Bacteria, Fungi and Archaea Domains in Rhizospheric Soil and Their Effects in Enhancing Agricultural Productivity
Abstract
1. Introduction
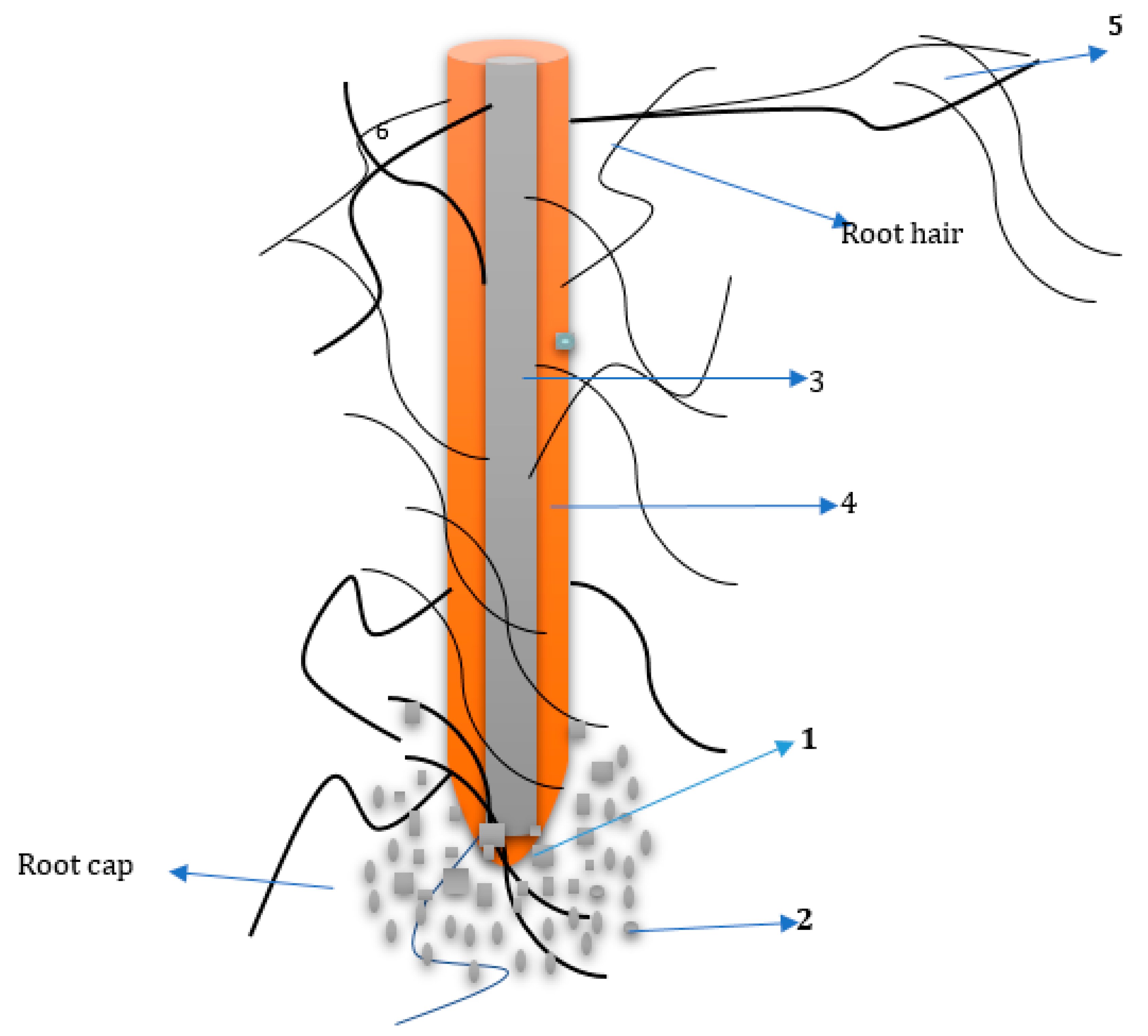
2. The Rhizosphere
3. The Effects of Rhizosphere
4. Root Exudation Mechanism
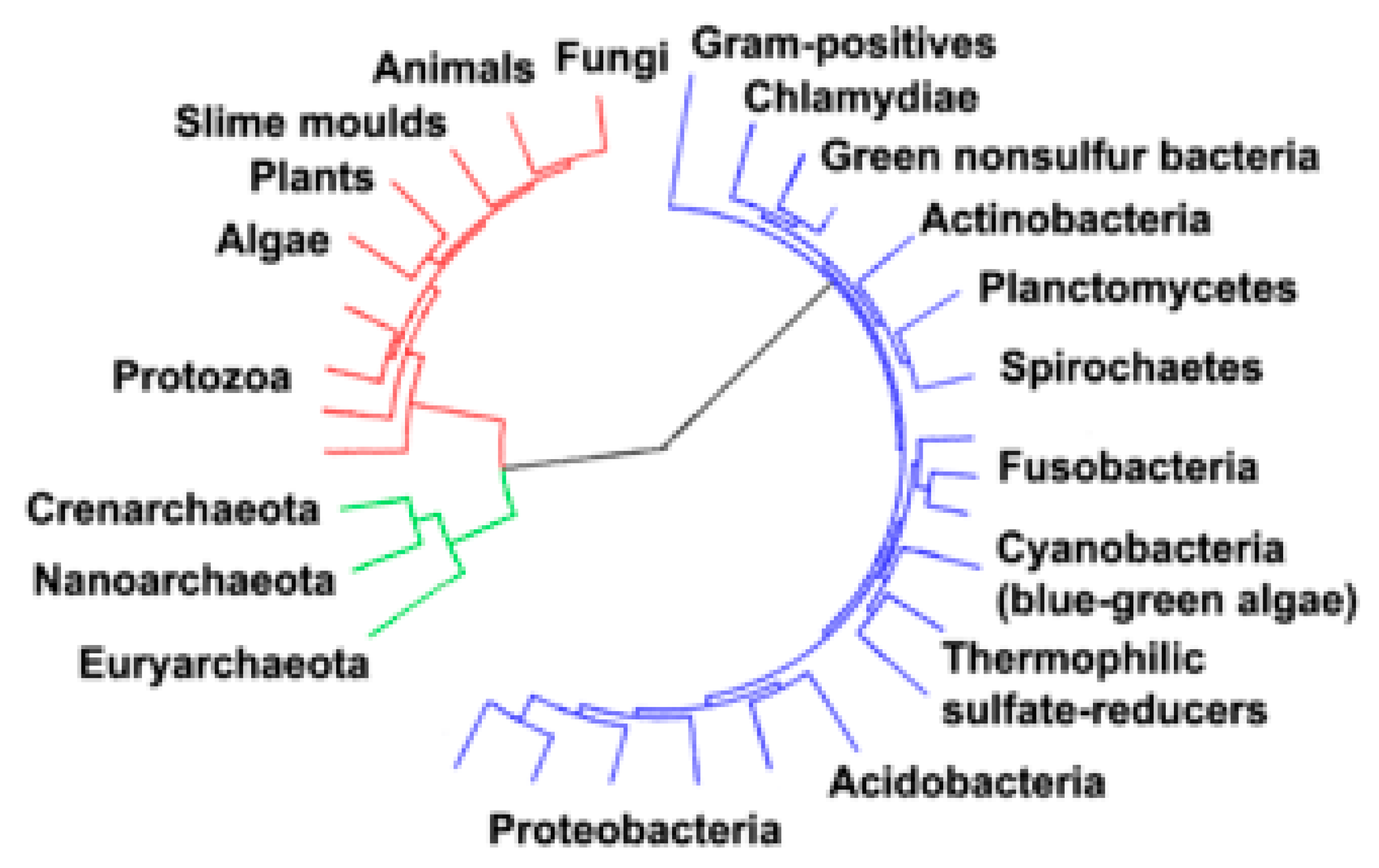
5. Archaea, Bacteria and Fungi Domains and Their Comparisons
6. Relationship among the Three Microbial Domains in the Rhizosphere
7. The Effects of Bacteria in Making Nutrients Available for Plants
8. Plant Growth Promoting Rhizobacteria (PGPR) Effects on Growth and Development of Plants
- Bioprotectants: This comprises strains of PGPR acting as biocontrol agents in order to suppress the pathogens and thus prevent plants from diseases or infections [77]. The same mode of action is required by the plant to develop resistance against bacterial [78], fungal [79], viral phytopthogens [80], insects [81] and also nematodes [82]. The ability of PGPR to produce and discharge metabolites which can ameliorate pathogens’ microbial loads and their activities or rhizosphere microflora that are deleterious is another major type of action found in several strains of PGPR [83,84]. For instance, siderophore compounds are produced which cleave ferric iron, causing it to be unavailable or rarely accessible to the inhabiting pathogenic microbes, diffusible antibiotic compounds, volatile organic compounds (VOCs), lytic enzymatic compounds, biosurfactant compounds and toxic compounds [85,86]. Competition for survival of the strains of PGPR with other phytopathogens by competing with the very few nutrients available and the little space in the rhizospheric environment is similarly a common and potential yardstick to checking the unwanted microbes’ growth, inhabiting the rhizospheric environment [87].
- Biofertilizers: These comprise of the strains of PGPR which enhance the uptake of nutrients by plants thus promoting the germination of seed and seedling development; this leads to crop yield improvement [86,88]. Several and different actions that involved PGPR as biofertilizers are fixation of N2 [89], enhancing the availability of phosphorous to plants (through solubilizing the inorganic phosphate and organic phosphate mineralization) [90] and discharging organic acids which aid to make nutrients like zinc and other vital elements available in a usable forms [91].
- Biostimulants: PGPR that are capable of producing phytohormone compounds, such as secondary metabolites like cytokinins, auxins, indole acetic acid (IAA), vitamins and riboflavin [91] are described as biostimulants. Moreover, some PGPR have the ability to degrade some complex chemicals like pesticides, herbicides and insecticides which have been found to be phytotoxic. This is a vital trait of the PGPR, for instance, P. aeruginosa PS1 ameliorate the toxicity of some herbicides used on leguminous plants such as clodinafop and quizalafop-p-ethyl [92]. In the presence of some insecticides, plant growth enhancing compounds have been reportedly produced and this includes pyriproxyfen and fipronil in some plants [93]. Likewise, reports have shown the E. asburiae PS2 strain possessing some vital plant growth enhancing activities such as indole acetic acid, solubilization of phosphate, siderophores production, hydrogen cyanide, exopolysaccharides and ammonium compounds in the presence of some herbicides like glyphosate, quizalafop-p-ethyl, metribuzin and clodinafop [94]. The strain of rhizobial MRL3 is another PGPR that has been shown to possess plant growth promoting traits when the organism was used to treat soil contaminated with insecticides such as pyriproxyfen and fipronil in plant lentil [95]. Hence, it could be concluded that strains of PGPR can be applied for the enhancement of the growth of plants even in soils that have been contaminated for a long period of time with different types of inorganic substances. From these novel traits of PGPR, plant eco-friendly rhizosphere microbes can be classified as either host plant growth enhancing microbes (HPGEM), which have a direct impact on the enhancement of the growth of plant, or as bio-control agents (BCA) which have a significant influence on the health of a plant by inhibiting phytopathogens, and hence have an indirect effect on its growing ability [96]. Thus, PGPR have direct and indirect mechanisms with significant traits that improve plant nutrition and health.
9. Bacterial Colonization and Their Systemic Inductive Resistance
10. Mycorrhizal Fungi Interaction with Plants
11. General Significance of Archaean Microbe
12. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Lal, R. Soil science and the carbon civilization. Soil Sci. Soc. Am. J. 2007, 71, 1425–1437. [Google Scholar] [CrossRef]
- Doran, J.W.; Stamatiadis, S.; Haberern, J. Soil health as an indicator of sustainable management. Agric. Ecosyst. Environ. 2002, 2, 107–110. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828. [Google Scholar] [CrossRef] [PubMed]
- Knoepp, J.D.; Coleman, D.C.; Crossley, D., Jr.; Clark, J.S. Biological indices of soil quality: An ecosystem case study of their use. For. Ecol. Manag. 2000, 138, 357–368. [Google Scholar] [CrossRef]
- Arshad, M.A.; Martin, S. Identifying critical limits for soil quality indicators in agro-ecosystems. Agric. Ecosyst. Environ. 2002, 88, 153–160. [Google Scholar] [CrossRef]
- Hartmann, A.; Schmid, M.; Van Tuinen, D.; Berg, G. Plant-driven selection of microbes. Plant Soil 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Morrissey, J.P.; Dow, J.M.; Mark, G.L.; O’Gara, F. Are microbes at the root of a solution to world food production? Embo Rep. 2004, 5, 922–926. [Google Scholar] [CrossRef]
- McNear, D.H., Jr. The rhizosphere-roots, soil and everything in between. Nat. Educ. Knowl. 2013, 4, 1. [Google Scholar]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef]
- Dijkstra, F.A.; Carrillo, Y.; Pendall, E.; Morgan, J.A. Rhizosphere priming: A nutrient perspective. Front. Microbiol. 2013, 4, 216. [Google Scholar] [CrossRef]
- Pinton, R.; Varanini, Z.; Nannipieri, P. The Rhizosphere: Biochemistry and Organic Substances at the Soil-Plant Interface; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Boer, W.d.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef] [PubMed]
- Gunawardena, U.; Zhao, X.; Hawes, M.C. Roots: Contribution to the rhizosphere. eLS 2001. [Google Scholar]
- Barea, J.-M.; Pozo, M.J.; Azcon, R.; Azcon-Aguilar, C. Microbial co-operation in the rhizosphere. J. Exp. Bot. 2005, 56, 1761–1778. [Google Scholar] [CrossRef] [PubMed]
- Gobat, J.-M.; Aragno, M.; Matthey, W. The Living Soil: Fundamentals of Soil Science and Soil Biology; Science Publishers: Enfield, NH, USA, 2004. [Google Scholar]
- Morgan, J.A.W.; Whipps, J.M. Methodological approaches to the study of rhizosphere carbon flow and microbial population dynamics. Rhizosphere Biochem. Org. Subst. Soil-Plant Interface 2000, 373. [Google Scholar]
- Aneja, K. Experiments in Microbiology, Plant Pathology and Biotechnology; New Age International: New Delhi, India, 2007. [Google Scholar]
- Antoun, H.; Prévost, D. Ecology of plant growth promoting rhizobacteria. In PGPR: Biocontrol and Biofertilization; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–38. [Google Scholar]
- Berg, G.; Opelt, K.; Zachow, C.; Lottmann, J.; Götz, M.; Costa, R.; Smalla, K. The rhizosphere effect on bacteria antagonistic towards the pathogenic fungus Verticillium differs depending on plant species and site. FEMS Microbiol. Ecol. 2006, 56, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Nie, M.; Zhang, X.-d.; Wang, J.-q.; Jiang, L.-f.; Yang, J.; Quan, Z.-x.; Cui, X.-h.; Fang, C.-m.; Li, B. Rhizosphere effects on soil bacterial abundance and diversity in the Yellow River Deltaic ecosystem as influenced by petroleum contamination and soil salinization. Soil Biol. Biochem. 2009, 41, 2535–2542. [Google Scholar] [CrossRef]
- Buyer, J.S.; Roberts, D.P.; Russek-Cohen, E. Soil and plant effects on microbial community structure. Can. J. Microbiol. 2002, 48, 955–964. [Google Scholar] [CrossRef]
- Gomes, N.C.M.; Fagbola, O.; Costa, R.; Rumjanek, N.G.; Buchner, A.; Mendona-Hagler, L.; Smalla, K. Dynamics of fungal communities in bulk and maize rhizosphere soil in the tropics. Appl. Environ. Microbiol. 2003, 69, 3758–3766. [Google Scholar] [CrossRef]
- Somers, E.; Vanderleyden, J.; Srinivasan, M. Rhizosphere bacterial signalling: A love parade beneath our feet. Crit. Rev. Microbiol. 2004, 30, 205–240. [Google Scholar] [CrossRef]
- Schloter, M.; Lebuhn, M.; Heulin, T.; Hartmann, A. Ecology and evolution of bacterial microdiversity. FEMS Microbiol. Rev. 2000, 24, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Oburger, E.; Jones, D.L. Sampling root exudates–mission impossible? Rhizosphere 2018, 6, 116–133. [Google Scholar] [CrossRef]
- Kochian, L.V.; Pineros, M.A.; Hoekenga, O.A. The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. In Root Physiology: From Gene to Function; Springer: Berlin/Heidelberg, Germany, 2005; pp. 175–195. [Google Scholar]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Miransari, M. Arbuscular mycorrhizal fungi and nitrogen uptake. Arch. Microbiol. 2011, 193, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Miranda, V.; Rothen, C.; Yela, N.; Aranda-Rickert, A.; Barros, J.; Calcagno, J.; Fracchia, S. Subterranean Desert Rodents (Genus Ctenomys) Create Soil Patches Enriched in Root Endophytic Fungal Propagules. Microb. Ecol. 2019, 77, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Brimecombe, M.J.; De Leij, F.; Lynch, J.M. Rhizodeposition and microbial populations. In The Rhizosphere Biochemistry and Organic Susbstances at the Soil-Plant Interface; CRC Press: Boca Raton, FL, USA, 2007; pp. 73–109. [Google Scholar]
- Conrad, K.A.; Dalal, R.C.; Dalzell, S.A.; Allen, D.E.; Fujinuma, R.; Menzies, N.W. Soil nitrogen status and turnover in subtropical leucaena-grass pastures as quantified by δ15N natural abundance. Geoderma 2018, 313, 126–134. [Google Scholar] [CrossRef]
- Narasimhan, K.; Basheer, C.; Bajic, V.B.; Swarup, S. Enhancement of plant-microbe interactions using a rhizosphere metabolomics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol. 2003, 132, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Uren, N.C. Types, amounts, and possible functions of compounds released into the rhizosphere by soil-grown plants. In The Rhizosphere; CRC Press: Boca Raton, FL, USA, 2000; pp. 35–56. [Google Scholar]
- Rudrappa, T.; Quinn, W.J.; Stanley-Wall, N.R.; Bais, H.P. A degradation product of the salicylic acid pathway triggers oxidative stress resulting in down-regulation of Bacillus subtilis biofilm formation on Arabidopsis thaliana roots. Planta 2007, 226, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Bertin, C.; Yang, X.; Weston, L.A. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 2003, 256, 67–83. [Google Scholar] [CrossRef]
- Singh, G.; Mukerji, K.G. Root exudates as determinant of rhizospheric microbial biodiversity. In Microbial Activity in the Rhizoshere; Springer: Berlin/Heidelberg, Germany, 2006; pp. 39–53. [Google Scholar]
- Kuzyakov, Y. Factors affecting rhizosphere priming effects. J. Plant Nutr. Soil Sci. 2002, 165, 382–396. [Google Scholar] [CrossRef]
- Hanson, P.; Edwards, N.; Garten, C.T.; Andrews, J. Separating root and soil microbial contributions to soil respiration: A review of methods and observations. Biogeochemistry 2000, 48, 115–146. [Google Scholar] [CrossRef]
- Miller, A.; Cramer, M. Root nitrogen acquisition and assimilation. In Root Physiology: From Gene to Function; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–36. [Google Scholar]
- Wichern, F.; Eberhardt, E.; Mayer, J.; Joergensen, R.G.; Müller, T. Nitrogen rhizodeposition in agricultural crops: Methods, estimates and future prospects. Soil Biol. Biochem. 2008, 40, 30–48. [Google Scholar] [CrossRef]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Hinsinger, P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: A review. Plant Soil 2001, 237, 173–195. [Google Scholar] [CrossRef]
- Vázquez, M.M.; César, S.; Azcón, R.; Barea, J.M. Interactions between arbuscular mycorrhizal fungi and other microbial inoculants (Azospirillum, Pseudomonas, Trichoderma) and their effects on microbial population and enzyme activities in the rhizosphere of maize plants. Appl. Soil Ecol. 2000, 15, 261–272. [Google Scholar] [CrossRef]
- Zhi, X.-Y.; Zhao, W.; Li, W.-J.; Zhao, G.-P. Prokaryotic systematics in the genomics era. Antonie Van Leeuwenhoek 2012, 101, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Howland, J.L. The Surprising Archaea: Discovering Another Domain of Life; Oxford University Press on Demand: Oxford, UK, 2000. [Google Scholar]
- Cavicchioli, R. Archaea—timeline of the third domain. Nat. Rev. Microbiol. 2011, 9, 51. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Deppenmeier, U. The unique biochemistry of methanogenesis. Prog. Nucleic Acid Res. Mol. Biol. 2002, 71, 223–283. [Google Scholar]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N. The COG database: An updated version includes eukaryotes. Bmc Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef]
- Makarova, K.S.; Aravind, L.; Wolf, Y.I.; Tatusov, R.L.; Minton, K.W.; Koonin, E.V.; Daly, M.J. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 2001, 65, 44–79. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.L. Were Gram-positive rods the first bacteria? Trends Microbiol. 2003, 11, 166–170. [Google Scholar] [CrossRef]
- Hugenholtz, P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002, 3, reviews0003-1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koonin, E.V. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005, 39, 309–338. [Google Scholar] [CrossRef] [PubMed]
- Beiko, R.G.; Harlow, T.J.; Ragan, M.A. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 14332–14337. [Google Scholar] [CrossRef]
- Ciccarelli, F.D.; Doerks, T.; Von Mering, C.; Creevey, C.J.; Snel, B.; Bork, P. Toward automatic reconstruction of a highly resolved tree of life. Science 2006, 311, 1283–1287. [Google Scholar] [CrossRef]
- Gupta, R.S. The natural evolutionary relationships among prokaryotes. Crit. Rev. Microbiol. 2000, 26, 111–131. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 2002, 52, 7–76. [Google Scholar] [CrossRef]
- Gupta, R.S. Molecular sequences and the early history of life. Microb. Phylogeny Evol. Concepts Controv. 2005, 160–183. [Google Scholar]
- Valas, R.E.; Bourne, P.E. The origin of a derived superkingdom: How a gram-positive bacterium crossed the desert to become an archaeon. Biol. Direct 2011, 6, 16. [Google Scholar] [CrossRef]
- Babalola, O.O. Beneficial bacteria of agricultural importance. Biotechnol. Lett. 2010, 32, 1559–1570. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.; Jastrow, J. Mycorrhizal fungi influence soil structure. In Arbuscular Mycorrhizas: Physiology and Function; Springer: Berlin/Heidelberg, Germany, 2000; pp. 3–18. [Google Scholar]
- Johnson, D.; Martin, F.; Cairney, J.W.; Anderson, I.C. The importance of individuals: Intraspecific diversity of mycorrhizal plants and fungi in ecosystems. New Phytol. 2012, 194, 614–628. [Google Scholar] [CrossRef] [PubMed]
- Salvioli, A.; Bonfante, P. Systems biology and “omics” tools: A cooperation for next-generation mycorrhizal studies. Plant Sci. 2013, 203, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Gaby, J.C.; Buckley, D.H. A global census of nitrogenase diversity. Environ. Microbiol. 2011, 13, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Geurts, R.; Lillo, A.; Bisseling, T. Exploiting an ancient signalling machinery to enjoy a nitrogen fixing symbiosis. Curr. Opin. Plant Biol. 2012, 15, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.L.; Connolly, E.L. Time to pump iron: Iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 2008, 11, 530–535. [Google Scholar] [CrossRef]
- Lemanceau, P.; Bauer, P.; Kraemer, S.; Briat, J.-F. Iron dynamics in the rhizosphere as a case study for analyzing interactions between soils, plants and microbes. Plant Soil 2009, 321, 513–535. [Google Scholar] [CrossRef]
- Shirley, M.; Avoscan, L.; Bernaud, E.; Vansuyt, G.; Lemanceau, P. Comparison of iron acquisition from Fe–pyoverdine by strategy I and strategy II plants. Botany 2011, 89, 731–735. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, Y.; Xie, X.; Kim, M.S.; Dowd, S.E.; Paré, P.W. A soil bacterium regulates plant acquisition of iron via deficiency—Inducible mechanisms. Plant J. 2009, 58, 568–577. [Google Scholar] [CrossRef]
- Marschner, P.; Crowley, D.; Rengel, Z. Rhizosphere interactions between microorganisms and plants govern iron and phosphorus acquisition along the root axis–model and research methods. Soil Biol. Biochem. 2011, 43, 883–894. [Google Scholar] [CrossRef]
- Rousk, J.; Bååth, E. Fungal and bacterial growth in soil with plant materials of different C/N ratios. FEMS Microbiol. Ecol. 2007, 62, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Collignon, C.; Uroz, S.; Turpault, M.; Frey-Klett, P. Seasons differently impact the structure of mineral weathering bacterial communities in beech and spruce stands. Soil Biol. Biochem. 2011, 43, 2012–2022. [Google Scholar] [CrossRef]
- Mapelli, F.; Marasco, R.; Balloi, A.; Rolli, E.; Cappitelli, F.; Daffonchio, D.; Borin, S. Mineral–microbe interactions: Biotechnological potential of bioweathering. J. Biotechnol. 2012, 157, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Viveros, O.; Jorquera, M.; Crowley, D.; Gajardo, G.; Mora, M. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant Nutr. 2010, 10, 293–319. [Google Scholar] [CrossRef]
- Ajilogba, C.F.; Babalola, O.O.; Ahmad, F. Antagonistic effects of Bacillus species in biocontrol of tomato Fusarium wilt. Stud. Ethno-Med. 2013, 7, 205–216. [Google Scholar] [CrossRef]
- Cameron, R.K.; Dixon, R.A.; Lamb, C.J. Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J. 1994, 5, 715–725. [Google Scholar] [CrossRef]
- Hu, X.; Wansha, L.; Chen, Q.; Yang, Y. Early signals transduction linking the synthesis of jasmonic acid in plant. Plant Signal. Behav. 2009, 4, 696–697. [Google Scholar] [CrossRef]
- Murphy, J.F.; Zehnder, G.W.; Schuster, D.J.; Sikora, E.J.; Polston, J.E.; Kloepper, J.W. Plant growth-promoting rhizobacterial mediated protection in tomato against Tomato mottle virus. Plant Dis. 2000, 84, 779–784. [Google Scholar] [CrossRef]
- Van Loon, L. Plant responses to plant growth-promoting rhizobacteria. In New Perspectives and Approaches in Plant Growth-Promoting Rhizobacteria Research; Springer: Berlin/Heidelberg, Germany, 2007; pp. 243–254. [Google Scholar]
- Burkett-Cadena, M.; Kokalis-Burelle, N.; Lawrence, K.S.; Van Santen, E.; Kloepper, J.W. Suppressiveness of root-knot nematodes mediated by rhizobacteria. Biol. Control 2008, 47, 55–59. [Google Scholar] [CrossRef]
- Adeniji, A.A.; Babalola, O.O. Bacillus velezensis: Phylogeny, useful applications, and avenues for exploitation. Appl. Microbiol. Biotechnol. 2019, 103, 3669–3682. [Google Scholar] [CrossRef] [PubMed]
- Radzki, W.; Mañero, F.G.; Algar, E.; García, J.L.; García-Villaraco, A.; Solano, B.R. Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. Antonie Van Leeuwenhoek 2013, 104, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307. [Google Scholar] [CrossRef] [PubMed]
- Berg, G. Plant–microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, V.; Viswanathan, R.; Raguchander, T.; Prakasam, V.; Samiyappan, R. Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop Prot. 2001, 20, 1–11. [Google Scholar] [CrossRef]
- Enebe, M.C.; Babalola, O.O. The influence of plant growth-promoting rhizobacteria in plant tolerance to abiotic stress: A survival strategy. Appl. Microbiol. Biotechnol. 2018, 102, 7821–7835. [Google Scholar] [CrossRef]
- Tilak, K.; Ranganayaki, N.; Pal, K.; De, R.; Saxena, A.; Nautiyal, C.S.; Mittal, S.; Tripathi, A.; Johri, B. Diversity of plant growth and soil health supporting bacteria. Curr. Sci. 2005, 136–150. [Google Scholar]
- Richardson, A.E.; Barea, J.-M.; McNeill, A.M.; Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Ahemad, M.; Khan, M.S. Phosphate-solubilizing and plant-growth-promoting Pseudomonas aeruginosa PS1 improves greengram performance in quizalafop-p-ethyl and clodinafop amended soil. Arch. Environ. Contam. Toxicol. 2010, 58, 361–372. [Google Scholar] [CrossRef]
- Bhattacharyya, P.; Goswami, M.; Bhattacharyya, L. Perspective of beneficial microbes in agriculture under changing climatic scenario: A review. J. Phytol. 2016, 26–41. [Google Scholar] [CrossRef]
- Mahanty, T.; Bhattacharjee, S.; Goswami, M.; Bhattacharyya, P.; Das, B.; Ghosh, A.; Tribedi, P. Biofertilizers: A potential approach for sustainable agriculture development. Environ. Sci. Pollut. Res. 2017, 24, 3315–3335. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Sathya, A.; Vijayabharathi, R.; Varshney, R.K.; Gowda, C.L.; Krishnamurthy, L. Plant growth promoting rhizobia: Challenges and opportunities. 3 Biotech 2015, 5, 355–377. [Google Scholar] [CrossRef] [PubMed]
- Avis, T.J.; Gravel, V.; Antoun, H.; Tweddell, R.J. Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol. Biochem. 2008, 40, 1733–1740. [Google Scholar] [CrossRef]
- Kamilova, F.; Validov, S.; Azarova, T.; Mulders, I.; Lugtenberg, B. Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ. Microbiol. 2005, 7, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Nihorimbere, V.; Ongena, M.; Smargiassi, M.; Thonart, P. Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol. Agron. Soc. Environ. 2011, 15, 327–337. [Google Scholar]
- Raaijmakers, J.M.; Vlami, M.; De Souza, J.T. Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 2002, 81, 537. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Zamioudis, C.; Pieterse, C.M. Modulation of host immunity by beneficial microbes. Mol. Plant-Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef]
- van de Mortel, J.E.; de Vos, R.C.; Dekkers, E.; Pineda, A.; Guillod, L.; Bouwmeester, K.; van Loon, J.J.; Dicke, M.; Raaijmakers, J.M. Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 2012, 160, 2173–2188. [Google Scholar] [CrossRef]
- Niu, D.-D.; Liu, H.-X.; Jiang, C.-H.; Wang, Y.-P.; Wang, Q.-Y.; Jin, H.-L.; Guo, J.-H. The plant growth–promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate-and jasmonate/ethylene-dependent signaling pathways. Mol. Plant-Microbe Interact. 2011, 24, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Schikora, A. Quorum sensing of bacteria and trans-kingdom interactions of N-acyl homoserine lactones with eukaryotes. J. Chem. Ecol. 2012, 38, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Cartieaux, F.; Contesto, C.; Gallou, A.; Desbrosses, G.; Kopka, J.; Taconnat, L.; Renou, J.-P.; Touraine, B. Simultaneous interaction of Arabidopsis thaliana with Bradyrhizobium sp. strain ORS278 and Pseudomonas syringae pv. tomato DC3000 leads to complex transcriptome changes. Mol. Plant-Microbe Interact. 2008, 21, 244–259. [Google Scholar] [CrossRef]
- Weston, D.J.; Pelletier, D.A.; Morrell-Falvey, J.L.; Tschaplinski, T.J.; Jawdy, S.S.; Lu, T.-Y.; Allen, S.M.; Melton, S.J.; Martin, M.Z.; Schadt, C.W. Pseudomonas fluorescens induces strain-dependent and strain-independent host plant responses in defense networks, primary metabolism, photosynthesis, and fitness. Mol. Plant-Microbe Interact. 2012, 25, 765–778. [Google Scholar] [CrossRef] [PubMed]
- Weston, L.A.; Ryan, P.R.; Watt, M. Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere. J. Exp. Bot. 2012, 63, 3445–3454. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhang, L.; Feng, G.; Christie, P.; Tian, C.; Li, X. Diversity of arbuscular mycorrhizal fungi associated with desert ephemerals growing under and beyond the canopies of Tamarisk shrubs. Chin. Sci. Bull. 2006, 51, 132–139. [Google Scholar] [CrossRef][Green Version]
- Harrison, M.J. Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 2005, 59, 19–42. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Duplessis, S.; Ditengou, F.; Lagrange, H.; Voiblet, C.; Lapeyrie, F. Developmental cross talking in the ectomycorrhizal symbiosis: Signals and communication genes. New Phytol. 2001, 151, 145–154. [Google Scholar] [CrossRef]
- Kosuta, S.; Chabaud, M.; Lougnon, G.; Gough, C.; Dénarié, J.; Barker, D.G.; Bécard, G. A Diffusible Factor from Arbuscular Mycorrhizal Fungi Induces Symbiosis-Specific MtENOD11 Expression in Roots ofMedicago truncatula. Plant Physiol. 2003, 131, 952–962. [Google Scholar] [CrossRef]
- Maillet, F.; Poinsot, V.; André, O.; Puech-Pagès, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011, 469, 58. [Google Scholar] [CrossRef]
- Rillig, M.C.; Mummey, D.L. Mycorrhizas and soil structure. New Phytol. 2006, 171, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Selosse, M.-A.; Richard, F.; He, X.; Simard, S.W. Mycorrhizal networks: Des liaisons dangereuses? Trends Ecol. Evol. 2006, 21, 621–628. [Google Scholar] [CrossRef]
- Klironomos, J.N. Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 2003, 84, 2292–2301. [Google Scholar] [CrossRef]
- Cabello, P.; Roldan, M.D.; Moreno-Vivian, C. Nitrate reduction and the nitrogen cycle in archaea. Microbiology 2004, 150, 3527–3546. [Google Scholar] [CrossRef] [PubMed]
- Mehta, M.P.; Baross, J.A. Nitrogen fixation at 92 C by a hydrothermal vent archaeon. Science 2006, 314, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Coolen, M.J.; Abbas, B.; Van Bleijswijk, J.; Hopmans, E.C.; Kuypers, M.M.; Wakeham, S.G.; Sinninghe Damste, J.S. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: A basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ. Microbiol. 2007, 9, 1001–1016. [Google Scholar] [CrossRef] [PubMed]
- Francis, C.A.; Beman, J.M.; Kuypers, M.M. New processes and players in the nitrogen cycle: The microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 2007, 1, 19. [Google Scholar] [CrossRef]
- Leininger, S.; Urich, T.; Schloter, M.; Schwark, L.; Qi, J.; Nicol, G.W.; Prosser, J.I.; Schuster, S.; Schleper, C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 2006, 442, 806. [Google Scholar] [CrossRef]
- Chelius, M.; Triplett, E. The Diversity of Archaea and Bacteria in Association with the Roots of Zea mays L. Microb. Ecol. 2001, 41. [Google Scholar] [CrossRef]
- Simon, H.; Dodsworth, J.; Goodman, R. Erratum: Crenarchaeota colonize terrestrial plant roots (Environmental Microbiology 2: 5 (495–505)). Environ. Microbiol. 2001, 3, 354. [Google Scholar] [CrossRef]
- Wegley, L.; Yu, Y.; Breitbart, M.; Casas, V.; Kline, D.I.; Rohwer, F. Coral-associated archaea. Mar. Ecol. Prog. Ser. 2004, 273, 89–96. [Google Scholar] [CrossRef]
- Shand, R.F.; Leyva, K.J. Archaeal antimicrobials: An undiscovered country. Archaea New Models Prokaryotic Biol. 2008, 2008. [Google Scholar]
| Complex Exudates | Compound Constituents |
|---|---|
| Organic compounds | Succinic acid, l-aspartic acid, Acetic acid, l-glutamic acid, salicylic acid, malic acid, isocitric acid, chorismic acid, shikimic acid, sinapic acid, shikimic acid, p-hydroxybenzoic acid, gallic acid, caffeic acid, protocatacheuic acid, p-coumaric acid, tartaric acid, ferulic acid, oxalic acid, citric acid, piscidic acid, mugineic acid |
| Complex carbohydrate | Glucose, arabinose, galactose, sucrose, fructose, pentose, raffinose, rhamnose, ribose, xylose and mannitol |
| Amino acids | Complete 20 protein genic amino acids, l-hydroxyproline, mugineic acid, amino butyric acid, homoserine |
| Coumarins | Umbelliferone |
| Flavonols | Kaempferol, quercitin, naringenin, naringin, rutin, myricetin, strigolactone, genistein and their derivative sugars |
| Lignins | Benzoic acid, nicotinic acid, catechol, cinnamic acid, gallic acid, phloroglucinol, syringic acid, sinapoyl aldehyde, ferulic acid, coumaric acid, vanillin, chlorogenic acid, quinic acid, pyroglutamic acid, sinapyl alcohol |
| Anthocyanins | Delphinidin, pelargonidin, cyanidin and their derivatives sugar molecules |
| Aurones | Sinapoyl choline, benzyl aurones synapates |
| Glucosinolates | Desuphoguconapin, desulphoprogoitrin, cyclobrassinone, desulphoglucoalyssin, desulphonapoleiferin |
| Sterols | Sitosterol, stigmasterol, campestrol |
| Anthocyanins | Delphinidin, pelargonidin, cyanidin and their derivative sugar molecules |
| Fatty acids | Oleic acid, linoleic acid, stearic acid, palmitic acid |
| Indole compounds | Brassitin, sinalexin, indole-3-acetic acid, methyl indole carboxylate, camalexin glucoside, brassilexin |
| Proteins and enzymes | Lectins, proteases, PR proteins, peroxidases, phosphatases, lipase, hydrolases |
| Allomones | Sorgoleone, 5,7,4′-trihydroxy-3′, jugulone, DIMBOA, 5′-dimethoxyflavone, DIBOA |
| Property | Bacteria | Archaea | Fungi |
|---|---|---|---|
| Cell Membrane | Made up of peptidoglycan and lipids are linked via ester molecule, | Made up of pseudo-peptidoglycan and lipids are linked via ether molecule | Made up of different structures and lipids are linked via ester molecule |
| Gene Structure and Configuration | Chromosomes are circular, translation and transcription are unique | Chromosomes are circular, translation and transcription are similar to eukaryotes (fungi) | Chromosomes are multiple and linear, translation and transcription are similar to archaea |
| Structure of Internal Cell | The nucleus or organelles has no membrane bound | The nucleus or organelles has no membrane bound | There is membrane bound nucleus and organelles |
| Metabolic Reaction | There are several, including aerobic and anaerobic respiration, photosynthetic, autotrophic reactions and fermentation | There are several with methanogenic reaction specifically unique to this domain | Cellular respiration, fermentation and photosynthetic reaction |
| Reproduction | Reproduction is asexual and transfer of genes is horizontal | Reproduction is asexual and transfer of genes is horizontal | Reproduction is sexual and asexual |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Odelade, K.A.; Babalola, O.O. Bacteria, Fungi and Archaea Domains in Rhizospheric Soil and Their Effects in Enhancing Agricultural Productivity. Int. J. Environ. Res. Public Health 2019, 16, 3873. https://doi.org/10.3390/ijerph16203873
Odelade KA, Babalola OO. Bacteria, Fungi and Archaea Domains in Rhizospheric Soil and Their Effects in Enhancing Agricultural Productivity. International Journal of Environmental Research and Public Health. 2019; 16(20):3873. https://doi.org/10.3390/ijerph16203873
Chicago/Turabian StyleOdelade, Kehinde Abraham, and Olubukola Oluranti Babalola. 2019. "Bacteria, Fungi and Archaea Domains in Rhizospheric Soil and Their Effects in Enhancing Agricultural Productivity" International Journal of Environmental Research and Public Health 16, no. 20: 3873. https://doi.org/10.3390/ijerph16203873
APA StyleOdelade, K. A., & Babalola, O. O. (2019). Bacteria, Fungi and Archaea Domains in Rhizospheric Soil and Their Effects in Enhancing Agricultural Productivity. International Journal of Environmental Research and Public Health, 16(20), 3873. https://doi.org/10.3390/ijerph16203873






