Deep Insight into the Transcriptome of the Single Silk Gland of Bombyx mori
Abstract
:1. Introduction
2. Results
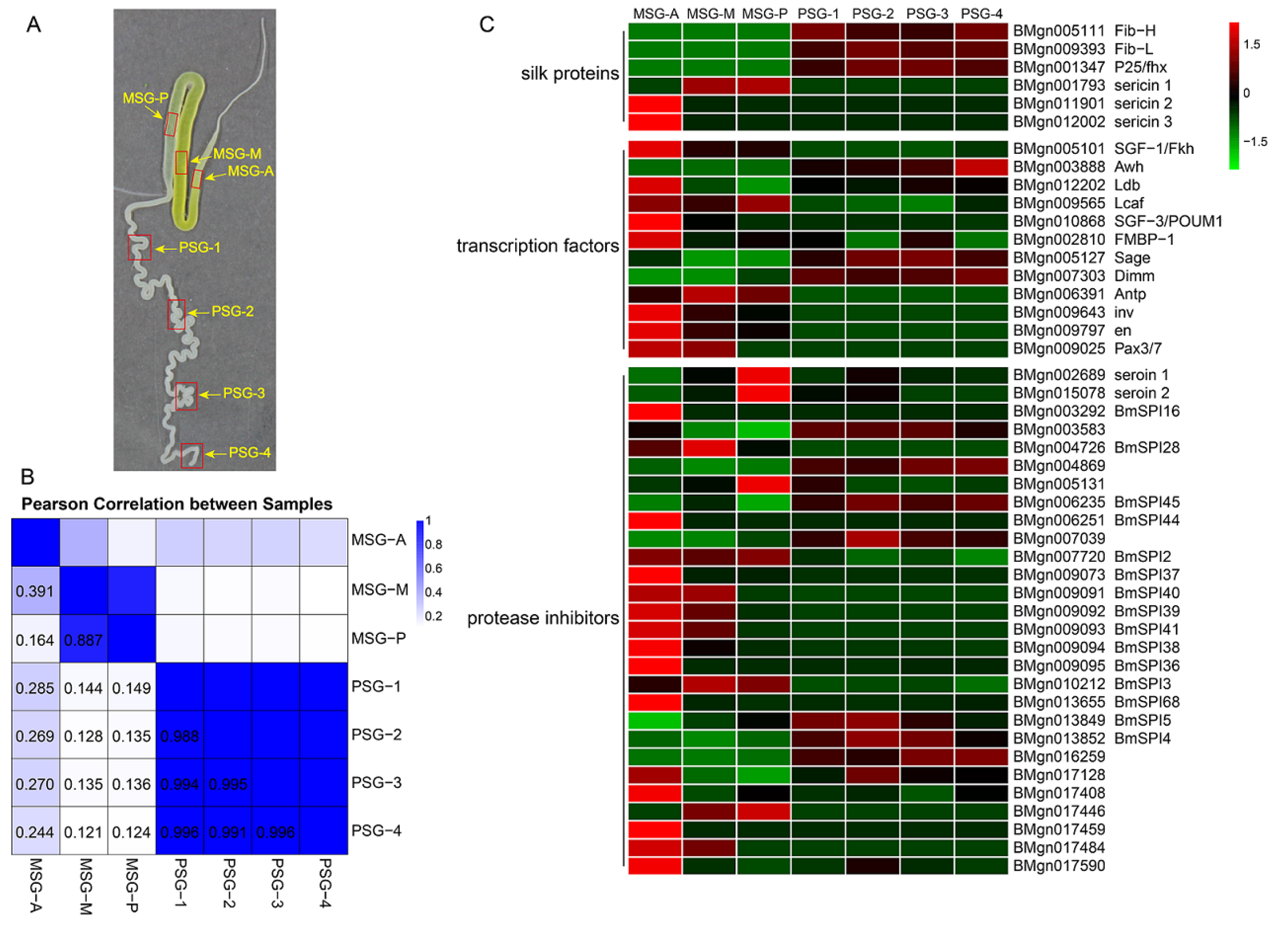
2.1. Overall Structure and Analysis of the Transcriptome Data
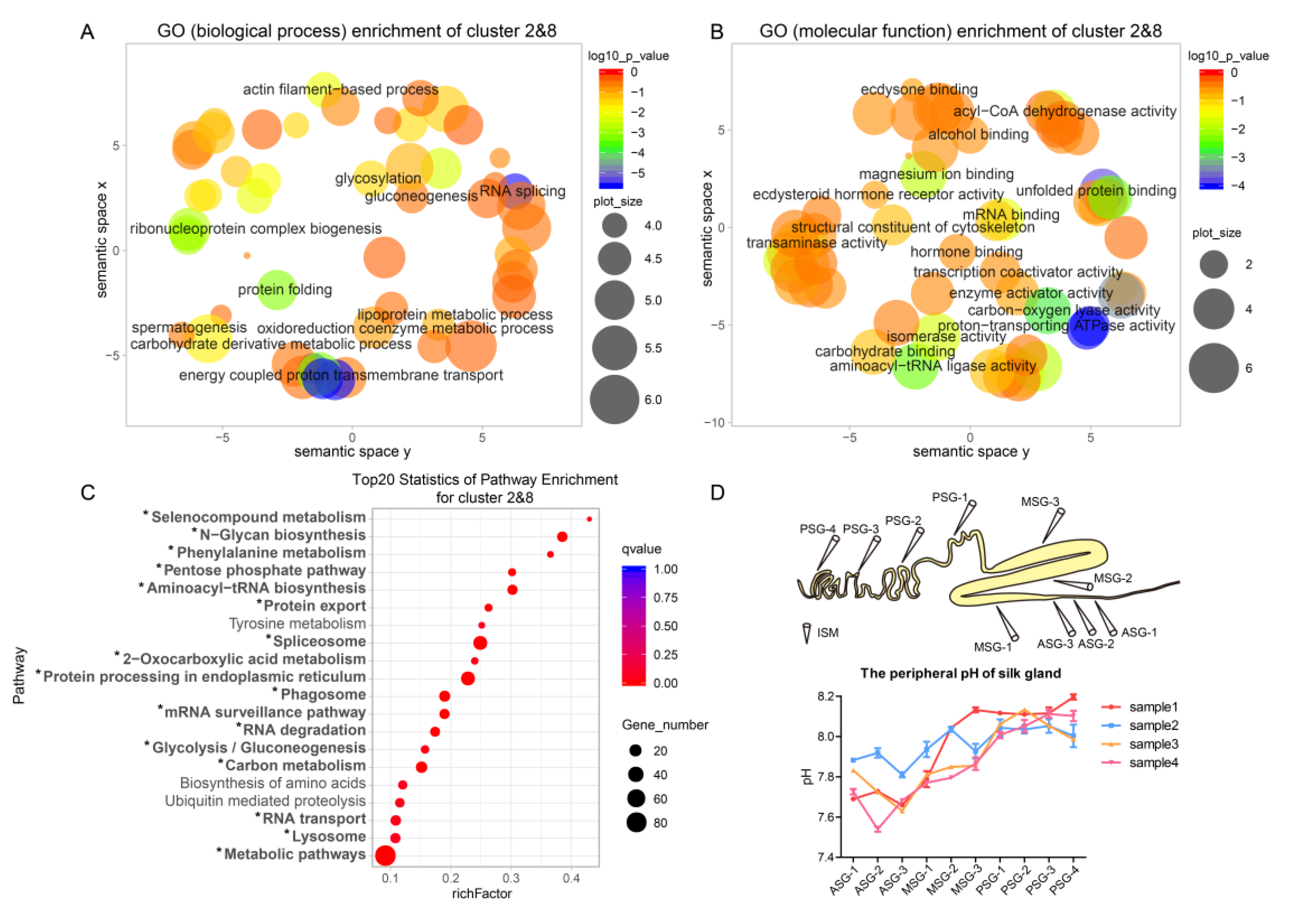
2.2. Divergent Functions in Each Segment are Related to Silk Protein Synthesis
2.3. The MSG is Functionally More Divergent than the PSG
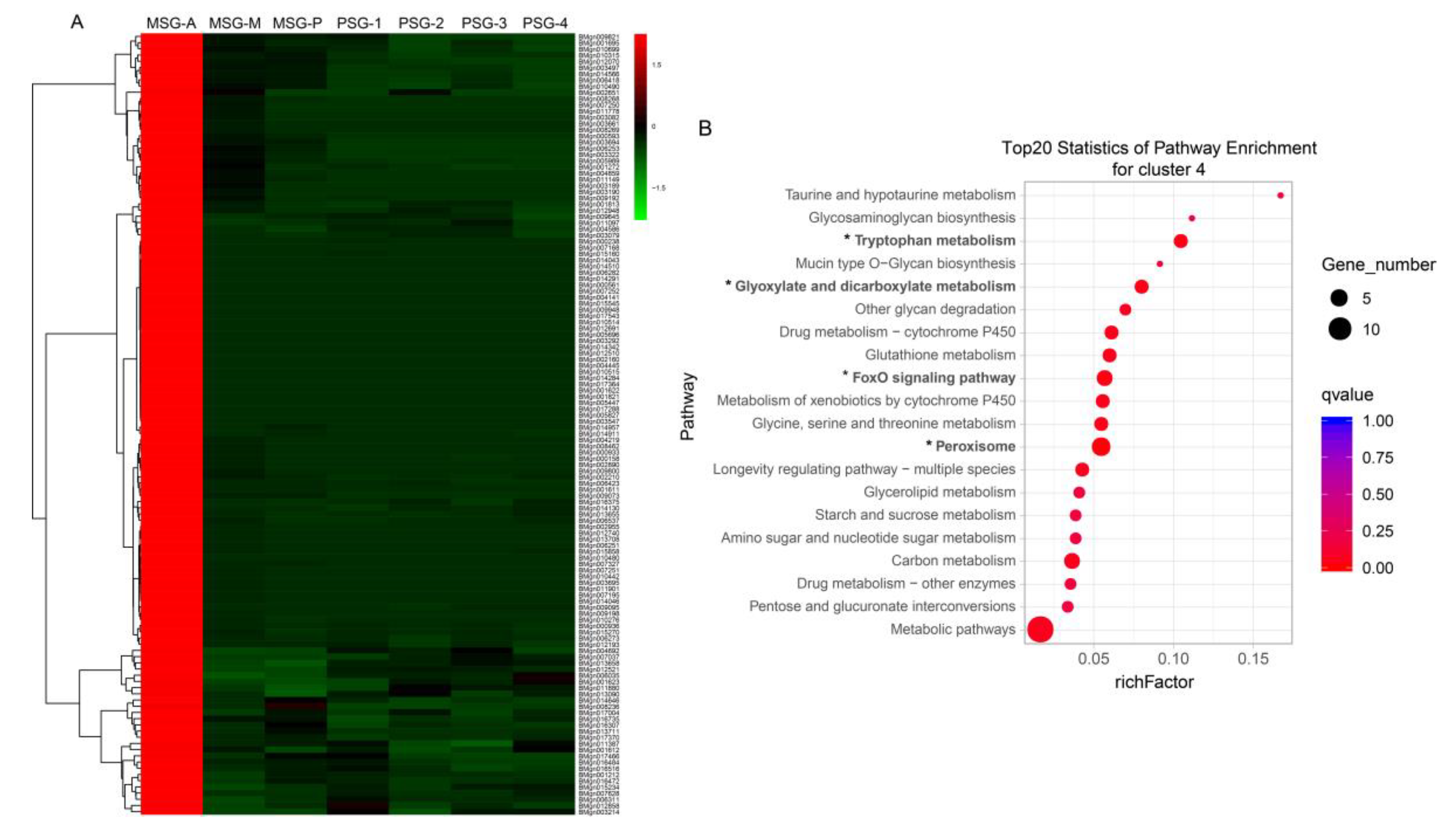
2.4. Specific Genes Contribute to the Distinct Functions of the Anterior MSG
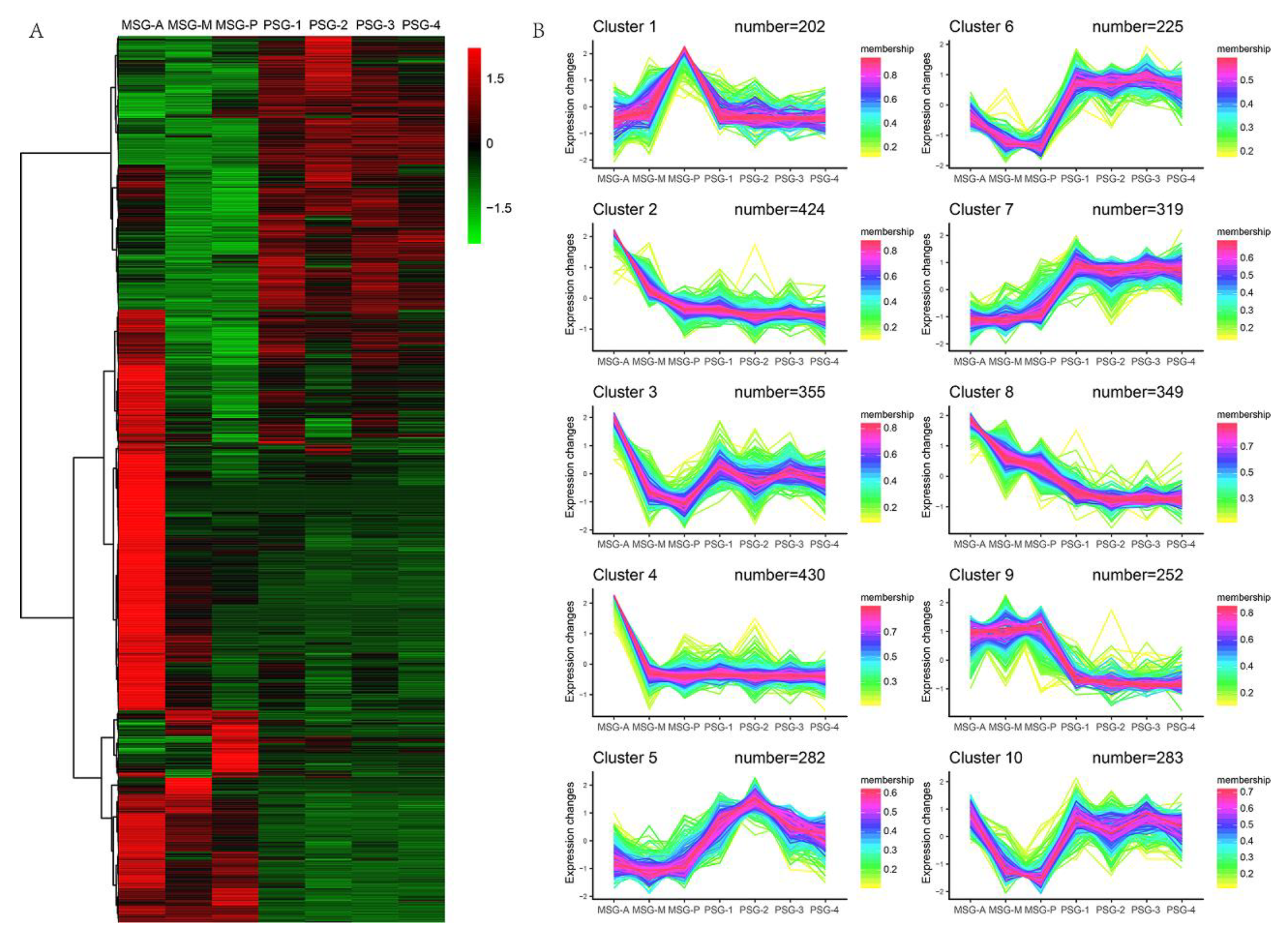
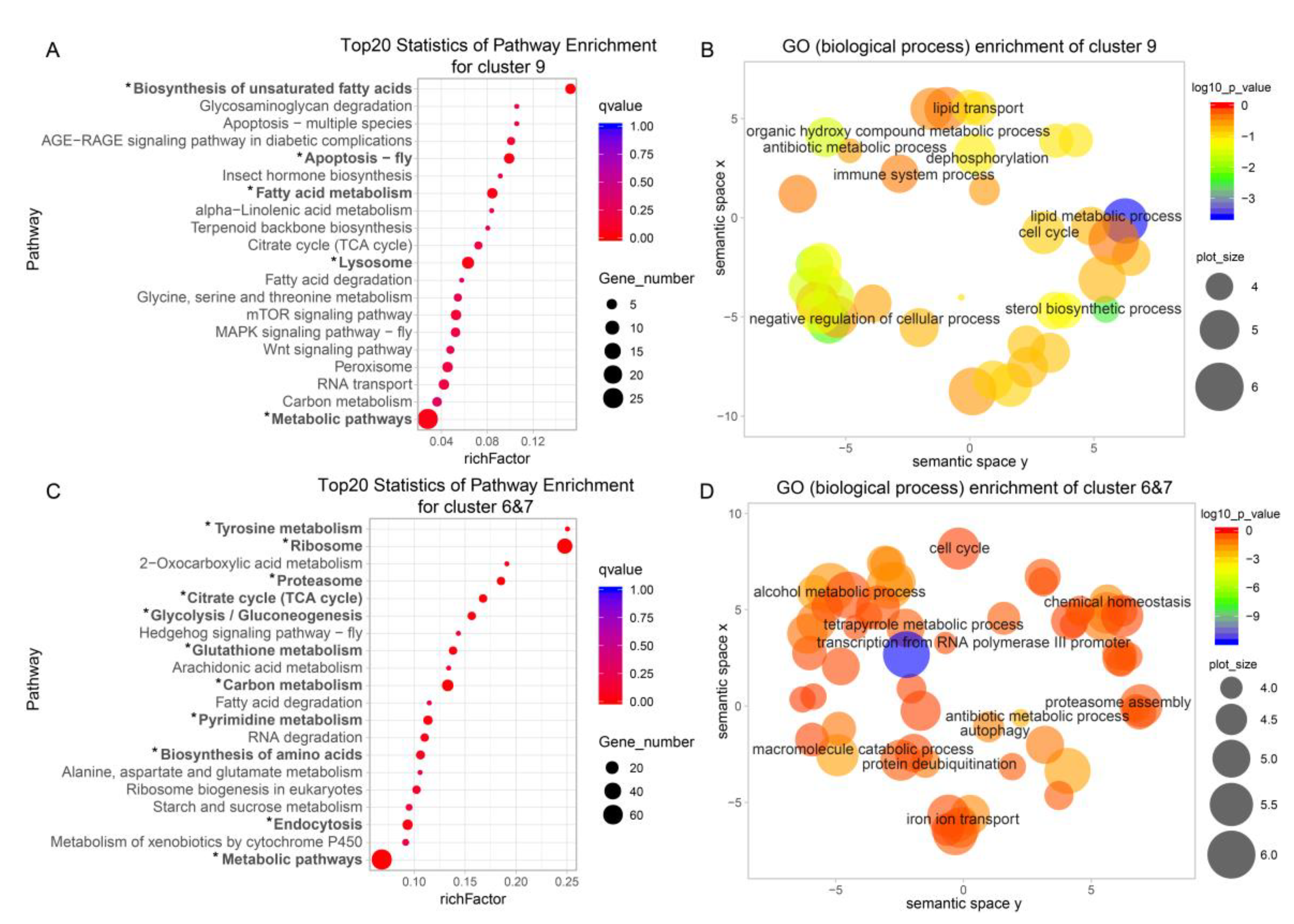
2.5. Gradually Upregulated Genes Contribute to Silk Protein Storage and Silk Formation
3. Discussion
4. Materials and Methods
4.1. Silkworm
4.2. Silk Gland Dissection and Sample Collection
4.3. RNA Preparation and Transcriptome Sequencing
4.4. Sequencing Data Processing
4.5. Differential Expression Analysis
4.6. Quantitative Reverse Transcription PCR (qRT-PCR)
4.7. pH Measurement of the Silk Gland Peripheral
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ASG | Anterior silk gland |
| DEG | Differentially expressed gene |
| FDR | False discovery rate |
| MSG | Middle silk gland |
| PSG | Posterior silk gland |
References
- Sutherland, T.D.; Young, J.H.; Weisman, S.; Hayashi, C.Y.; Merritt, D.J. Insect silk: One name, many materials. Annu. Rev. Entomol. 2010, 55, 171–188. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Li, S.; Feng, Q. Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu. Rev. Entomol. 2014, 59, 513–536. [Google Scholar] [CrossRef]
- Ma, S.; Shi, R.; Wang, X.; Liu, Y.; Chang, J.; Gao, J.; Lu, W.; Zhang, J.; Zhao, P.; Xia, Q. Genome editing of BmFib-H gene provides an empty Bombyx mori silk gland for a highly efficient bioreactor. Sci. Rep. 2014, 4, 6867. [Google Scholar] [CrossRef]
- Lin, Z.; Huang, W.; Zhang, J.; Fan, J.S.; Yang, D. Solution structure of eggcase silk protein and its implications for silk fiber formation. Proc. Natl. Acad. Sci. USA 2009, 106, 8906–8911. [Google Scholar] [CrossRef] [PubMed]
- Gamo, T.; Inokuchi, T.; Laufer, H. Polypeptides of fibroin and sericin secreted from the different sections of the silk gland in Bombyx mori. Insect Biochem. 1977, 7, 285–295. [Google Scholar] [CrossRef]
- Inoue, S.; Tanaka, K.; Arisaka, F.; Kimura, S.; Ohtomo, K.; Mizuno, S. Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain, and P25, with a 6:6:1 molar ratio. J. Biol. Chem. 2000, 275, 40517–40528. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Mori, K.; Mizuno, S. Immunological identification of the major disulfide-linked light component of silk fibroin. J. Biochem. 1993, 114, 1–4. [Google Scholar] [CrossRef]
- Takasu, Y.; Hata, T.; Uchino, K.; Zhang, Q. Identification of ser2 proteins as major sericin components in the non-cocoon silk of Bombyx mori. Insect Biochem. Mol. Biol. 2010, 40, 339–344. [Google Scholar] [CrossRef]
- Takasu, Y.; Yamada, H.; Tsubouchi, K. Isolation of three main sericin components from the cocoon of the silkworm, Bombyx mori. Biosci. Biotechnol. Biochem. 2002, 66, 2715–2718. [Google Scholar] [CrossRef]
- Shao, Z.; Vollrath, F. Surprising strength of silkworm silk. Nature 2002, 418, 741. [Google Scholar] [CrossRef] [PubMed]
- He, Y.X.; Zhang, N.N.; Li, W.F.; Jia, N.; Chen, B.Y.; Zhou, K.; Zhang, J.; Chen, Y.; Zhou, C.Z. N-terminal domain of Bombyx mori fibroin mediates the assembly of silk in response to pH decrease. J. Mol. Biol. 2012, 418, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Greving, I.; Cai, M.; Vollrath, F.; Schniepp, H.C. Shear-induced self-assembly of native silk proteins into fibrils studied by atomic force microscopy. Biomacromolecules 2012, 13, 676–682. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, L.; Guo, X.; Guo, T.; Wang, S.; Lu, C. Analysis of tissue-specific region in sericin 1 gene promoter of Bombyx mori. Biochem. Biophys. Res. Commun. 2006, 342, 273–279. [Google Scholar] [CrossRef]
- Takasu, Y.; Yamada, H.; Tamura, T.; Sezutsu, H.; Mita, K.; Tsubouchi, K. Identification and characterization of a novel sericin gene expressed in the anterior middle silk gland of the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2007, 37, 1234–1240. [Google Scholar] [CrossRef]
- Kludkiewicz, B.; Takasu, Y.; Fedic, R.; Tamura, T.; Sehnal, F.; Zurovec, M. Structure and expression of the silk adhesive protein ser2 in Bombyx mori. Insect Biochem. Mol. Biol. 2009, 39, 938–946. [Google Scholar] [CrossRef]
- Dong, Z.; Zhao, P.; Zhang, Y.; Song, Q.; Zhang, X.; Guo, P.; Wang, D.; Xia, Q. Analysis of proteome dynamics inside the silk gland lumen of Bombyx mori. Sci. Rep. 2016, 6, 21158. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, P.; Dong, Z.; Wang, D.; Guo, P.; Guo, X.; Song, Q.; Zhang, W.; Xia, Q. Comparative proteome analysis of multi-layer cocoon of the silkworm, Bombyx mori. PLoS ONE 2015, 10, e0123403. [Google Scholar] [CrossRef]
- Chang, H.; Cheng, T.; Wu, Y.; Hu, W.; Long, R.; Liu, C.; Zhao, P.; Xia, Q. Transcriptomic analysis of the anterior silk gland in the domestic silkworm (Bombyx mori) - insight into the mechanism of silk formation and spinning. PLoS ONE 2015, 10, e0139424. [Google Scholar] [CrossRef]
- Li, J.Y.; Ye, L.P.; Che, J.Q.; Song, J.; You, Z.Y.; Yun, K.C.; Wang, S.H.; Zhong, B.X. Comparative proteomic analysis of the silkworm middle silk gland reveals the importance of ribosome biogenesis in silk protein production. J. Proteom. 2015, 126, 109–120. [Google Scholar] [CrossRef]
- Dong, Z.; Zhao, P.; Wang, C.; Zhang, Y.; Chen, J.; Wang, X.; Lin, Y.; Xia, Q. Comparative proteomics reveal diverse functions and dynamic changes of Bombyx mori silk proteins spun from different development stages. J. Proteome Res. 2013, 12, 5213–5222. [Google Scholar] [CrossRef]
- Kimoto, M.; Kitagawa, T.; Kobayashi, I.; Nakata, T.; Kuroiwa, A.; Takiya, S. Inhibition of the binding of msg-intermolt-specific complex, MIC, to the sericin-1 gene promoter and sericin-1 gene expression by POU-M1/SGF-3. Dev. Genes Evol. 2012, 222, 351–359. [Google Scholar] [CrossRef]
- Ohno, K.; Sawada, J.; Takiya, S.; Kimoto, M.; Matsumoto, A.; Tsubota, T.; Uchino, K.; Hui, C.C.; Sezutsu, H.; Handa, H.; et al. Silk gland factor-2, involved in fibroin gene transcription, consists of LIM homeodomain, LIM-interacting, and single-stranded DNA-binding proteins. J. Biol. Chem. 2013, 288, 31581–31591. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Zhao, P.; Dong, Z.; Duan, J.; Wang, G.; Wang, L.; Li, Y.; Xiang, Z.; Xia, Q. Genome-wide identification and immune response analysis of serine protease inhibitor genes in the silkworm, Bombyx mori. PLoS ONE 2012, 7, e31168. [Google Scholar] [CrossRef]
- Sun, W.; Shen, Y.H.; Qi, D.W.; Xiang, Z.H.; Zhang, Z. Molecular cloning and characterization of ecdysone oxidase and 3-dehydroecdysone-3alpha-reductase involved in the ecdysone inactivation pathway of silkworm, Bombyx mori. Int. J. Biol. Sci. 2012, 8, 125–138. [Google Scholar] [CrossRef]
- Azuma, M.; Ohta, Y. Changes in H+-translocating vacuolar-type ATPase in the anterior silk gland cell of Bombyx mori during metamorphosis. J. Exp. Biol. 1998, 201, 479–486. [Google Scholar]
- Goo, T.W.; Park, S.; Jin, B.R.; Yun, E.Y.; Kim, I.; Nho, S.K.; Kang, S.W.; Kwon, O.Y. Endoplasmic reticulum stress response of Bombyx mori calreticulin. Mol. Biol. Rep. 2005, 32, 133–139. [Google Scholar] [CrossRef]
- Hou, Y.; Xia, Q.; Zhao, P.; Zou, Y.; Liu, H.; Guan, J.; Gong, J.; Xiang, Z. Studies on middle and posterior silk glands of silkworm (Bombyx mori) using two-dimensional electrophoresis and mass spectrometry. Insect Biochem. Mol. Biol. 2007, 37, 486–496. [Google Scholar] [CrossRef]
- Mach, V.; Takiya, S.; Ohno, K.; Handa, H.; Imai, T.; Suzuki, Y. Silk gland factor-1 involved in the regulation of Bombyx sericin-1 gene contains fork head motif. J. Biol. Chem. 1995, 270, 9340–9346. [Google Scholar] [CrossRef]
- Julien, E.; Bordeaux, M.C.; Garel, A.; Couble, P. Fork head alternative binding drives stage-specific gene expression in the silk gland of Bombyx mori. Insect Biochem. Mol. Biol. 2002, 32, 377–387. [Google Scholar] [CrossRef]
- Kimoto, M.; Tsubota, T.; Uchino, K.; Sezutsu, H.; Takiya, S. LIM-homeodomain transcription factor Awh is a key component activating all three fibroin genes, fibH, fibL and fhx, in the silk gland of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2015, 56, 29–35. [Google Scholar] [CrossRef]
- Takiya, S.; Kokubo, H.; Suzuki, Y. Transcriptional regulatory elements in the upstream and intron of the fibroin gene bind three specific factors POU-M1, Bm Fkh and FMBP-1. Biochem J. 1997, 321, 645–653. [Google Scholar] [CrossRef]
- Zhao, X.M.; Liu, C.; Li, Q.Y.; Hu, W.B.; Zhou, M.T.; Nie, H.Y.; Zhang, Y.X.; Peng, Z.C.; Zhao, P.; Xia, Q.Y. Basic helix-loop-helix transcription factor Bmsage is involved in regulation of fibroin h-chain gene via interaction with sgf1 in Bombyx mori. PLoS ONE 2014, 9, e94091. [Google Scholar] [CrossRef]
- Zhao, X.M.; Liu, C.; Jiang, L.J.; Li, Q.Y.; Zhou, M.T.; Cheng, T.C.; Mita, K.; Xia, Q.Y. A juvenile hormone transcription factor Bmdimm-fibroin H chain pathway is involved in the synthesis of silk protein in silkworm, Bombyx mori. J. Biol. Chem. 2015, 290, 972–986. [Google Scholar] [CrossRef]
- Hu, W.; Liu, C.; Cheng, T.; Li, W.; Wang, N.; Xia, Q. Histomorphometric and transcriptomic features characterize silk glands’ development during the molt to intermolt transition process in silkworm. Insect Biochem. Mol. Biol. 2016, 76, 95–108. [Google Scholar] [CrossRef]
- Kimoto, M.; Tsubota, T.; Uchino, K.; Sezutsu, H.; Takiya, S. Hox transcription factor Antp regulates sericin-1 gene expression in the terminal differentiated silk gland of Bombyx mori. Dev. Biol. 2014, 386, 64–71. [Google Scholar] [CrossRef]
- Hui, C.C.; Matsuno, K.; Ueno, K.; Suzuki, Y. Molecular characterization and silk gland expression of Bombyx engrailed and invected genes. Proc. Natl. Acad. Sci. USA 1992, 89, 167–171. [Google Scholar] [CrossRef]
- Zurovec, M.; Yang, C.; Kodrik, D.; Sehnal, F. Identification of a novel type of silk protein and regulation of its expression. J. Biol. Chem. 1998, 273, 15423–15428. [Google Scholar] [CrossRef]
- Singh, C.P.; Vaishna, R.L.; Kakkar, A.; Arunkumar, K.P.; Nagaraju, J. Characterization of antiviral and antibacterial activity of Bombyx mori seroin proteins. Cell Microbiol. 2014, 16, 1354–1365. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, P.; Liu, S.; Dong, Z.; Chen, J.; Xiang, Z.; Xia, Q. A novel protease inhibitor in Bombyx mori is involved in defense against Beauveria bassiana. Insect Biochem. Mol. Biol. 2012, 42, 766–775. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, P.; Liu, H.; Guo, X.; He, H.; Zhu, R.; Xiang, Z.; Xia, Q. TIL-type protease inhibitors may be used as targeted resistance factors to enhance silkworm defenses against invasive fungi. Insect Biochem. Mol. Biol. 2015, 57, 11–19. [Google Scholar] [CrossRef]
- Guo, P.C.; Dong, Z.; Xiao, L.; Li, T.; Zhang, Y.; He, H.; Xia, Q.; Zhao, P. Silk gland-specific proteinase inhibitor serpin16 from the Bombyx mori shows cysteine proteinase inhibitory activity. Biochem. Biophys. Res. Commun. 2015, 457, 31–36. [Google Scholar] [CrossRef]
- Sakudoh, T.; Sezutsu, H.; Nakashima, T.; Kobayashi, I.; Fujimoto, H.; Uchino, K.; Banno, Y.; Iwano, H.; Maekawa, H.; Tamura, T.; et al. Carotenoid silk coloration is controlled by a carotenoid-binding protein, a product of the yellow blood gene. Proc. Natl. Acad. Sci. USA 2007, 104, 8941–8946. [Google Scholar] [CrossRef]
- Sakudoh, T.; Iizuka, T.; Narukawa, J.; Sezutsu, H.; Kobayashi, I.; Kuwazaki, S.; Banno, Y.; Kitamura, A.; Sugiyama, H.; Takada, N.; et al. A CD36-related transmembrane protein is coordinated with an intracellular lipid-binding protein in selective carotenoid transport for cocoon coloration. J. Biol. Chem. 2010, 285, 7739–7751. [Google Scholar] [CrossRef]
- Hossain, M.S.; Liu, Y.; Zhou, S.; Li, K.; Tian, L.; Li, S. 20-hydroxyecdysone-induced transcriptional activity of FoxO upregulates brummer and acid lipase-1 and promotes lipolysis in Bombyx fat body. Insect Biochem. Mol. Biol. 2013, 43, 829–838. [Google Scholar] [CrossRef]
- Yamamoto, K.; Banno, Y.; Fujii, H.; Miake, F.; Kashige, N.; Aso, Y. Catalase from the silkworm, Bombyx mori: Gene sequence, distribution, and overexpression. Insect Biochem. Mol. Biol. 2005, 35, 277–283. [Google Scholar] [CrossRef]
- Ai, J.; Zhu, Y.; Duan, J.; Yu, Q.; Zhang, G.; Wan, F.; Xiang, Z.H. Genome-wide analysis of cytochrome p450 monooxygenase genes in the silkworm, Bombyx mori. Gene 2011, 480, 42–50. [Google Scholar] [CrossRef]
- Niwa, R.; Matsuda, T.; Yoshiyama, T.; Namiki, T.; Mita, K.; Fujimoto, Y.; Kataoka, H. CYP306A1, a cytochrome p450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J. Biol. Chem. 2004, 279, 35942–35949. [Google Scholar] [CrossRef]
- Foo, C.W.P.; Bini, E.; Hensman, J.; Knight, D.P.; Lewis, R.V.; Kaplan, D.L. Role of pH and charge on silk protein assembly in insects and spiders. Appl. Phys. A 2006, 82, 223–233. [Google Scholar] [CrossRef]
- Andersson, M.; Chen, G.; Otikovs, M.; Landreh, M.; Nordling, K.; Kronqvist, N.; Westermark, P.; Jornvall, H.; Knight, S.; Ridderstrale, Y.; et al. Carbonic anhydrase generates CO2 and H+ that drive spider silk formation via opposite effects on the terminal domains. Plos Biol. 2014, 12, e1001921. [Google Scholar] [CrossRef]
- Domigan, L.J.; Andersson, M.; Alberti, K.A.; Chesler, M.; Xu, Q.; Johansson, J.; Rising, A.; Kaplan, D.L. Carbonic anhydrase generates a pH gradient in Bombyx mori silk glands. Insect Biochem. Mol. Biol. 2015, 65, 100–106. [Google Scholar] [CrossRef]
- Maxson, M.E.; Sergio, G. The vacuolar-type H+-ATPase at a glance - more than a proton pump. J. Cell Sci. 2014, 127, 4987–4993. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Peng, L.; Chen, H.; Xia, Q.; Zhao, P. Comparative transcriptome analysis of Bombyx mori spinnerets and filippi’s glands suggests their role in silk fiber formation. Insect Biochem. Mol. Biol. 2016, 68, 89–99. [Google Scholar] [CrossRef]
- Lu, P.; Xia, H.; Gao, L.; Pan, Y.; Wang, Y.; Cheng, X.; Lu, H.; Lin, F.; Chen, L.; Yao, Q.; et al. V-ATPase is involved in silkworm defense response against Bombyx mori nucleopolyhedrovirus. PLoS ONE 2013, 8, e64962. [Google Scholar]
- Michalak, M.; Corbett, E.F.; Mesaeli, N.; Nakamura, K.; Opas, M. Calreticulin: One protein, one gene, many functions. Biochem. J. 1999, 344, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Voeltz, G.K.; Rapoport, T.A. Rough sheets and smooth tubules. Cell 2006, 126, 435–439. [Google Scholar] [CrossRef]
- Wakefield, S.; Tear, G. The Drosophila reticulon, Rtnl-1, has multiple differentially expressed isoforms that are associated with a sub-compartment of the endoplasmic reticulum. Cell. Mol. Life Sci. Cmls 2006, 63, 2027–2038. [Google Scholar] [CrossRef] [PubMed]
- Macours, N.; Hens, K. Zinc-metalloproteases in insects: ACE and ECE. Insect Biochem. Mol. Biol. 2004, 34, 501–510. [Google Scholar] [CrossRef]
- Heeneman, S.; Sluimer, J.C.; Daemen, M.J. Angiotensin-converting enzyme and vascular remodeling. Circ. Res. 2007, 101, 441–454. [Google Scholar] [CrossRef]
- Yu, G.C.; Wang, L.G.; Han, Y.Y.; He, Q.Y. Clusterprofiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Kolde, R.; Kolde, M.R. Package ‘pheatmap’. R Package 2015. R Package Version 1.0.12. Available online: https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf (accessed on 4 Jan 2019).
- Mengjun, L.G. Tcseq: Time course sequencing data analysis. 2017. Available online: https://rdrr.io/bioc/TCseq/f/inst/doc/TCseq.pdf (accessed on 2 May 2019).

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, R.; Ma, S.; He, T.; Peng, J.; Zhang, T.; Chen, X.; Wang, X.; Chang, J.; Xia, Q.; Zhao, P. Deep Insight into the Transcriptome of the Single Silk Gland of Bombyx mori. Int. J. Mol. Sci. 2019, 20, 2491. https://doi.org/10.3390/ijms20102491
Shi R, Ma S, He T, Peng J, Zhang T, Chen X, Wang X, Chang J, Xia Q, Zhao P. Deep Insight into the Transcriptome of the Single Silk Gland of Bombyx mori. International Journal of Molecular Sciences. 2019; 20(10):2491. https://doi.org/10.3390/ijms20102491
Chicago/Turabian StyleShi, Run, Sanyuan Ma, Ting He, Jian Peng, Tong Zhang, Xiaoxu Chen, Xiaogang Wang, Jiasong Chang, Qingyou Xia, and Ping Zhao. 2019. "Deep Insight into the Transcriptome of the Single Silk Gland of Bombyx mori" International Journal of Molecular Sciences 20, no. 10: 2491. https://doi.org/10.3390/ijms20102491
APA StyleShi, R., Ma, S., He, T., Peng, J., Zhang, T., Chen, X., Wang, X., Chang, J., Xia, Q., & Zhao, P. (2019). Deep Insight into the Transcriptome of the Single Silk Gland of Bombyx mori. International Journal of Molecular Sciences, 20(10), 2491. https://doi.org/10.3390/ijms20102491





