Genome-Wide Identification of MIKCc-Type MADS-Box Family Gene and Floral Organ Transcriptome Characterization in Ma Bamboo (Dendrocalamus latiflorus Munro)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, cDNA Synthesis and Transcriptome Sequencing
2.2. RNA-seq Bioinformatics Analysis
2.3. Sequence Search and Identification of MIKCc-Type MADS-Box Genes
2.4. Phylogenetic Tree Construction
2.5. Genome Synteny and Gene Synteny Analysis
2.6. Prediction of Cis-Regulatory Elements of Promoter Region
2.7. RT-qPCR Validation
3. Results
3.1. MIKCc-Type MADS-Box Genes Identification and Phylogenetic Tree Construction
3.2. Hormone-Related Promoter Cis-Regulatory Elements
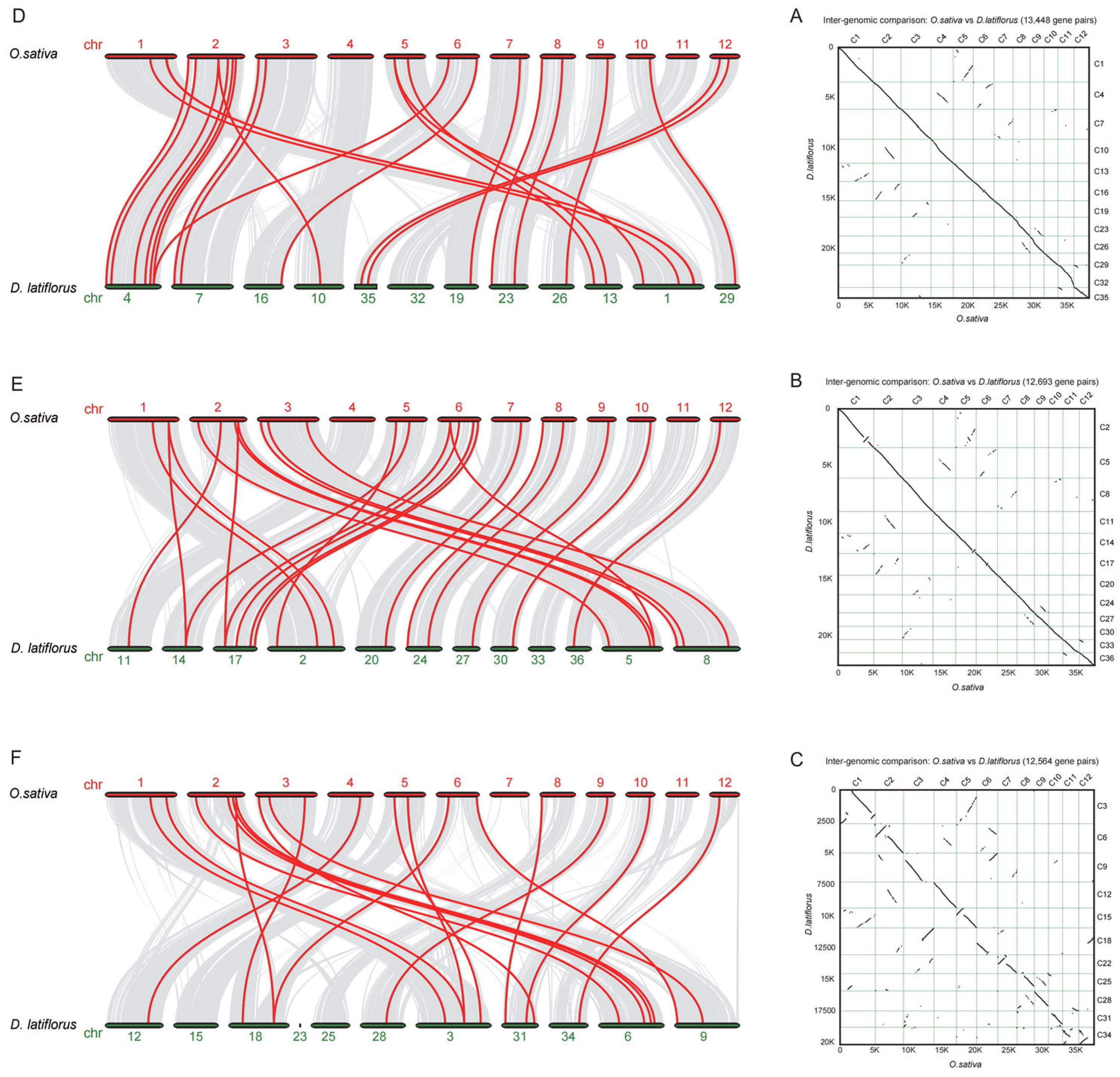
3.3. Synteny Analysis between D. latiflflorus and O. sativa
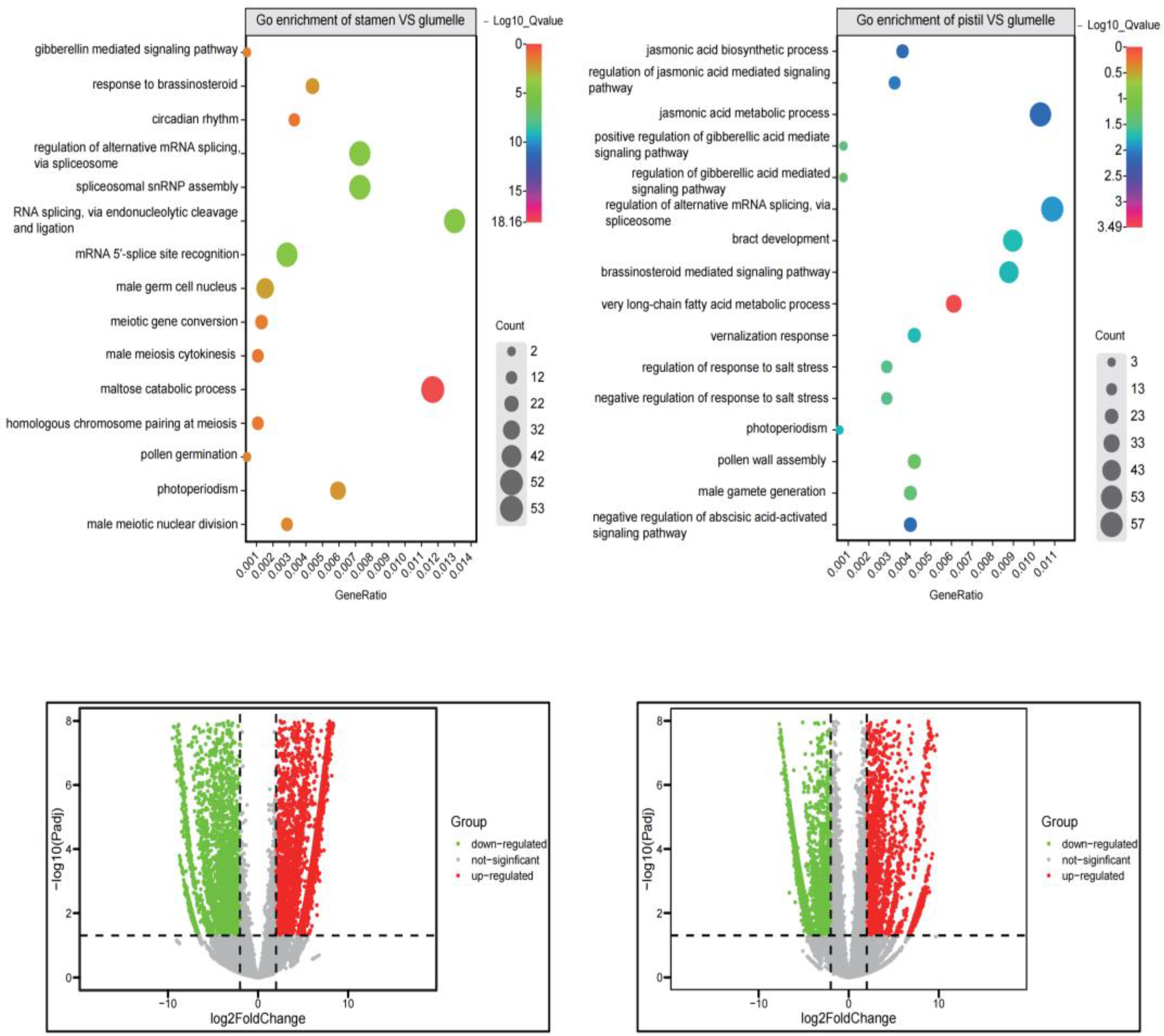
3.4. Transcriptome-Scale Analysis of Floral Organ
3.5. Go Enrichment Analysis
3.6. Expression of MIKCc-Type MADS-Box Gene in Flower Organs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, G.; Meng, C.; Jiang, P.; Xu, Q. Review of Carbon Fixation in Bamboo Forests in China. Bot. Rev. 2011, 77, 262–270. [Google Scholar] [CrossRef]
- Janzen, D.H. Why Bamboos Wait So Long to Flower. Annu. Rev. Ecol. Syst. 1976, 7, 341–391. [Google Scholar] [CrossRef]
- Cho, L.; Yoon, J.; An, G. The control of flowering time by environmental factors. Plant J. 2017, 90, 708–719. [Google Scholar] [CrossRef]
- Mizuki, I.; Sato, A.; Matsuo, A.; Suyama, Y.; Suzuki, J.; Makita, A. Clonal structure, seed set, and self-pollination rate in mass-flowering bamboo species during off-year flowering events. PLoS ONE 2014, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Yang, G.; Zhao, L.; Zhang, X.; Li, D.; Guo, Z. Complementary Transcriptome and Proteome Analyses Provide Insight into the Floral Transition in Bamboo (Dendrocalamus latiflorus Munro). Int. J. Mol. Sci. 2020, 21, 8430. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.; Chou, M.; Yue, J.; Hsu, C.; Chang, W.; Ko, S.; Liao, D.; Huang, Y.; Chen, J.J.W.; Yuan, J.; et al. BeMADS1 is a key to delivery MADSs into nucleus in reproductive tissues-De novo characterization of Bambusa edulis transcriptome and study of MADS genes in bamboo floral development. BMC Plant Biol. 2014, 14, 1–16. [Google Scholar] [CrossRef]
- Zheng, X.; Lin, S.; Fu, H.; Wan, Y.; Ding, Y. The Bamboo Flowering Cycle Sheds Light on Flowering Diversity. Front. Plant Sci. 2020, 11, 1–20. [Google Scholar] [CrossRef]
- Shrestha, R.; Gómez-Ariza, J.; Brambilla, V.; Fornara, F. Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 2014, 114, 1445–1458. [Google Scholar] [CrossRef]
- Kai, C.; Kaiqiang, H.; Feihu, X.; Huihui, W.; Markus, V.K.; Pengfei, G.; Jiakai, L.; Wentao, W.; Xuqing, L.; Hangxiao, Z.; et al. High-Efficient and Transient Transformation of Moso Bamboo (Phyllostachys edulis) and ma Bamboo (Dendrocalamus latiflorus Munro). J. Plant Biol. 2021. [Google Scholar] [CrossRef]
- Zheng, Y.; Yang, D.; Rong, J.; Chen, L.; Zhu, Q.; He, T.; Chen, L.; Ye, J.; Fan, L.; Gao, Y.; et al. Allele-aware chromosome-scale assembly of the allopolyploid genome of hexaploid ma bamboo (Dendrocalamus latiflorus Munro). J. Integr. Plant Biol. 2022, 64, 649–670. [Google Scholar] [CrossRef]
- Ye, S.; Chen, G.; Kohnen, M.V.; Wang, W.; Cai, C.; Ding, W.; Wu, C.; Gu, L.; Zheng, Y.; Ma, X.; et al. Robust CRISPR/Cas9 mediated genome editing and its application in manipulating plant height in the first generation of hexaploid ma bamboo (Dendrocalamus latiflorus Munro). Plant Biotechnol. J. 2020, 18, 1501–1503. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Winter, K.U.; Meyer, B.; Saedler, H.; Theissen, G. MADS-Box gene diversity in seed plants 300 million years ago. Mol. Biol. Evol. 2000, 17, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Martel, C.; Vrebalov, J.; Tafelmeyer, P.; Giovannoni, J.J. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 2011, 157, 1568–1579. [Google Scholar] [CrossRef] [PubMed]
- Gramzow, L.; Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, 214. [Google Scholar] [CrossRef]
- Kong, X.; Wang, F.; Geng, S.; Guan, J.; Tao, S.; Jia, M.; Sun, G.; Wang, Z.; Wang, K.; Ye, X.; et al. The wheat AGL6-like MADS-box gene is a master regulator for floral organ identity and a target for spikelet meristem development manipulation. Plant Biotechnol. J. 2021, 20, 75–88. [Google Scholar] [CrossRef]
- Su, Z.; Ma, X.; Guo, H.; Sukiran, N.L.; Guo, B.; Assmann, S.M.; Ma, H. Flower development under drought stress: Morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. Plant Cell 2013, 25, 3785–3807. [Google Scholar] [CrossRef]
- Fabio, F.; Amaury, D.M.; George, C. SnapShot: Control of Flowering in Arabidopsis. Cell 2010, 141, 550. [Google Scholar]
- Schultz, E.A.; Haughn, G.W. LEAFY, a Homeotic Gene That Regulates Inflorescence Development in Arabidopsis. The Plant cell 1991, 3, 771–781. [Google Scholar] [CrossRef]
- Yuan, J.; Yue, J.; Gu, X.; Lin, C. Flowering of Woody Bamboo in Tissue Culture Systems. Front. Plant Sci. 2017, 8, 1–8. [Google Scholar] [CrossRef]
- Han, Y.; Chen, Z.; Lv, S.; Ning, K.; Ji, X.; Liu, X.; Wang, Q.; Liu, R.; Fan, S.; Zhang, X. MADS-Box Genes and Gibberellins Regulate Bolting in Lettuce (Lactuca sativa L.). Front. Plant Sci. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Yang, X.; Wu, F.; Lin, X.; Du, X.; Chong, K.; Gramzow, L.; Schilling, S.; Becker, A.; Theissen, G.; Meng, Z. Live and let die - the B(sister) MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa). PLoS ONE 2012, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.S.; Jang, S.; Lee, S.; Nam, J.; Kim, C.; Lee, S.H.; Chung, Y.Y.; Kim, S.R.; Lee, Y.H.; Cho, Y.G.; et al. Leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 2000, 12, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Kater, M.M.; Dreni, L.; Colombo, L. Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 2006, 57, 3433–3444. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Geng, X.; Yang, L.; Chen, Y.; Zhao, Z.; Shi, W.; Kang, L.; Wu, R.; Lu, C.; Gao, J. Total and Mitochondrial Transcriptomic and Proteomic Insights into Regulation of Bioenergetic Processes for Shoot Fast-Growth Initiation in Moso Bamboo. Cells 2022, 11, 1240. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wang, H.; Cai, D.; Gao, Y.; Zhang, H.; Wang, Y.; Lin, C.; Ma, L.; Gu, L. Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis). Plant J. 2017, 91, 684–699. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 1–11. [Google Scholar] [CrossRef]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER Web Server for Protein Sequence Similarity Search. Curr. Protoc. Bioinform. 2017, 60, 3.15.1–3.15.23. [Google Scholar] [CrossRef]
- Jawad, A.A.; Ayyez, H.N.; Klaif, S.F. Sequencing and phylogeny of sulphonamide resistant genes by using MEGA6 software program. Int. J. Res. Pharm. Sci. 2018, 9. [Google Scholar] [CrossRef]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef] [PubMed]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Lydia, G.; Günter, T. Phylogenomics of MADS-Box Genes in Plants—Two Opposing Life Styles in One Gene Family. Biology 2013, 2, 1150–1164. [Google Scholar]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.; Singh, V.; Tyagi, A.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef] [PubMed]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef]
- Weiss, J.; Alcantud-Rodriguez, R.; Toksöz, T.; Egea-Cortines, M. Meristem maintenance, auxin, jasmonic and abscisic acid pathways as a mechanism for phenotypic plasticity in Antirrhinum majus. Sci. Rep. UK 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Manchado-Rojo, M.; Delgado-Benarroch, L.; Roca, M.J.; Weiss, J.; Egea-Cortines, M. Quantitative levels of Deficiens and Globosa during late petal development show a complex transcriptional network topology of B function. Plant J. 2012, 72, 294–307. [Google Scholar] [CrossRef]
- Marcelo, C.D.; Camila, M.P.; Gerco, C.A.; Richard, G.H.I. MADS: The missing link between identity and growth? Trends Plant Sci. 2010, 16, 89–97. [Google Scholar]
- Ge, W.; Zhang, Y.; Cheng, Z.; Hou, D.; Li, X.; Gao, J. Main regulatory pathways, key genes and microRNAs involved in flower formation and development of moso bamboo (Phyllostachys edulis). Plant Biotechnol. J. 2017, 15, 82–96. [Google Scholar] [CrossRef]
- Li, L.; Mu, S.; Cheng, Z.; Cheng, Y.; Zhang, Y.; Miao, Y.; Hou, C.; Li, X.; Gao, J. Characterization and expression analysis of the WRKY gene family in moso bamboo. Sci. Rep. UK 2017, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fahad, A.; Muhammad, Z.I.; Abdul, K.; Saddam, H.; Ihsanullah, D.; Shah, F.; Wajid, N. Gibberellin-sensitive Rht alleles confer tolerance to heat and drought stresses in wheat at booting stage. J. Cereal Sci. 2016, 70, 72–78. [Google Scholar]
- He, Y.; Liu, C.; Zhu, L.; Fu, M.; Sun, Y.; Zeng, H. Jasmonic Acid Plays a Pivotal Role in Pollen Development and Fertility Regulation in Different Types of P(T)GMS Rice Lines. Int. J. Mol. Sci. 2021, 22, 7926. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Goossens, A.; Lacchini, E. Jasmonate: A hormone of primary importance for plant metabolism. Curr. Opin. Plant Biol. 2022, 67, 102197. [Google Scholar] [CrossRef]
- Zhan, P.; Ma, S.; Xiao, Z.; Li, F.; Wei, X.; Lin, S.; Wang, X.; Ji, Z.; Fu, Y.; Pan, J.; et al. Natural variations in grain length 10(GL10) regulate rice grain size. J. Genet. Genom. 2022, 49, 405–413. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, F.; Hong, Y.; Yao, J.; Ren, Z.; Shi, H.; Zhu, J.K. The Flowering Repressor SVP Confers Drought Resistance in Arabidopsis by Regulating Abscisic Acid Catabolism. Mol. Plant 2018, 11, 1184–1197. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, D.; Yang, J.; Wan, J.; Xu, Y.; Li, L.; Rong, J.; Chen, L.; He, T.; Zheng, Y. Genome-Wide Identification of MIKCc-Type MADS-Box Family Gene and Floral Organ Transcriptome Characterization in Ma Bamboo (Dendrocalamus latiflorus Munro). Genes 2023, 14, 78. https://doi.org/10.3390/genes14010078
Yang D, Yang J, Wan J, Xu Y, Li L, Rong J, Chen L, He T, Zheng Y. Genome-Wide Identification of MIKCc-Type MADS-Box Family Gene and Floral Organ Transcriptome Characterization in Ma Bamboo (Dendrocalamus latiflorus Munro). Genes. 2023; 14(1):78. https://doi.org/10.3390/genes14010078
Chicago/Turabian StyleYang, Deming, Jing Yang, Jiayi Wan, Yanping Xu, Lei Li, Jundong Rong, Lingyan Chen, Tianyou He, and Yushan Zheng. 2023. "Genome-Wide Identification of MIKCc-Type MADS-Box Family Gene and Floral Organ Transcriptome Characterization in Ma Bamboo (Dendrocalamus latiflorus Munro)" Genes 14, no. 1: 78. https://doi.org/10.3390/genes14010078
APA StyleYang, D., Yang, J., Wan, J., Xu, Y., Li, L., Rong, J., Chen, L., He, T., & Zheng, Y. (2023). Genome-Wide Identification of MIKCc-Type MADS-Box Family Gene and Floral Organ Transcriptome Characterization in Ma Bamboo (Dendrocalamus latiflorus Munro). Genes, 14(1), 78. https://doi.org/10.3390/genes14010078





