucrose, lactose, maltose, starch | 0.1–6.0/ 1.0 | S0; Thio | AA, LA, CO2, H2 | [72] | |
| Thermotoga maritima | Geotermal vent | 55–90/ 80 | 5.5–9.0/ 6.5 | 1.5–11.0 by 0.6 | ribose, xylose, glucose, sucrose, maltose, lactose, galactose, starch, glycogen | 0.2–3.8/ 2.7 | Fe (III) S0; Thio | AA, LA, CO2, H2, ALA, EPS, AABA | [3] | |
| Thermotoga profunda | Hot spring, Japan | 50–72/ 60 | 6.0–8.6/ 7.4 | 0.8–2.1 by 0.4 | glucose, trehalose, cellobiose, arabinose, xylose, ribose, pyruvate | n. d | S0; Thio | n. d | [73] | |
| Thermotoga caldifontis | Hot spring, Japan | 55–85/ 70 | 6.0–8.6/ 7.4 | 1.2–3.5 by 0.5 | glucose, maltose, trehalose, cellobiose, arabinose, xylose, ribose, pyruvate, starch | n. d | Thio | n. d | [73] | |
| Thermotoga neapolitana | Submarine thermal vent | 55–95/ 77 | 6.0–9.0/ 7.5 | 1.5–11.0 by 0.6 | fructose, fucose, galactose, mannose, rhamnose, pyruvate, glucosamine, lactulose, turanose, glycerol, dextrin, ribose, xylose, glucose, sucrose, maltose, lactose, starch, glycogen | 0.2–6.0/ 2.0 | S0 | AA, ALA, CO2, H2 | [74] | |
| Pseudothermotoga | Pseudothermotoga lettingae | Thermophilic bioreactor | 50–75/ 65 | 6.0–8.5/ 7.0 | 2.0–3.0 by 0.5–1.0 | glucose, EtOH, acetate, formate | 0.0–2.8/ 1.0 | S0; Thio; AQDS; Fe(III) | AA, ALA, LA, EtOH, AA, BA, CO2, H2 | [75] |
| Pseudothermotoga elfii | Oil reservoir | 50–72/ 66 | 5.5–7.5/ 7.5 | 2.0–3.0 by 0.5–1.0 | glucose, arabinose, fructose, lactose, maltose, mannose, ribose, sucrose, xylose | 0.0–2.8/ 1.0 | Thio | AA, CO2, H2 | [76] | |
| Pseudothermotoga hypogea | Oil reservoir, Africa | 56–90/ 70 | 6.1–9.1/ 7.3–7.4 | 2.0–3.0 by 0.5–1.0 | fructose, galactose, glucose, lactose, maltose, mannose, sucrose, xylose, xylan | 0.0–1.5/ 0.2 | Thio | AA, ALA, CO2, H2, EtOH | [77] | |
| Pseudothermotoga | Pseudothermotoga subterranea | Oil reservoir, Paris | 50–75/ 70 | 6.0–8.5/ 7.0 | 3.0–10.0 by 0.5 | YE, peptone, tryptone, casein | 0.0–2.4/ 1.2 | Cys, Thio | n.d. | [78] |
| Pseudothermotoga thermarum | Hot spring, Africa | 55–84/ 70 | 6.0–9.0/ 7.0 | 1.5–11.0 by 0.6 | starch, glucose, maltose | 0.2–0.5/ 0.35 | S0 | n.d. | [6] | |
| Fervidobacterium | Fervidobacterium nodosum | Hot spring, New Zealand | 40–80/ 65–70 | 6.0–8.0/ 7.0 | 1.0–2.5 by 0.5–0.55 | glucose, sucrose, starch and lactose | n.d./<1.0 | S0 | AA, LA, CO2, H2, EtOH, But, Val | [5] |
| Fervidobacterium pennavorans | Hot spring, Portugal | 50–80/ 70 | 5.5–8.0/ 6.5 | 2.0–20.0 by 0.5 | cellobiose, starch, glycogen, pullulan, glucose, fructose, maltose, xylose, native feathers | 0.0–4.0/ 0.4 | S0; Thio | AA, CO2, ALA, Glu, EtOH, But, H2, BuOH | [79] | |
| Fervidobacterium islandicum | Icelandic Hot spring | 50–80/ 65 | 6.0–8.0/ 7.2 | 1.0–4.0 by 0.6 | pyruvate, ribose, glucose, maltose, raffinose, starch, cellulose | 0.0–1.0/ 0.2 | S0; Thio | LA, AA, H2, EtOH, CO2, iBut, iVal | [80] | |
| Fervidobacterium riparium | Hot spring, Russia | 46–80/ 65 | 5.7–7.9/ 7.8 | 1.0–3.0 by 0.4–0.5 | peptone, YE, pyruvate, glucose, xylose, fructose, maltose, sucrose, cellobiose, starch, xylan, CMC, cellulose, filter paper | 0.0–1.0/ 0.0 | S0 | H2, AA, CO2, PPA, iBut, But | [81] | |
| Fervidobacterium gondwanense | Hot spring, Australia | 45–80/ 65–68 | 5.5–8.5/ 7.0 | 4.0–40.0 by 0.5–0.6 | cellobiose, amylopectin, maltose, starch, dextrin, xylose, glucose, pyruvate, lactose, fructose, mannose, CMC, galactose | 0.0–0.6/ 0.1 | S0 | EtOH, AA, LA, CO2, H2 | [82] | |
| Fervidobacterium thailandese | Hot spring, Thailand | 60–88/ 78–80 | 6.5–8.5/ 7.5 | 1.1–2.5 by 0.5–0.6 | glucose, maltose, sucrose, fructose, cellobiose, CMC, cellulose, starch | <0.5/0.5 | S0 | n.d. | [83] | |
| Fervidobacterium changbaicum | Hot spring, China | 55–90/ 75–80 | 6.3–8.5/ 7.5 | 1.0–8.0 by 0.5–0.6 | glucose, lactose, fructose, sucrose, maltose, starch, sorbitol, cellobiose, trehalose, galactose, melibiose, pyruvate, glycerin | 0.0–1.0/ 0.0 | S0 | n.d. | [84] | |
| Thermosipho | Thermosipho africanus | Hot spring, Africa | 53–77/ 75 | 6.0–8.0/ 7.2 | 3.0–4.0 by 0.5 | glucose, ribose, maltose, starch, galactose, fructose, sucrose | 0.11–3.6 | S0; Thio | AA, H2, CO2, EtOH, LA | [85] |
| Thermosipho japonicus | Hydrothermal vent, Japan | 45–80/ 72 | 5.3–9.3/ 7.2–7.6 | 3.0–4.0 by 0.5 | YE, peptone, and tryptone, maltose, glucose, galactose, starch, sacharose, ribose, casein | 0.7–7.9/ 4.0 | S0; Thio | n.d. | [86] | |
| Thermosipho geolei | Oil reservoir, Russia | 45–75/ 70 | 6.0–9.4/ 7.5 | 2.0–3.0 by 0.4–0.6 | Glucose, peptone, beef extract, YE | 0.5–7.0/ 2.0–3.0 | S0 | H2, AA, ALA, CO2, iVal | [87] | |
| Thermosipho | Thermosipho affectus | Hydrothermal vent, Atlantic Ocean | 37–75/ 70 | 5.6–8.2/ 6.6 | 1.2–6.0 by 0.4–0.9 | YE, beef extract, glucose, maltose, sucrose, starch, dextrin, CMC, cellulose | 1.0–5.5/ 2.0 | S0 | AA, H2, CO2, EtOH | [88] |
| Thermosipho globiformans | Hydrothermal vent | 40–75/ 68 | 5.0–8.2/ 6.8 | 2.0–4.0 by 0.5 | YE, tryptone, starch | 0.2–5.2/ 2.5 | S0; Fe2O3 | n.d. | [89] | |
| Thermosipho melanesiensis | Hydrothermal vent, Pacific Ocean | 50–75/ 70 | 4.5–8.5/ 6.5–7.5 | 1.0–3.5 by 0.4–0.6 | BHI, malt extract, tryptone, sucrose, starch, glucose, maltose, lactose, cellobiose, galactose | 1.0–6.0/ 3.0 | S0 | H2, AA, ALA, CO2 | [90] | |
| Thermosipho activus | Riftia sheath, Guaymas Basin | 44–75/ 65 | 5.5–8.0/ 6.0 | 1.5–10.0 by 0.3–0.8 | glucose, maltose, cellobiose, cellulose, filter paper, chitin, xylan, pectin, xanthan gum, YE, beef extract, tryptone, casein, keratin, arabinose, xylose, gelatin | 0.3–6.0/ 2.5 | S0, Fe (III) | AA, H2, CO2 | [91] | |
| Thermosipho atlanticus | Hydrothermal vent, Atlantic Ocean | 45–80/ 65 | 5.0–9.0/ 6.0 | 1.0–2.6 by 0.2–0.6 | cellobiose, xylose, starch, LA, maltose, mannose, trehalose, lactose, arabinose, galactose, mannitol, peptone, casamino acids, gelatin, BHI, YE, glucose | 1.5–4.6/ 2.3 | S0, Thio, Cys | AA, iVal, H2, Gly, ALA, Pro | [92] | |
| Geotoga | Geotoga subterranea | Oilfields, USA | 30–60/ 45 | 5.5–9.0/ 6.5 | 4.0– 7.5 by 0.5 | mannose, starch, maltodextrins, glucose, lactose, sucrose, galactose, maltose | 0.5-10/ 4.0 | S0 | H2, CO2, AA, EtOH | [10] |
| Geotoga petraea | Oilfields, USA | 30–55/ 50 | 5.5–9.0/ 6.5 | 3.0– 20.0 by 0.6 | mannose, starch, maltodextrins, glucose, lactose, sucrose, galactose, maltose | 0.5–10/ 3.0 | S0 | H2, CO2, AA, EtOH | [10] | |
| Petrotoga | Petrotoga miotherma | Oilfields, USA | 35–65/ 55 | 5.5–9.0/ 6.5 | 2.0– 7.5 by 0.6 | mannose, starch, maltodextrins, glucose, lactose, sucrose, galactose, maltose, maltodexstrins, xylose | 0.5–10/ 2.0 | S0 | H2, CO2, AA, EtOH | [10] |
| Petrotoga olearia | Oil reservoir, Russia | 37–60/ 55 | 6.5–8.5/ 7.5 | 0.9–2.5 by 0.3–0.6 | arabinose, xylose, cellobiose, dextrin, sucrose, glucose, fructose, maltose, ribose, trehalose, xylan, pyruvate, peptone, starch | 0.5–8.0/ 2.0 | S0 | H2, AA, LA, ALA, EtOH | [93] | |
| Petrotoga sibirica | Oil reservoir, Russia | 37–55/ 55 | 6.5–9.4/ 8.0 | 0.9–2.5 by 0.3–0.6 | sucrose, glucose, fructose, maltose, ribose, trehalose, xylan, pyruvate, peptone, galactose | 0.5–7.0/ 1.0 | S0 | H2, AA, LA, ALA, EtOH | [93] | |
| Petrotoga | Petrotoga mobilis | Oilfield, North Sea | 40–65/ 58–60 | 5.5–8.5/ 6.5–7.0 | 1.0–50.0 by 0.5–1.5 | starch, xylan, maltodextrin, maltose, cellobiose, sucrose, lactose, glucose, galactose, fructose, arabinose, xylose, ribose, rhamnose | 0.5–9.0/ 3.0–4.0 | S0, Thio | H2, CO2, AA, EtOH | [94] |
| Petrotoga halophila | Offshore oil, Africa | 45–65/ 60 | 5.6–7.8/ 6.7–7.2 | 2.0–45.0 by 0.5–0.7 | arabinose, cellobiose, fructose, galactose, glucose, lactose, maltose, rhamnose, ribose, starch, sucrose, xylose, xylan, pyruvate | 0.5–9.0/ 4.0–6.0 | S0 | AA, LA, ALA, H2, CO2 | [95] | |
| Petrotoga mexicana | Offshore oil, Africa | 25–65/ 55 | 5.8–8.5/ 6.6 | 1.0–30.0 by 0.5–0.7 | arabinose, cellobiose, fructose, galactose, glucose, lactose, maltose, mannose, raffinose, rhamnose, ribose, starch, sucrose, xylose, xylan, pyruvate. | 1.0–20.0/ 3.0 | S0, Thio, Sulfite | AA, LA, H2, CO2, ALA | [96] | |
| Petrotoga japonica | Oil reservoir, Japan | 40–65/ 60 | 6.0–9.0/ 7.5 | 2.5–7.0 by 0.25–0.75 | starch, xylan, maltose, cellobiose, sucrose, lactose, glucose, galactose, fructose, casamino acids, mannose, arabinose, xylose, ribose | 0.5–9.0/ 0.5–1.0 | S0, Thio | AA, H2, CO2, ALA | [97] | |
| Marinitoga | Marinitoga piezophila | Hydrothermal chimney, Pacific Ocean | 45–70/ 65 | 5.0–8.0/ 6.0 | 1.0–1.5 by 0.5 | starch, fructose, glucose, galactose, maltose, cellobiose, ribose, acetate | 1.0–5.0/ 3.0 | S0, Thio, Cys | n.d. | [98] |
| Marinitoga litoralis | Hot spring, Indian Ocean | 45–70/ 65 | 5.5–7.5/ 6.0 | 1.0–7.0 by 0.8–1.0 | cellobiose, galactose, glucose, glycogen, lactose, maltose, ribose, starch, BHI, casamino acids, casein, peptone, pyruvate, tryptone, YE | 0.8–4.6/ 2.6 | S0 | n.d. | [99] | |
| Marinitoga okinawensis | Hydrothermal field, Okinawa | 30–70/ 55–60 | 5.5–7.4/ 5.5–5.8 | 1.5–5.0 by 0.5–0.8 | YE, tryptone, peptone, starch, glucose, glycerol | 1.0–5.5/ 3.0–3.5 | S0, Cys | n.d. | [100] | |
| Marinitoga hydrogenitolerans | Hydrothermal chimney, Atlantic Ocean | 35–65/ 60 | 4.5–8.5/ 6.0 | 1.5–5.0 by 0.5–0.8 | glucose, starch, glycogen, chitin, YE, BHI, peptone, casein, pyruvate, maltose | 1.0–6.5/ 3.0–4.0 | S0, Thio, Cys | AA, EtOH, Fo, H2, CO2 | [101] | |
| Marinitoga artica | Hydrothermal chimney, Norwegian | 45–70/ 65 | 5.0–7.5/ 5.5 | 1.0–5.0 by 0.5–0.8 | glucose, trehalose, maltose, sucrose, maltodextrin, starch, pectin, meat extract, tryptone, YE, pyruvate, fructose, mannose, cellobiose, cellulose, peptone | 1.5–5.5/ 2.5 | S0, Cys | n.d. | [102] | |
| Marinitoga camini | Hydrothermal chimney, Atlantic Ridge | 25–65/ 55 | 5.0–9.0/ 7.0 | 2.0–3.0 by 0.5–1.0 | BHI, gluten, peptone, tryptone, pyruvate, glucose, fructose, maltose, cellobiose, sucrose, starch, cellulose, CMC, pectin, chitin | 1.0–4.5/ 2.0 | S0, Cys | AA, iBut, iVal, H2, 3-IAA, LA CO2, HPA, PA | [11] | |
| Oceanotoga | Oceanotoga teriensis | Offshore oil, India | 25–70/ 55– 58 | 5.5–9.0/ 7.5 | 1.5–1.7 by 0.5–0.7 | glucose, fructose, cellobiose, arabinose, raffinose, rhamnose, sucrose, xylose, ribose, starch, EtOH, formate, acetate, BHI, YE, bio–trypticase | 0.0–12/ 4.3 | S0, Thio | AA, H2, CO2, EtOH | [12] |
| Defluviitoga | Defluviitog tunisiensis | Mesothermic digester | 37–65/ 55 | 6.7–7.9/ 6.9 | 3.0–30.0 by 1.0 | arabinose, cellobiose, fructose, galactose, glucose, lactose, maltose, mannose, raffinose, ribose, sucrose, xylose, cellulose, xylan | 0.2–3.0/ 0.5 | S0, Thio | AA, H2, CO2 | [9] |
| Mesotoga | Mesotoga infera | Deep aquifer, France | 30–50/ 45 | 6.2–7.9/ 7.4 | 2.0–4.0 by 1.0–2.0 | arabinose, cellobiose, fructose, galactose, glucose, lactose, LA, mannose, maltose, raffinose, ribose, sucrose, xylose | 0.0–1.5/ 0.2 | S0 | AA, CO2 | [26] |
| Mesotoga prima | Sediment, USA | 20–50/ 37 | 6.5–8.0/ 7.5 | 1.0 by 0.2 | xylose, fructose, ribose, sucrose, mannose, galactose, maltose, lactose, peptone, tryptone, casamino acids, glucose, arabinose, cellobiose, casein, pyruvate | 2.0–6.0/ 4.0 | S0, Thio, Sulfite | AA, But, iBut, iVal, 2–MeBu | [8] | |
| Kosmotoga | Kosmotoga arenicorallina | Hot spring, Japan | 50–65/ 60 | 6.2–8.0/ 7.1 | 1.1–2.7 by 1.1–1.9 | xylose, maltose, glycerol | 1.0–6.0/ 3.0 | S0, Cys | n.d. | [103] |
| Kosmotoga pacifica | Hydrothermal field, Pacific Ocean | 33–78/ 70 | 6.2–8.0/ 7.1 | 1.0 by 0.6 | maltose, YE, peptone, BHI, glycerol, tryptone, xylose, glucose, fructose, cellobiose, trehalose, LA, propionate, glutamate | 0.5–6.0/ n.d. | S0, Cys | n.d. | [104] | |
| Kosmotoga olearia | Fluid, North Sea | 20–80/ 65 | 5.5–8.0/ 6.8 | 0.8–1.2 by 0.4–0.7 | maltose, ribose, sucrose, starch, casamino acids, tryptone, pyruvate | 1.0–6.0/ 2.5–3.0 | Thio | H2, CO2, AA, EtOH, PPA | [7] | |
| Kosmotoga shengliensis | Oilfield, China | 45–75/ 65 | 6.0–8.0/ 7.0 | 0.7–0.9 | glucose, acetate, mEtOH, galactose, fructose, xylose, sucrose, maltose, sorbitol, lactose, xylan, arabinose, formate, rhamnose, glycerol, pyruvate, starch, LA | 0.0–4.0/ 1.5 | S0, Thio, Sulfate | AA, LA, ALA, CO2, H2 | [15] | |
| Athalassatoga | Athalassatoga saccharophila | Hot spring, Japan | 30–60/ 55 | 4.5–7.5/ 5.5–6.0 | 0.8–2.0 by 0.7–0.8 | arabinose, fructose, glucose, lactose, maltose, mannose, ribose, sucrose, xylose, starch, glycogen, peptone, YE | <1/0.0 | Fe (III), Thio, Cys | AA, iBut, iVal | [14] |
| Mesoaciditoga | Mesoaciditoga lauensis | Hydrothermal vent, Pacific Ocean | 45–65/ 57–60 | 4.1–6.0/ 5.5–5.7 | 0.8–1.0 by 0.4 | YE, peptone, maltose, sucrose, glucose, xylose, ribose, starch, tryptone | 0.5–6.0/ 3.0 | S0; Thio, Cys | n.d. | [13] |
| Parameter | Organism | T (°C) | Culture Type | Mixing Speed (rpm) | Reactor/Working Volume (L) | Substrate Loaded (mmol/L) | Operational Parameter | Substrate Consumed (mmol/L) | Products | Ref. | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H2 yielda | AA (mmol/L) | LA (mmol/L) | ALA (mmol/L) | But (mmol/L) | ||||||||||
| PH2 (mbar) | T. maritima | 80 | B | 350 | 1.4/0.1 | Glucose (28) | PH2 = 7.1 ± 0.4 | 19.8 ± 1.1 | 2.34 | 25.0 ± 1.4 | 10.5 ± 0.5 | [107] | ||
| PH2 = 71.4 ± 2.1 | 19.7 ± 1.4 | 2.44 | 24.6 ± 2.4 | 11.0 ± 0.6 | ||||||||||
| PH2 = 178.5 ± 3.5 | 17.2 ± 0.9 | 2.32 | 20.1 ± 1.0 | 9.4 ± 0.5 | ||||||||||
| PH2 = 606.9 ± 18.7 | 13.4 ± 0.7 | n. d. | 13.0 ± 0.7 | 11.0 ± 0.6 | ||||||||||
| Stirring Speed (rpm) | T. neapolitana | 75 | CSABR | 300 | 3.0/1.0 | Xylose (33.3) | 300 | 31.43 | 2.13 ± 0.11 | 41.8 ± 2.16 | 1.78 ± 0.11 | [113] | ||
| 400 | 400 | 32.56 | 2.94 ± 0.15 | 50.12 ± 2.5 | 4.0 ± 0.22 | |||||||||
| 500 | 500 | 32.03 | 2.31 ± 0.12 | 44.62 ± 2.16 | 4.84 ± 0.22 | |||||||||
| 600 | 600 | 31.87 | 2.24 ± 0.11 | 41.12 ± 2.0 | 1.89 ± 0.11 | |||||||||
| T. neapolitana subsp. capnolactica | 80 | CSTR | 300 | 3.0/2.0 | Glucose (28) | 300 | 22.9 ± 2.7 | 3.0 ± 0.0 | 32.3 ± 4.3 | 10.0 ± 1.0 | 1.1 ± 0.1 | [69] | ||
| 500 | 500 | 24.8 ± 0.4 | 3.2 ± 0.1 | 37.7 ± 2.7 | 8.1 ± 0.2 | 1.0 ± 0.1 | ||||||||
| 300 | 300 + GaR | 24.7 ± 0.2 | 3.5 ± 0.2 | 39.2 ± 1.2 | 4.4 ± 0.1 | 0.9 ± 0.0 | ||||||||
| 500 | 500 + GaR | 24.9 ± 0.2 | 3.3 ± 0.1 | 38.7 ± 2.2 | 5.1 ± 0.5 | 0.8 ± 0.0 | ||||||||
| Gas sparging | T. neapolitana | 80 | B | 250 | 3.8/1.0 | Glucose (28) | N2 | 25.9 ± 1.3 | 2.8 | 44.8 ± 5.4 | 12.5 ± 2.9 | 1.3 ± 0.4 | [31] | |
| CO2 | 26.1 ± 1.2 | 2.8 | 35.6 ± 5.8 | 20.0 ± 6.1 | 2.7 ± 0.5 | |||||||||
| 75 | SB | no | 0.12/0.04 | Glycerol (108.6) | w/o | 13 ±0.6 | 1.24 ± 0.06 | 8.71 ± 0.35 | 0.36 ± 0.02 | [115] | ||||
| N2 | 14 ± 0.7 | 2.06 ± 0.09 | 10.04 ± 0.5 | 0.34 ± 0.02 | ||||||||||
| N2 plus pH control | 18 ± 0.9 | 1.98 ± 0.1 | 12.62 ± 0.53 | 0.25 ± 0.01 | ||||||||||
| Gas sparging | T. neapolitana | 77 | SB | 150 | 0.12/0.04 | Glucose (39) | w/o | - | 1.82 ± 0.09 | 64.28 ± 2.83 | 33.48 ± 1.47 | [110] | ||
| N2 | - | 3.24 ± 0.14 | 81.42 ± 3.49 | 36.77 ± 2.04 | ||||||||||
| Xylose (27) | w/o | - | 1.14 ± 0.07 | 40.30 ± 3.5 | 37.68 ± 1.7 | |||||||||
| N2 | - | 2.20 ± 0.13 | 71.94 ± 3.66 | 50.62 ± 2.38 | ||||||||||
| T. neapolitana subsp. capnolactica | 80 | SB | no | 0.12/0.03 | Glucose (28) | N2 | 25.7 ± 0.1 | 2.5 ± 0.06 | 27.3 ± 0.8 | 8.6 ± 0.2 | 2.5 ± 0.2 | [70] | ||
| CO2 | 28.3 ± 1.0 | 2.9 ± 0.1 | 22.1 ± 0.9 | 11.3 ± 0.1 | 3.0 ± 0.3 | |||||||||
| T. neapolitana | 80 | SB | no | 0.12/0.03 | Glucose (28) | N2 | 21.7 ± 0.6 | 2.5 ± 0.03 | 30.2 ± 0.4 | 2.2 ± 0.02 | 1.9 ± 0.3 | |||
| CO2 | 20.8 ± 2.3 | 1.9 ± 0.1 | 20.8 ± 0.1 | 1.2 ± 0.06 | 2.4 ± 0.3 | |||||||||
| T. maritima | 80 | SB | no | 0.12/0.03 | Glucose (28) | N2 | 23.2 ± 1.0 | 1.9± 0.06 | 25.5 ± 0.5 | 5.3 ± 0.8 | 2.4 ± 0.06 | |||
| CO2 | 19.9 ± 0.6 | 2.0 ± 0.1 | 18.3 ± 0.3 | 1.6 ± 0.2 | 2.3 ± 0.3 | |||||||||
| T. naphtophila | 80 | SB | no | 0.12/0.04 | Glucose (28) | N2 | 13.30 ± 1.10 | 2.20 ± 0.20 | 15.70 ± 0.10 | 1.40 ± 0.06 | 0.80 ±0.10 | |||
| CO2 | 20.80 ± 1.70 | 1.60 ± 0.20 | 19.20 ± 0.10 | 5.00 ± 0.02 | 1.80 ±0.05 | |||||||||
| T. petrophila | 80 | SB | no | 0.12/0.05 | Glucose (28) | N2 | 9.20 ± 1.30 | 3.00 ± 0.40 | 13.10 ± 0.05 | 2.00 ± 0.01 | 0.00 | |||
| CO2 | 14.20 ± 0.60 | 1.90 ± 0.10 | 12.60 ± 0.10 | 3.80 ± 0.02 | 0.30 ±0.10 | |||||||||
| T. caldifontis | 70 | SB | no | 0.12/0.05 | Glucose (28) | N2 | 10.90 ± 1.10 | 2.60 ± 0.10 | 16.70 ± 3.60 | 2.20 ± 0.50 | 3.20 ±0.90 | |||
| CO2 | 15.20 ± 0.90 | 1.80 ± 0.03 | 15.60 ± 1.50 | 2.30 ± 0.40 | 6.60 ±0.70 | |||||||||
| T. profunda | 60 | SB | no | 0.12/0.05 | Glucose (28) | N2 | 18.1 0 ±0.40 | 1.50 ± 0.20 | 15.90 ± 0.40 | 5.70 ± 0.10 | 1.40 ±0.06 | |||
| CO2 | 22.60 ± 1.70 | 0.70 ± 0.04 | 5.60 ± 0.20 | 2.3 ± 0.04 | 2.60 ±0.30 | |||||||||
| Pseudot. hypogea | 70 | SB | no | 0.12/0.05 | Glucose (28) | N2 | 8.80 ± 1.10 | 1.10 ± 0.30 | 6.40 ± 0.10 | 0.10 ± 0.00 | 2.90 ±0.10 | |||
| CO2 | 4.30 ± 0.10 | 0.50 ± 0.10 | 3.10 ± 0.20 | 0.10 ± 0.00 | 3.40 ±0.30 | |||||||||
| Pseudot. elfii | 70 | SB | no | 0.12/0.05 | Glucose (28) | N2 | 7.00 ± 0.90 | 2.00 ± 0.20 | 8.30 ± 0.06 | 0.20 ± 0.03 | 4.20 ±0.30 | [70] | ||
| CO2 | 6.70 ± 0.20 | 2.10 ± 0.10 | 7.80 ± 0.30 | 0.10 ± 0.01 | 10.0 ±0.30 | |||||||||
| Pseudot. lettingae | 70 | SB | no | 0.12/0.05 | Glucose (28) | N2 | 9.30 ± 0.50 | 1.20 ± 0.10 | 5.10 ± 0.05 | 0.20 ± 0.00 | 2.70 ±0.05 | |||
| CO2 | 8.10 ± 0.70 | 1.30 ± 0.30 | 4.40 ± 0.10 | 0.05 ± 0.01 | 3.70 ±0.20 | |||||||||
| Gas sparging | Pseudot. subterranea | 70 | SB | no | 0.12/0.05 | Glucose (28) | N2 | 23.10 ± 2.10 | 1.80 ± 0.20 | 30.60 ± 6.90 | 16.20 ± 4.60 | 9.50 ±0.40 | ||
| CO2 | 27.00 ± 1.40 | 1.40 ± 0.10 | 31.90 ± 7.90 | 10.70 ± 4.0 | 20.0 ± 8.0 | |||||||||
| Pseudot. thermarum | 80 | SB | no | 0.12/0.05 | Glucose (28) | N2 | Complete | 1.8 ± 0.02 | 30.00 ± 2.20 | 6.50 ± 0.20 | 1.10 ±0.07 | |||
| CO2 | Complete | 1.50 ± 0.10 | 24.80 ± 0.70 | 5.60 ± 0.60 | 2.20 ±0.20 | |||||||||
| Biomass (g CDW/L) | T. neapolitana | 80 | Flask | 300 | 0.25/0.2 | Glucose (28) | 0.46 | 3.2 ± 0.04 | 2.39 | 34.3 ± 0.6 | 10.9 ± 0.4 | [68] | ||
| 0.91 | 2.9 ± 0.06 | 2.44 | 32.9 ± 0.8 | 12.2 ± 0.8 | ||||||||||
| 1.33 | 3.4 ± 0.01 | 2.58 | 32.3 ± 0.2 | 11.5 ± 0.5 | ||||||||||
| 1.74 | 3.0 ± 0.04 | 2.37 | 31.4 ± 1.1 | 14.7 ± 0.7 | ||||||||||
| pH | T. neapolitana subsp. capnolactica | 80 | SB | no | 0.12/0.03 | Glucose (28) | w/o | 18.54 ± 0.15 | 1.78 ± 0.29 | 22.76 ± 0.40 | 11.35 ± 0.62 | [67] | ||
| 0.01M MOPS | 26.42 ± 0.05 | 3.27 ± 0.18 | 26.65 ± 0.87 | 14.23 ± 0.22 | ||||||||||
| 0.01M TRIS | 25.55 ± 0.06 | 3.10 ± 0.10 | 26.77 ± 0.29 | 12.08 ± 0.89 | ||||||||||
| 0.01M HEPES | 25.99 ± 0.03 | 2.85 ± 0.40 | 25.56 ± 0.49 | 13.58 ± 0.88 | ||||||||||
| 0.01M HCO3− | 25.62 ± 0.10 | 2.20 ± 0.30 | 22.82 ± 0.84 | 14.63 ± 3.23 | ||||||||||
| 0.01M phosphate | 26.17 ± 0.26 | 2.78 ± 0.40 | 24.70 ± 0.59 | 14.92 ± 0.25 | ||||||||||
| T. neapolitana | 75 | CSABR | 300 | 3.0/1.0 | Glucose (28) | w/o pH control | 21.98 ± 1.11 | 2.05 ± 0.1 | 30.81 ± 1.5 | 3.33 ± 0.22 | [113] | |||
| plus pH control | 27.47 ± 1.39 | 3.2 ± 0.16 | 38.3 ± 2.0 | 1.77 ± 0.11 | ||||||||||
| Xylose (33.3) | w/o pH control | 29.77 ± 1.46 | 1.84 ± 0.09 | 34.47 ± 1.66 | 3.77 ± 0.22 | |||||||||
| plus pH control | 31.83 ± 1.6 | 2.22 ± 0.11 | 41.8 ± 2.0 | 1.66 ± 0.11 | ||||||||||
| pH | T. neapolitana | 75 | CSABR | 300 | 3.0/1.0 | Sucrose (14.6) | w/o pH control | 13.78 ± 0.7 | 3.52 ± 0.18 | 33.13 ± 1.65 | 3.11 ± 0.11 | [113] | ||
| plus pH control | 14.69 ± 0.06 | 4.95 ± 0.25 | 35.47 ± 1.83 | 2.11 ± 0.11 | ||||||||||
| Xylose (33.3) | w/o pH control | 29.44 | 1.85 ± 0.09 | 34.97 ± 1.66 | 3.88 ±0.22 | |||||||||
| pH = 6.5 | 32.57 | 2.71 ± 0.14 | 49.62 ± 2.50 | 3.44 ± 0.11 | ||||||||||
| pH = 7.0 | 32.9 | 2.84 ±0.14 | 50.29 ± 2.50 | 4.00 ± 0.22 | ||||||||||
| pH = 7.5 | 31.77 | 2.23 ± 0.11 | 41.96 ± 2.16 | 1.89 ± 0.11 | ||||||||||
| 75 | SB | no | 0.04/ 0.12 | Glycerol (108.6) | w/o HEPES | 16.96 ± 0.8 | 1.23 ± 0.06 | 9.14 ± 0.45 | [116] | |||||
| 0.05 M HEPES | 28.26 ± 1.4 | 2.73 ± 0.14 | 22.35 ± 1.05 | |||||||||||
| T. neapolitana | 80 | B | 250 | 3.8/1.0 | Glucose (28) | w/o NaHCO3 | 25.9 ± 1.3 | 2.8 | 44.5 ± 5.4 | 12.5 ± 2.69 | [31] | |||
| NaHCO3 14 mM | 25.4 ± 2.1 | 1.7 | 30.5 ± 4.9 | 18.0 ± 0.6 | ||||||||||
| NaHCO3 20 mM | 23.2 ± 1.9 | 1.0 | 44.4 ± 8.2 | 9.2 ± 2.7 | ||||||||||
| pH | NaHCO3 40 mM | 6.2 ± 0.8 | 2.7 | 18.0 ± 4.3 | 0.7 ± 1.5 | |||||||||
| 75 | B | no | 0.12/0.04 | Glycerol (108.6) | w/o IA | - | 438 ± 22 b | 7.49 ± 0.33 | 3.55 ± 0.22 * | [122] | ||||
| 1.5 g/L IA | - | 619 ± 30 b | 11.49 ± 0.5 | 1.66 ± 0.0 * | ||||||||||
| Temp. (°C) | T. neapolitana | 60 | SB | 75 | 0.26/0.05 | Glucose (14) | 60 | 2.2 * | 2.04 ± 0.05 | 2.0 | n. d | [65] | ||
| 65 | 65 | 5.0 * | 3.09 ± 0.3 | 7.0 | 0.05 | |||||||||
| 70 | 70 | 8.5 * | 3.18 ± 0.02 | 11.5 | 0.45 | |||||||||
| 77 | 77 | 11.0 ± 0.5 * | 3.85 ± 0.28 | 16.5 | 0.85 ± 0.1 | |||||||||
| 85 | 85 | 11.0 ± 0.5 * | 3.75 ± 0.49 | 18.0 ± 1.0 | 1.25 ± 0.05 | |||||||||
| Oxygen | T. maritima | 80 | B | 150 | 2.30/1.53 | Glucose (20) | w/o O2 | 17.41 | 38.09 b | 18.05 | 4.36 | 1.60 ± 0.2 | [129] | |
| with O2 | 19.30 | 31.75 b | 18.27 | 5.45 | 1.30 ± 0.2 | |||||||||
| Parameter | Organism | T (°C) | Culture Type | Mixing Speed (rpm) | Reactor/Working Volume (L) | Substrate Loaded (mmol/L) | Operational Parameter | Substrate Consumed (mmol/L) | Products | Ref. | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H2 | AA (mmol/L) | LA (mmol/L) | ALA (mmol/L) | ||||||||||
| Nitrogen sources (g/L) | Pseudot. elfii | 65 | B | 100 | 3.0/1.0 | no | w/o YE | - | 40 a | [108] | |||
| CA + V | - | 4 a | |||||||||||
| CA + V + aa | - | 6 a | |||||||||||
| 65 | B | 100 | 3.0/1.0 | Glucose (22.4) | YE (5) | n.d. | 100 a | ||||||
| CA + V | n.d. | 14 a | |||||||||||
| CA +V + aa | n.d. | 14 a | |||||||||||
| 65 | B | 100 | 3.0/1.0 | no | YE (2) -Tryp (0) | - | 13.9 b | 3.5 | |||||
| YE (2) -Tryp (2) | - | 14.8 b | 3.4 | ||||||||||
| YE (5) -Tryp (0) | - | 14.0 b | 0.0 | ||||||||||
| YE (5) -Tryp (5) | - | 28.8 b | 4.9 | ||||||||||
| 65 | B | 100 | 3.0/1.0 | Glucose (56) | YE (2) -Tryp (0) | 10.3 | 25.8 b | 10.7 | |||||
| YE (2) -Tryp (2) | 18.3 | 78.5 b | 19.7 | ||||||||||
| YE (5) -Tryp (0) | 13.1 | 84.9 b | 26.3 | ||||||||||
| YE (5) -Tryp (5) | 17.9 | 82.5 b | 21.2 | ||||||||||
| T. neapolitana | 80 | SB | no | 0.12/0.05 | Glucose (28) | YE (0.5) | 26.6 * | 260 *c | 15 * | [64] | |||
| YE (1.0) | 26 * | 320 *c | 22.5 * | ||||||||||
| YE (2.0) | 25.5 * | 360 *c | 26.6 * | ||||||||||
| YE (4.0) | 25 * | 430 *c | 30 * | ||||||||||
| YE (6.0) | 25 * | 430 *c | 33.3 * | ||||||||||
| T. maritima | 80 | SB | no | 0.12/0.05 | Glucose (28.00) | YE (0.5) | 25.5 * | 190 *c | 0.0 * | ||||
| YE (1.0) | 25 * | 260 *c | 20.8 * | ||||||||||
| YE (2.0) | 25 * | 270 *c | 23 * | ||||||||||
| Nitrogen sources (g/L) | T. maritima | 80 | SB | no | 0.12/0.05 | Glucose (28.00) | YE (4.0) | 25 * | 335 *c | 27.5 * | [64] | ||
| YE (6.0) | 24 * | 390 *c | 28 * | ||||||||||
| T. neapolitana | 77 | B | 75 | 0.12/0.05 | Glucose (28) | no YE | 23 * | 9 *b | 4.2 * | [136] | |||
| YE (0.5) | Completed * | 16 *b | 7.2 * | ||||||||||
| NaCl (g/L) | T. neapolitana subsp. capnolactica | 80 | SB | no | 0.12/0.03 | Glucose (28) | w/o | 25.62 ± 0.07 | 2.30 ± 0.50 d | 20.66 ± 0.27 | 2.80 ± 0.26 | 1.28 ± 0.9 | [67] |
| NaCl (5) | 26.00 ± 0.14 | 2.50 ± 1.20 d | 24.59 ± 0.95 | 6.23 ± 3.26 | 1.61 ± 0.58 | ||||||||
| NaCl (10) | 26.12 ± 0.16 | 3.10 ± 0.80 d | 26.05 ± 4.69 | 11.61 ± 2.42 | 2.46 ± 0.24 | ||||||||
| NaCl (20) | 25.96 ± 0.11 | 3.30 ± 0.20 d | 25.58 ± 1.03 | 13.44 ± 0.94 | 2.41 ± 0.09 | ||||||||
| NaCl (30) | 25.68 ± 0.25 | 2.91 ± 0.37 d | 23.22 ± 0.81 | 21.63 ± 6.15 | 2.38 ± 0.10 | ||||||||
| Organism | Carbon Source (mM) | Sulfur Source (mM) | Substrate Consumed (mmol/L) | Products mmol/L Culture | Ref. | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H2 | AA | LA | ALA | EtOH | iVal | H2S | Glu | |||||
| T. maritima | Glucose (25) | w/o | 7.1 ± 0.4 | 21.3 ± 2.1 | 10.1 ± 0.8 | 0.8 ± 0.1 | - | [107] | ||||
| DMSO ** | 9.2 ± 0.5 | 28.7 ± 2.9 | 13.3 ± 1.1 | 0.8 ± 0.1 | - | |||||||
| S0 ** | 16.6 ± 0.8 | 46.1 ± 4.6 | 23.8 ± 1.9 | 3.4 ± 0.3 | - | |||||||
| Met ** | 18.3 ± 0.9 | 53.3 ± 5.3 | 26.5 ± 2.1 | 3.1 ± 0.3 | - | |||||||
| Thio ** | 17.5 ± 0.9 | 47.3 ± 4.7 | 24.1 ± 1.9 | 6.3 ± 0.6 | - | |||||||
| Cys ** | 20.4 ± 1.0 | 58.5 ± 5.8 | 30.5 ± 2.4 | 4.1 ± 0.4 | - | |||||||
| Na2S ** | 20.4 ± 1.0 | 54.9 ± 5.5 | 30.7 ± 2.5 | 4.7 ± 0.5 | - | |||||||
| Glucose (60) | w/o Thio | 17.7 ± 1.9 | 25.0 ± 2.2 | 12.8 ± 1.0 | 5.4 ± 0.6 | 1.39 ± 0.2 | ||||||
| Thio (0.01) | 20.0 ± 1.1 | 31.0 ± 2.3 | 16.0 ± 0.8 | 10.2 ± 1.1 | - | |||||||
| Thio (0.03) | 28.0 ± 1.5 | 57.9 ± 4.8 | 30.6 ± 1.9 | 8.2 ± 0.7 | - | |||||||
| Thio (0.06) | 38.5 ± 2.0 | 73.3 ± 5.9 | 38.2 ± 2.4 | 18.1 ± 1.8 | - | |||||||
| Thio (0.12) | 45.7 ± 2.5 | 99.7 ± 8.3 | 52.4 ± 3.3 | 15.4 ± 1.6 | 3.8 ± 0.3 | |||||||
| Thio (0.18) | 45.4 ± 2.2 | 86.9 ± 8.2 | 45.0 ± 2.2 | 23.4 ± 2.3 | - | |||||||
| Thio (0.24) | 43.8 ± 2.2 | 88.6 ± 8.9 | 46.1 ± 3.3 | 26.4 ± 1.4 | 3.8 ± 0.2 | |||||||
| Glucose (20) | w/o | 13.70 | 36.09 | 15.62 | 0.70 | n.d. | [145] | |||||
| Thio (20) | 13.55 | 4.02 | 15.99 | 0.80 | 14.45 | |||||||
| T. neapolitana | Glucose (20) | w/o | 14.00 | 31.67 | 18.27 | 0.87 | n.d. | [145] | ||||
| Thio (20) | 13.90 | 16.07 | 16.12 | 0.60 | 7.39 | |||||||
| Pseudot. lettingae | Methanol (20) | w/o | 19.70 | n. d. | 13.70 | - | - | [75] | ||||
| Thio (20) | 18.7 | n. d. | - | 5.8 | 11.2 | |||||||
| S0 (2%) | 10.6 | n. d. | - | 3.1 | 7.3 | |||||||
| Pseudot. hypogea | Glucose (20) | w/o | 8.60 | 29.03 | 4.49 | 1.71 | n. d. | [145] | ||||
| Thio (20) | 14.39 | 2.29 | 19.7 | 1.06 | 15.08 | |||||||
| Pseudot. hypogea | Glucose (20) | w/o | 7.0 | 9.4 a | 5.0 | 1.7 | 1.0 | 0.2 | [77] | |||
| Thio (20) | 13.0 | 0.9 a | 19.8 | 1.0 | 1.6 | 15.1 | ||||||
| Pseudot. hypogea | Xylose (20) | w/o | 12.9 | 19.0 a | 8.9 | 2.4 | 1.0 | 0.2 | [77] | |||
| Thio (20) | 12.0 | 1.8 a | 13.7 | 1.3 | 1.0 | 7.5 | ||||||
| Pseudot. elfii | Glucose (20) | w/o | 3.1 | 8.8 | 4.0 | 0.0 | [77] | |||||
| Thio (20) | 10.4 | 2.0 | 17.9 | 23.00 | ||||||||
| Glucose (20) | w/o | 2.75 | 7.70 | 3.49 | 1.05 | n. d. | [145] | |||||
| Thio (20) | 8.15 | n. d. | 12.63 | 0.41 | 14.55 | |||||||
| Ts. geolei | Glucose (0.28) | w/o | 7.0 b | 9.3 a | 8.5 b | 1.2 b | 0.5 b | [87] | ||||
| S0 (2%) | 6.0 b | 0.0 a | 7.5 b | 0.5 b | 12.5 b | |||||||
| Ms. Prima Phos Ac3 | Glucose (20) | w/o | 1.50 ± 0.20 | <1 c | 1.67 ± 0.21 | 1.05 ± 0.25 | [27] | |||||
| S0 | 6.57 ± 0.19 | <1 c | 9.21 ± 0.13 | 24.40 ± 0.30 | ||||||||
| Ms. Prima MesG1Ag4.2T | Fructose (20) | w/o | 1.00 ± 0.23 | <1 c | 0.70 ± 0.41 | 1.18 ± 0.41 | ||||||
| S0 | 3.27 ± 0.85 | <1 c | 8.48 ± 1.96 | 18.03 ± 5.16 | ||||||||
| Ts. africanus | Glucose (28) | w/o | 7.20 | 16.80 | 7.90 | <0.2 | 0.79 | n.d. | [145] | |||
| Thio (20) | 7.70 | 1.00 | 12.40 | - | - | 14.60 | ||||||
| Ts. atlanticus | Glucose (28) | w/o | 5.6 | 12.5 | 1.7 | 0.14 | - | [92] | ||||
| S0 (1%) | 6.0 | 7.5 | 1.9 | 0.15 | 1.3 | |||||||
| F. islandicum | Glucose (20) | w/o | 14.20 | 21.58 | 6.25 | 3.98 | n.d. | [145] | ||||
| Thio (20) | 16.20 | n. d. | 20.25 | 1.22 | 34.02 | |||||||
| F. pennavorans | Glucose (11) | w/o | - | 0.25 * | 6.7 * | 4.0 ± 0.5 * | 1.3 * | [32] | ||||
| Thio (20) | - | 0.2 * | 6.7 * | 4.50 * | No * | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lanzilli, M.; Esercizio, N.; Vastano, M.; Xu, Z.; Nuzzo, G.; Gallo, C.; Manzo, E.; Fontana, A.; d’Ippolito, G. Effect of Cultivation Parameters on Fermentation and Hydrogen Production in the Phylum Thermotogae. Int. J. Mol. Sci. 2021, 22, 341. https://doi.org/10.3390/ijms22010341
Lanzilli M, Esercizio N, Vastano M, Xu Z, Nuzzo G, Gallo C, Manzo E, Fontana A, d’Ippolito G. Effect of Cultivation Parameters on Fermentation and Hydrogen Production in the Phylum Thermotogae. International Journal of Molecular Sciences. 2021; 22(1):341. https://doi.org/10.3390/ijms22010341
Chicago/Turabian StyleLanzilli, Mariamichela, Nunzia Esercizio, Marco Vastano, Zhaohui Xu, Genoveffa Nuzzo, Carmela Gallo, Emiliano Manzo, Angelo Fontana, and Giuliana d’Ippolito. 2021. "Effect of Cultivation Parameters on Fermentation and Hydrogen Production in the Phylum Thermotogae" International Journal of Molecular Sciences 22, no. 1: 341. https://doi.org/10.3390/ijms22010341
APA StyleLanzilli, M., Esercizio, N., Vastano, M., Xu, Z., Nuzzo, G., Gallo, C., Manzo, E., Fontana, A., & d’Ippolito, G. (2021). Effect of Cultivation Parameters on Fermentation and Hydrogen Production in the Phylum Thermotogae. International Journal of Molecular Sciences, 22(1), 341. https://doi.org/10.3390/ijms22010341








