Transcriptome-Based Analysis Reveals a Crucial Role of BxGPCR17454 in Low Temperature Response of Pine Wood Nematode (Bursaphelenchus xylophilus)
Abstract
1. Introduction
2. Results
2.1. RNA Sequencing and General Transcription Patterns
2.2. B. xylophilus Genes Differentially Expressed in Response to Low Temperature
2.3. Validation of DEGs by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.4. Identification and Transcript Abundance Analysis of 6 Low-temperature-related BxGPCRs
2.5. RNAi Validation of Low-temperature-related BxGPCR
3. Discussion
4. Material and Methods
4.1. Sample Preparation
4.2. RNA Sequencing
4.3. Sequencing Data Analysis
4.4. Validation of DEGs by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.5. Identification of Low-temperature-related BxGPCRs
4.6. Transcript Abundance Analysis of 6 BxGPCRs under Low Temperature
4.7. RNAi Validation of Low-Temperature-Related BxGPCR
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mota, M.M.; Futai, K.; Vieira, P. Pine Wilt Disease and The Pinewood Nematode, Bursaphelenchus xylophilus. In Integrated Management of Fruit Crops Nematodes; Ciancio, A., Mukerji, K., Eds.; Springer: Dordrecht, The Netherlands, 2009; Volume 4, pp. 253–274. [Google Scholar]
- Wu, H.Y.; Tan, Q.Q.; Jiang, S.X. First report of pine wilt disease caused by Bursaphelenchus xylophilus on pinta thunbergii in the inland city of Zibo, Shandong, China. Plant Dis. 2013, 97, 1126. [Google Scholar] [CrossRef] [PubMed]
- Gruffudd, H.R.; Jenkins, T.A.R.; Evans, H.F. Using an evapo-transpiration model (ETpN) to predict the risk and expression of symptoms of pine wilt disease (PWD) across Europe. Boil. Invasions 2016, 18, 2823–2840. [Google Scholar] [CrossRef]
- Yu, Z.; Li, S.; Zhou, Y.; Zhou, R.; Wang, Y. Spatial estimation and prediction of suitable distribution of Bursaphelenchus xylophilus with different warming modes in China. J. Northeast For. Univ. 2018, 46, 85–91. [Google Scholar]
- Feng, Y.M.; Zhang, H.J.; Lü, Q.; Liang, J.; Zhang, X.Y. Quantification of suitability distribution region of Bursaphelenchus xylophilus in China. Scientia Silvae Sinicae 2009, 45, 65–71. [Google Scholar] [CrossRef]
- Devaney, E. Thermoregulation in the life cycle of nematodes. Int. J. Parasitol. 2006, 36, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J.A.; et al. Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef]
- Zhao, L.L.; Wei, W.; Zhang, X.Y.; Kulhavy, D.; Sun, J.H. Low temperature induces two growth-arrested stages and change of secondary metabolites in Bursaphelenchus xylophilus. Nematology 2007, 9, 663–670. [Google Scholar] [CrossRef]
- Mamiya, Y. The life history of the pine wood nematode, Bursaphelenchus lignicolus. Jpn. J. Nematol. 1975, 5, 16–25. [Google Scholar]
- Ohta, A.; Ujisawa, T.; Sonoda, S.; Kuhara, A. Light and pheromone-sensing neurons regulates cold habituation through insulin signalling in Caenorhabditis elegans. Nat. Commun. 2014, 5, 4412. [Google Scholar] [CrossRef]
- Mertens, I.; Clinckspoor, I.; Janssen, T.; Nachman, R.; Schoofs, L. FMRFamide related peptide ligands activate the Caenorhabditis elegans orphan GPCR Y59H11AL.1. Peptides 2006, 27, 1291–1296. [Google Scholar] [CrossRef]
- Murray, P.; Hayward, S.A.L.; Govan, G.G.; Gracey, A.Y.; Cossins, A.R. An explicit test of the phospholipid saturation hypothesis of acquired cold tolerance in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2007, 104, 5489–5494. [Google Scholar] [CrossRef]
- Wang, F.; Li, D.L.; Ma, L.; Wang, B.W.; Chen, Q.L.; Zhang, R.Z.; Su, D.; Kang, X.Y.; Zhai, W. Identification and rnai of a Bursaphelenchus xylophilus dauer formation gene: Bx-daf6. J. Beijing For. Univ. 2016, 38, 21–27. [Google Scholar]
- Wang, B.; Ma, L.; Wang, F.; Wang, B.; Hao, X.; Xu, J.; Ma, Y. Low Temperature Extends the Lifespan of Bursaphelenchus xylophilus through the cGMP Pathway. Int. J. Mol. Sci. 2017, 18, 2320. [Google Scholar] [CrossRef]
- Wang, B.; Liu, W.; Wang, F.; Ma, L.; Li, D.; Wang, B.; Hao, X.; Northeast Forestry University. Fat accumulation in Bursaphelenchus xylophilus by positively regulating Bx-SCD under low temperature. J. Northeast For. Univ. 2017, 7. [Google Scholar]
- Teng, M.S.; Dekkers, M.P.J.; Ng, B.L.; Rademakers, S.; Jansen, G.; Fraser, A.G.; McCafferty, J. Expression of mammalian GPCRs in C. elegans generates novel behavioural responses to human ligands. BMC Boil. 2006, 4, 22. [Google Scholar]
- Cao, P.; Sun, W.; Kramp, K.; Zheng, M.; Salom, D.; Jastrzebska, B.; Jin, H.; Palczewski, K.; Feng, Z. Light-sensitive coupling of rhodopsin and melanopsin to G(i/o) and G(q) signal transduction in Caenorhabditis elegans. FASEB J. 2012, 26, 480–491. [Google Scholar] [CrossRef] [PubMed]
- Pierce, K.L.; Premont, R.T.; Lefkowitz, R.J. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 2002, 3, 639–650. [Google Scholar] [CrossRef]
- Lagerström, M.C.; Schiöth, H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008, 7, 339–357. [Google Scholar] [CrossRef]
- Xiao, R.; Zhang, B.; Dong, Y.; Gong, J.; Xu, T.; Liu, J.; Xu, X.Z.S. A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell 2013, 152, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-J.; Xue, J.; Zhuo, J.-C.; Cheng, R.-L.; Xu, H.-J.; Zhang, C.-X. Comparative analysis of the transcriptional responses to low and high temperatures in three rice planthopper species. Mol. Ecol. 2017, 26, 2726–2737. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Hu, P.; Wang, T.; Tao, J.; Zong, S. Differential transcriptome analysis reveals genes related to cold tolerance in seabuckthorn carpenter moth, Eogystia hippophaecolus. PLoS ONE 2017, 12, e0187105. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.; Robertson, H.M.; Feder, J.L.; Varala, K.; Hudson, M.E.; Ragland, G.J.; Hahn, D.A.; Berlocher, S.H. Sympatric ecological speciation meets pyrosequencing: sampling the transcriptome of the apple maggot Rhagoletis pomonella. BMC Genom. 2009, 10, 633. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Neal, S.J.; Robertson, R.M.; Westwood, J.T.; Walker, V.K. Cold hardening and transcriptional change in Drosophila melanogaster. Insect Mol. Boil. 2005, 14, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Laayouni, H.; García-Franco, F.; Chávez-Sandoval, B.E.; Trotta, V.; Beltran, S.; Corominas, M.; Santos, M. Thermal evolution of gene expression profiles in Drosophila subobscura. BMC Evol. Boil. 2007, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, J.G.; Nielsen, M.M.; Loeschcke, V. Gene expression profile analysis of Drosophila melanogaster selected for resistance to environmental stressors. J. Evol. Boil. 2007, 20, 1624–1636. [Google Scholar] [CrossRef]
- Ohta, A.; Kuhara, A. Molecular mechanism for trimetric G protein-coupled thermosensation and synaptic regulation in the temperature response circuit of Caenorhabditis elegans. Neurosci. Res. 2013, 76, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Berchet, V.; Thomas, T.; Cavicchioli, R.; Russell, N.J.; Gounot, A. Structural analysis of the elongation factor G protein from the low-temperature-adapted bacterium Arthrobacter globiformis SI55. Extremophiles 2000, 4, 123–130. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 Confers Chilling Tolerance in Rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell 2002, 14, s165–s183. [Google Scholar] [CrossRef]
- Siebenaller, J.F.; Garrett, D.J. The effects of the deep-sea environment on transmembrane signaling. Comp. Biochem. Physiol. Part B Biochem. Mol. Boil. 2002, 131, 675–694. [Google Scholar] [CrossRef]
- Li, L.; Zhang, X. Differential inhibition of the TRPM8 ion channel by Gαq and Gα11. Channels 2013, 7, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Clapham, D.E. TRP channels as cellular sensors. Nature 2003, 426, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Bandell, M.; Story, G.M.; Hwang, S.W.; Viswanath, V.; Eid, S.R.; Petrus, M.J.; Earley, T.J.; Patapoutian, A. Noxious cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron 2004, 41, 849–857. [Google Scholar] [CrossRef]
- De Luca, F.; Di Vito, M.; Fanelli, E.; Reyes, A.; Greco, N.; De Giorgi, C. Characterization of the heat shock protein 90 gene in the plant parasitic nematode Meloidogyne artiellia and its expression as related to different developmental stages and temperature. Gene 2009, 440, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.D.; Caron, A.W.; Bourget, L.; Denis-Larose, C.; Massie, B. Role of the human heat shock protein hsp70 in protection against stress-induced apoptosis. Mol. Cell. Boil. 1997, 17, 5317–5327. [Google Scholar] [CrossRef]
- Wentz, J.M.; Mendenhall, A.R.; Bortz, D.M. Pattern Formation in the Longevity-Related Expression of Heat Shock Protein-16.2 in Caenorhabditis elegans. Bull. Math. Boil. 2018, 80, 2669–2697. [Google Scholar] [CrossRef]
- Brunquell, J.; Morris, S.; Lu, Y.; Cheng, F.; Westerheide, S.D. The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genom. 2016, 17, 559. [Google Scholar] [CrossRef]
- Xu, X.-L.; Wu, X.-Q.; Ye, J.-R.; Huang, L. Molecular Characterization and Functional Analysis of Three Pathogenesis-Related Cytochrome P450 Genes from Bursaphelenchus xylophilus (Tylenchida: Aphelenchoidoidea). Int. J. Mol. Sci. 2015, 16, 5216–5234. [Google Scholar] [CrossRef]
- Jia, K.; Albert, P.S.; Riddle, D.L. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development 2002, 129, 221–231. [Google Scholar]
- Mak, H.Y.; Ruvkun, G. Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development 2004, 131, 1777–1786. [Google Scholar] [CrossRef]
- Chatzigeorgiou, M.; Yoo, S.; Watson, J.D.; Lee, W.-H.; Spencer, W.C.; Kindt, K.S.; Hwang, S.W.; Miller, D.M., 3rd; Treinin, M.; Driscoll, M.; et al. Specific roles for DEG/ENaC and TRP channels in touch and thermosensation in C. elegans nociceptors. Nat. Neurosci. 2010, 13, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xiao, R.; Ronan, E.A.; He, Y.; Hsu, A.-L.; Liu, J.; Xu, X.S. Environmental temperature differentially modulates C. elegans longevity through a thermosensitive TRP channel. Cell Rep. 2015, 11, 1414–1424. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xiao, Y.; Ji, X.-L.; Zhang, K.-Q.; Zou, C.-G. The cAMP-PKA pathway-mediated fat mobilization is required for cold tolerance in C. elegans. Sci. Rep. 2017, 7, 638. [Google Scholar] [CrossRef] [PubMed]
- Svensk, E.; Stahlman, M.; Andersson, C.H.; Johansson, M.; Boren, J.; Pilon, M. PAQR-2 regulates fatty acid desaturation during cold adaptation in C. elegans. PLoS Genet. 2013, 9, e1003801. [Google Scholar] [CrossRef] [PubMed]
- Brock, T.J.; Browse, J.; Watts, J.L. Fatty Acid Desaturation and the Regulation of Adiposity in Caenorhabditis elegans. Genetics 2007, 176, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Boil. 2013, 14, R36. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Trzaskowski, B.; Latek, D.; Yuan, S.; Ghoshdastider, U.; Debinski, A.; Filipek, S. Action of Molecular Switches in GPCRs—Theoretical and Experimental Studies. Curr. Med. Chem. 2012, 19, 1090–1109. [Google Scholar] [CrossRef]
- Venkatakrishnan, A.J.; Deupi, X.; Lebon, G.; Tate, C.G.; Schertler, G.F.; Babu, M.M. Molecular signatures of G-protein-coupled receptors. Nature 2013, 494, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Z.; Li, D.; Chen, Q. Identification and Characterization of a Bursaphelenchus xylophilus (Aphelenchida: Aphelenchoididae) Thermotolerance-Related Gene: Bx-HSP90. Int. J. Mol. Sci. 2012, 13, 8819–8833. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Li, D.; Wang, F.; Zhang, R.; Ling, Y. Trehalose metabolism genes of Aphelenchoides besseyi (Nematoda: Aphelenchoididae) in hypertonic osmotic pressure survival. Boil. Open 2017, 6, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, F.; Li, D.; Zhang, R.; Ling, Y. Trehalose metabolism genes render rice white tip nematode Aphelenchoides besseyi (Nematoda: Aphelenchoididae) resistant to an anaerobic environment. J. Exp. Biol. 2018, 221, jeb171413. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Q.; Zhou, X. A 2-Cys peroxiredoxin in response to oxidative stress in the pine wood nematode, Bursaphelenchus xylophilus. Sci. Rep. 2016, 6, 27438. [Google Scholar] [CrossRef] [PubMed]
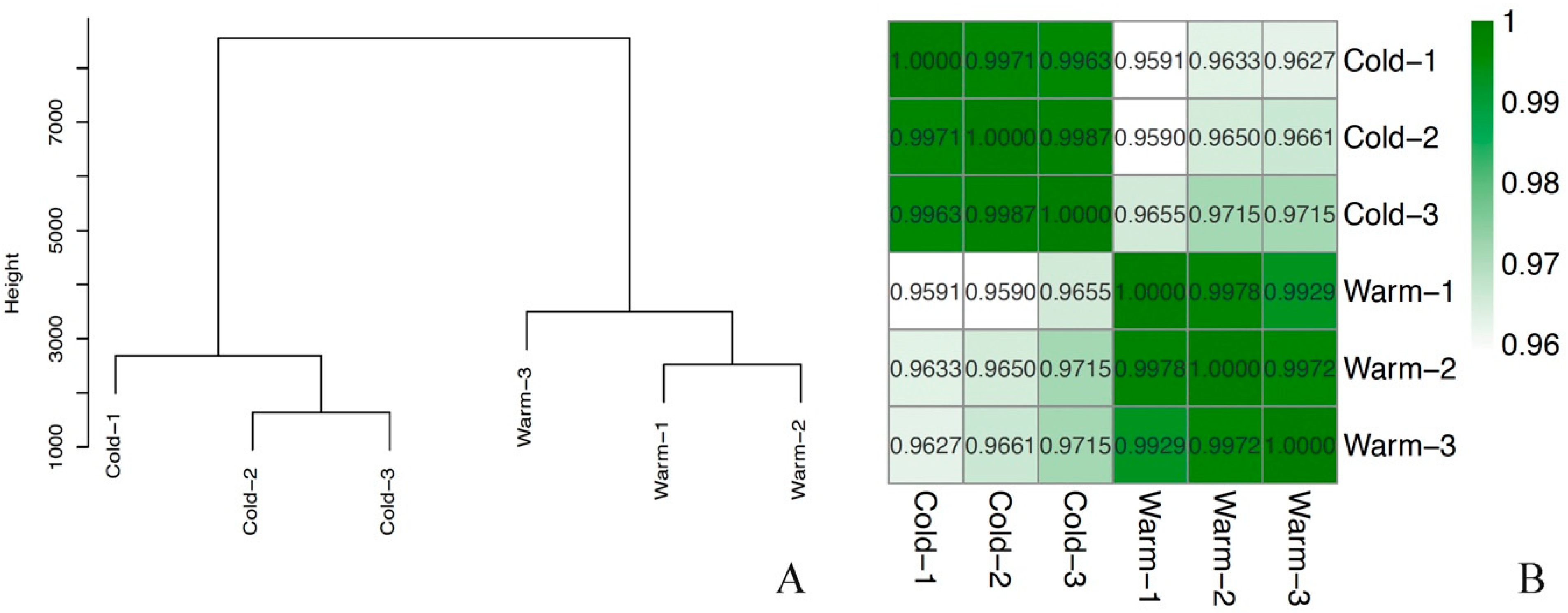
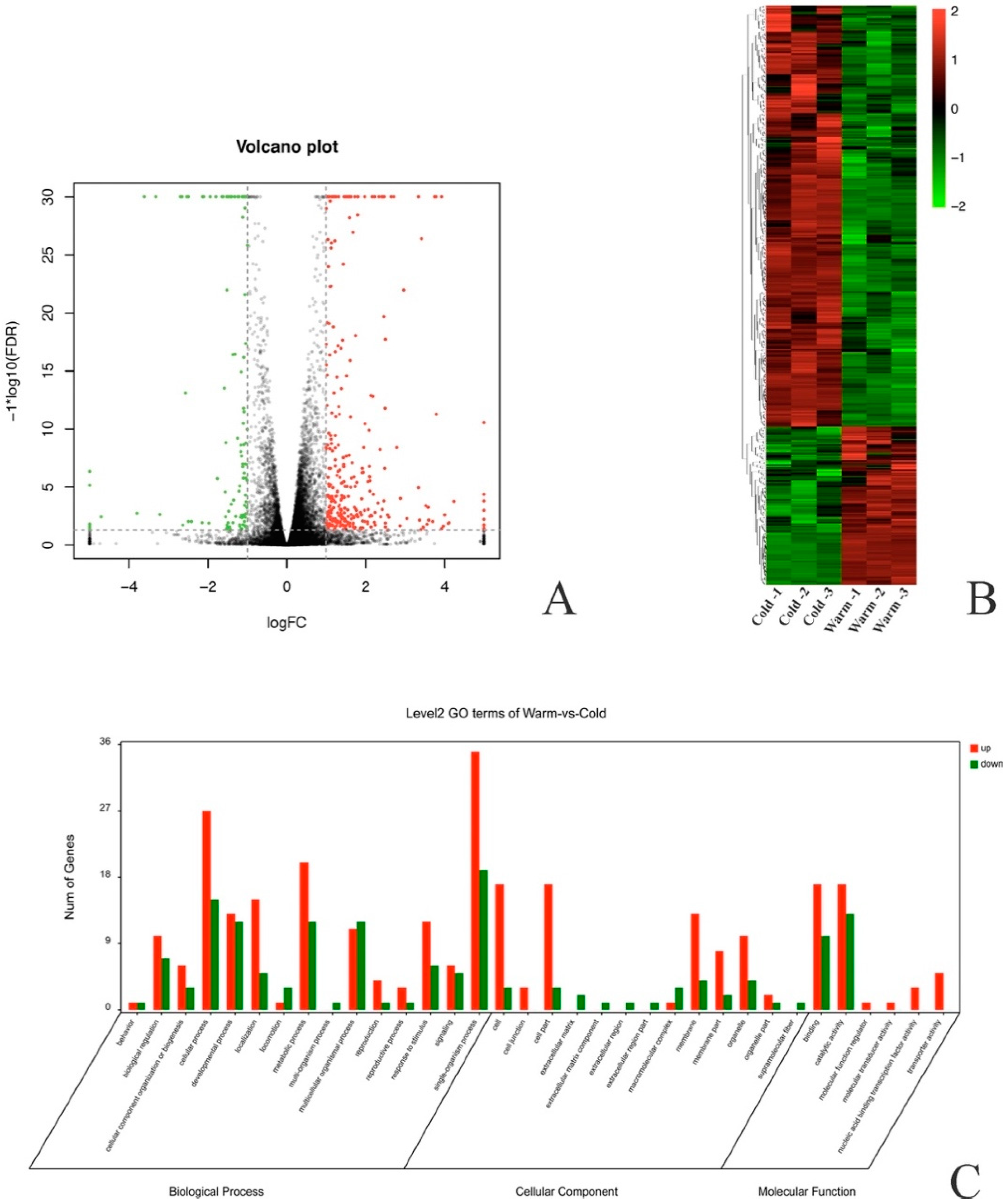
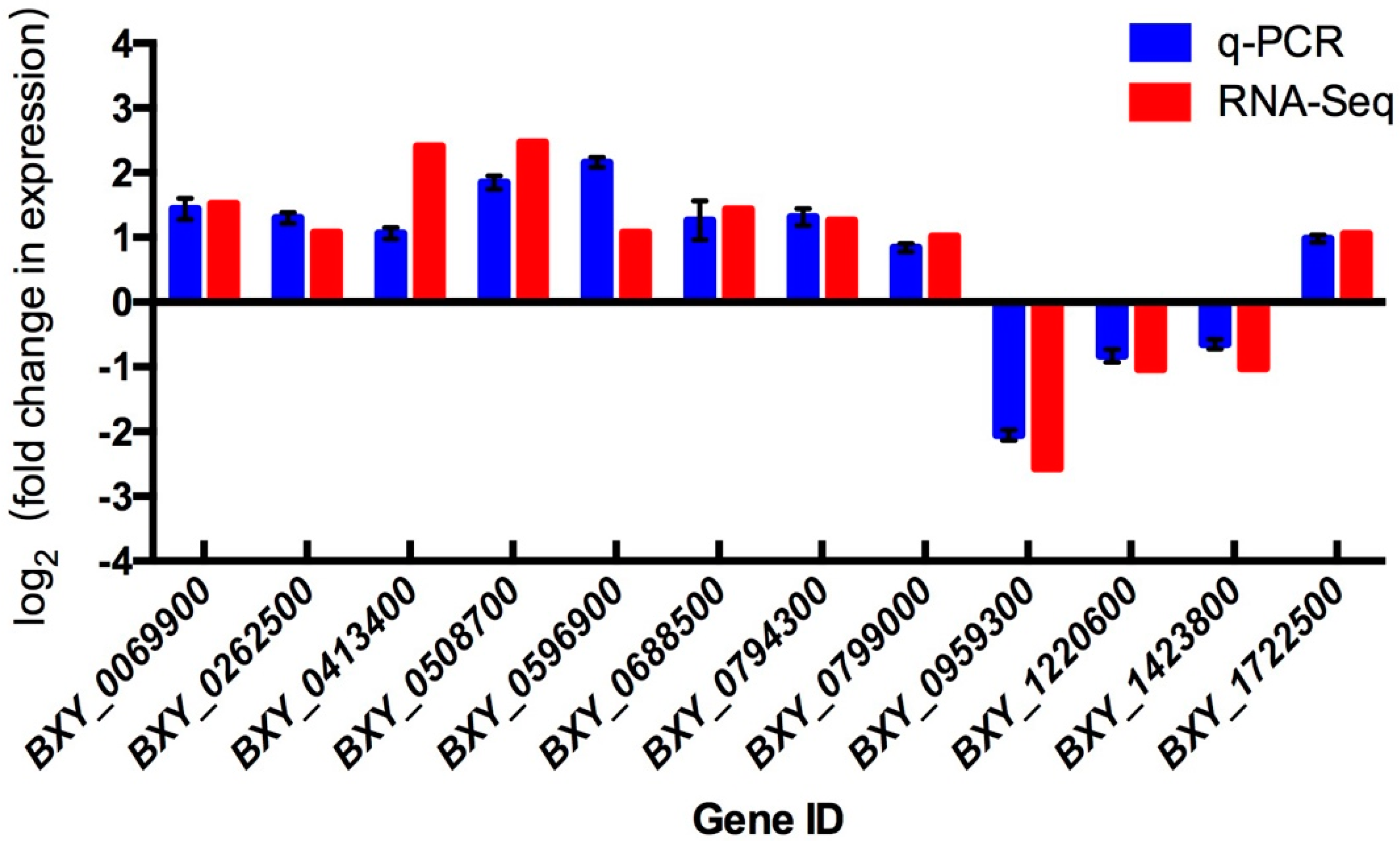
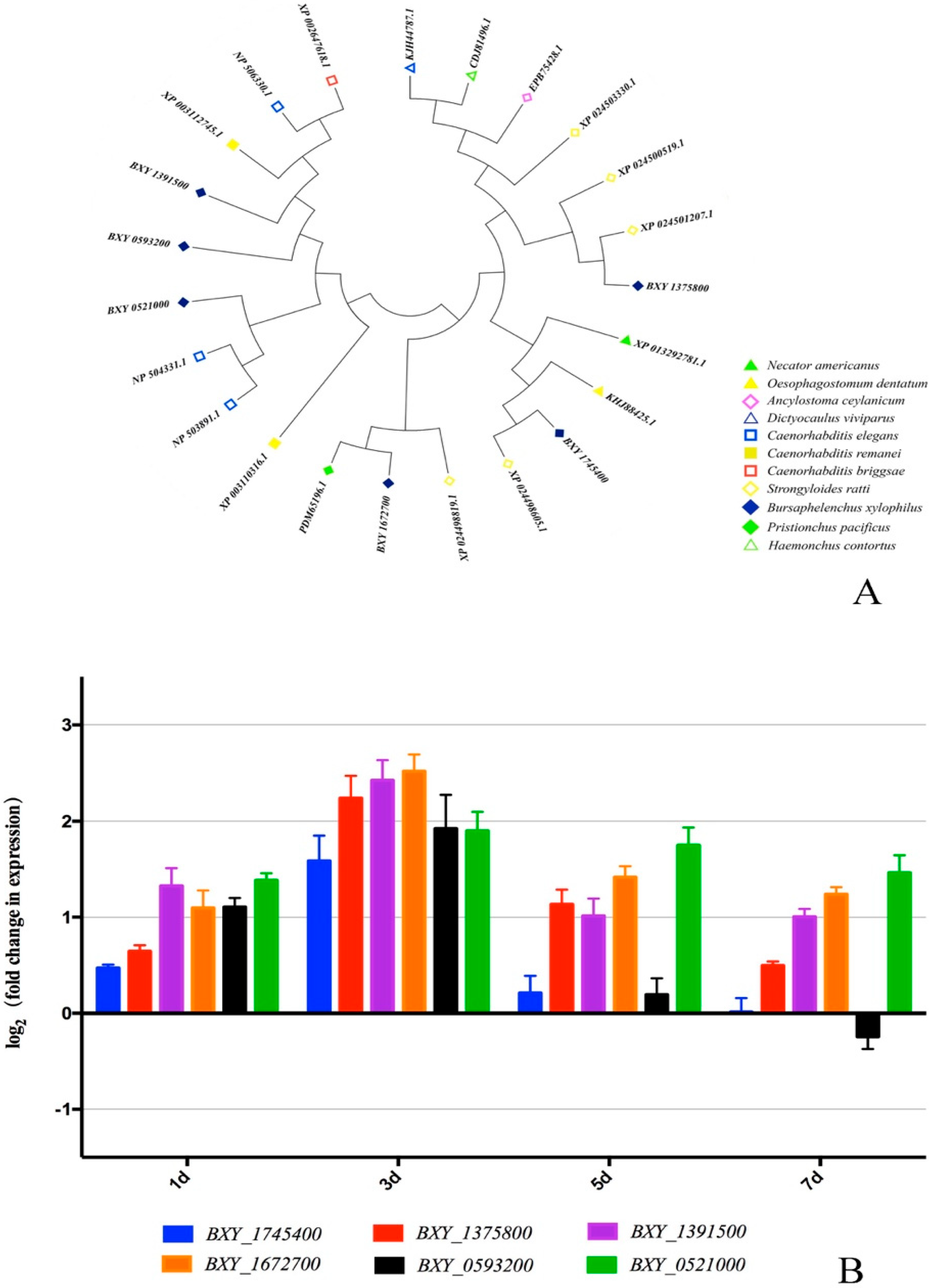

| Samples | Clean Reads | High Quality Clean Reads (%) | High Quality Clean Bases | Q20 (%) | Q30 (%) | GC (%) | Mapping Rate |
|---|---|---|---|---|---|---|---|
| Cold-1 | 51,107,560 | 47,519,530 (92.98%) | 6,850,534,848 | 95.08% | 86.34% | 48.70% | 75.24% |
| Cold-2 | 47,251,708 | 43,777,894 (92.65%) | 6,230,763,134 | 94.72% | 85.36% | 48.35% | 73.29% |
| Cold-3 | 43,275,876 | 40,225,612 (92.95%) | 5,760,600,645 | 94.92% | 85.93% | 48.62% | 75.36% |
| Warm-1 | 46,243,122 | 42,555,170 (92.02%) | 6,101,120,213 | 94.74% | 85.43% | 48.87% | 75.61% |
| Warm-2 | 45,717,826 | 42,541,694 (93.05%) | 6,065,096,300 | 94.87% | 85.83% | 48.74% | 75.56% |
| Warm-3 | 98,615,884 | 88,413,574 (89.65%) | 12,105,837,900 | 93.42% | 82.30% | 48.27% | 70.47% |
| Total | 332,211,976 | 305,033,474 | 43,113,953,040 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Hao, X.; Xu, J.; Ma, Y.; Ma, L. Transcriptome-Based Analysis Reveals a Crucial Role of BxGPCR17454 in Low Temperature Response of Pine Wood Nematode (Bursaphelenchus xylophilus). Int. J. Mol. Sci. 2019, 20, 2898. https://doi.org/10.3390/ijms20122898
Wang B, Hao X, Xu J, Ma Y, Ma L. Transcriptome-Based Analysis Reveals a Crucial Role of BxGPCR17454 in Low Temperature Response of Pine Wood Nematode (Bursaphelenchus xylophilus). International Journal of Molecular Sciences. 2019; 20(12):2898. https://doi.org/10.3390/ijms20122898
Chicago/Turabian StyleWang, Bowen, Xin Hao, Jiayao Xu, Yan Ma, and Ling Ma. 2019. "Transcriptome-Based Analysis Reveals a Crucial Role of BxGPCR17454 in Low Temperature Response of Pine Wood Nematode (Bursaphelenchus xylophilus)" International Journal of Molecular Sciences 20, no. 12: 2898. https://doi.org/10.3390/ijms20122898
APA StyleWang, B., Hao, X., Xu, J., Ma, Y., & Ma, L. (2019). Transcriptome-Based Analysis Reveals a Crucial Role of BxGPCR17454 in Low Temperature Response of Pine Wood Nematode (Bursaphelenchus xylophilus). International Journal of Molecular Sciences, 20(12), 2898. https://doi.org/10.3390/ijms20122898





