Hippocampal Changes Elicited by Metabolic and Inflammatory Stressors following Prenatal Maternal Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Experiments
2.2. RNA Sequencing, Mapping, and Analysis
2.3. Gene Ontology, Network Inference, and Transcription Factor Analysis
3. Results
3.1. Sequence and Annotation Statistics
3.2. Effects of Maternal Immune Activation, Postnatal Stress, and Sex on the Gene Pathways in the Hippocampus
3.3. Effects of Maternal Immune Activation, Postnatal Stress, and Sex on Gene Expression in the Hippocampus
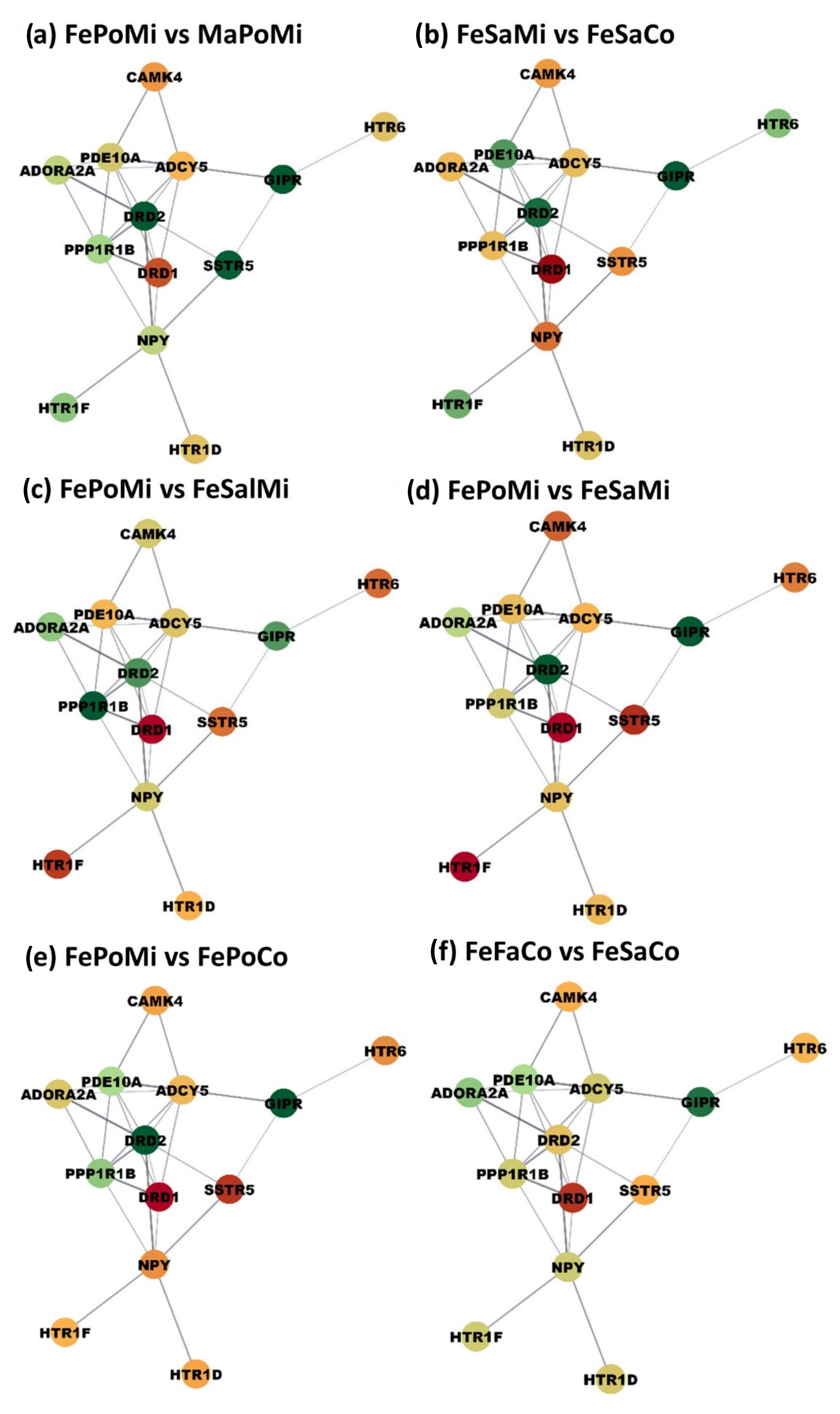
3.4. cAMP Signaling Network Profiles Impacted by Maternal Immune Activation, Postnatal Stress, and Sex
3.5. Regulatory Motifs and Transcription Factors Associated with Genes Impacted by Maternal Immune Activation, Postnatal Stress, and Sex in the Hippocampus
4. Discussion
4.1. Processes and Pathways Impacted by Maternal Immune Activation, Postnatal Stress, and Sex
4.2. Genes Impacted by Maternal Immune Activation, Postnatal Stress, and Sex
4.3. cAMP Signaling Network Profiles Impacted by Maternal Immune Activation, Secondary Stress, and Sex
4.4. Regulatory Motifs and Transcription Factors Impacted by Maternal Immune Activation, Postnatal Stress, and Sex
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Keever, M.R.; Zhang, P.; Bolt, C.R.; Antonson, A.M.; Rymut, H.E.; Caputo, M.P.; Houser, A.K.; Hernandez, A.G.; Southey, B.R.; Rund, L.A.; et al. Lasting and sex-dependent impact of maternal immune activation on molecular pathways of the amygdala. Front. Neurosci. 2020, 14, 774. [Google Scholar] [CrossRef] [PubMed]
- Rymut, H.E.; Bolt, C.R.; Caputo, M.P.; Houser, A.K.; Antonson, A.M.; Zimmerman, J.D.; Villamil, M.B.; Southey, B.R.; Rund, L.A.; Johnson, R.W.; et al. Long-lasting impact of maternal immune activation and interaction with a second immune challenge on pig behavior. Front. Vet. Sci. 2020, 7, 561151. [Google Scholar] [CrossRef] [PubMed]
- Rymut, H.E.; Rund, L.A.; Bolt, C.R.; Villamil, M.B.; Southey, B.R.; Johnson, R.W.; Rodriguez-Zas, S.L. The Combined Effect of Weaning Stress and Immune Activation during Pig Gestation on Serum Cytokine and Analyte Concentrations. Animals 2021, 11, 2274. [Google Scholar] [CrossRef] [PubMed]
- Couch, A.C.M.; Berger, T.; Hanger, B.; Matuleviciute, R.; Srivastava, D.P.; Thuret, S.; Vernon, A.C. Maternal immune activation primes deficiencies in adult hippocampal neurogenesis. Brain Behav. Immun. 2021, 97, 410–422. [Google Scholar] [CrossRef]
- Antonson, A.M.; Balakrishnan, B.; Radlowski, E.C.; Petr, G.; Johnson, R.W. Altered Hippocampal Gene Expression and Morphology in Fetal Piglets following Maternal Respiratory Viral Infection. Dev. Neurosci. 2018, 40, 104–119. [Google Scholar] [CrossRef]
- Southey, B.R.; Keever-Keigher, M.R.; Rymut, H.E.; Rund, L.A.; Johnson, R.W.; Rodriguez-Zas, S.L. Disruption of Alternative Splicing in the Amygdala of Pigs Exposed to Maternal Immune Activation. Immuno 2021, 1, 499–517. [Google Scholar] [CrossRef]
- Southey, B.R.; Zhang, P.; Keever, M.R.; Rymut, H.E.; Johnson, R.W.; Sweedler, J.V.; Rodriguez-Zas, S.L. Effects of maternal immune activation in porcine transcript isoforms of neuropeptide and receptor genes. J. Integr. Neurosci. 2021, 20, 21–31. [Google Scholar] [CrossRef]
- Rymut, H.E.; Rund, L.A.; Southey, B.R.; Johnson, R.W.; Rodriguez-Zas, S.L. Terpenoid Backbone Biosynthesis among Pig Hippocampal Pathways Impacted by Stressors. Genes 2022, 13, 814. [Google Scholar] [CrossRef]
- Keever-Keigher, M.R.; Zhang, P.; Bolt, C.R.; Rymut, H.E.; Antonson, A.M.; Corbett, M.P.; Houser, A.K.; Hernandez, A.G.; Southey, B.R.; Rund, L.A.; et al. Interacting impact of maternal inflammatory response and stress on the amygdala transcriptome of pigs. G3 2021, 11, jkab113. [Google Scholar] [CrossRef]
- Southey, B.R.; Bolt, C.R.; Rymut, H.E.; Keever, M.R.; Ulanov, A.V.; Li, Z.; Rund, L.A.; Johnson, R.W.; Rodriguez-Zas, S.L. Impact of weaning and maternal immune activation on the metabolism of pigs. Front. Mol. Biosci. 2021, 8, 660764. [Google Scholar] [CrossRef]
- Rymut, H.E.; Rund, L.A.; Bolt, C.R.; Villamil, M.B.; Bender, D.E.; Southey, B.R.; Johnson, R.W.; Rodriguez-Zas, S.L. Biochemistry and Immune Biomarkers Indicate Interacting Effects of Pre- and Postnatal Stressors in Pigs across Sexes. Animals 2021, 11, 987. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 August 2022).
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Janky, R.; Verfaillie, A.; Imrichova, H.; Van de Sande, B.; Standaert, L.; Christiaens, V.; Hulselmans, G.; Herten, K.; Sanchez, M.N.; Potier, D.; et al. iRegulon: From a Gene List to a Gene Regulatory Network Using Large Motif and Track Collections. PLoS Comp. Biol. 2014, 10, e1003731. [Google Scholar] [CrossRef]
- Baik, S.-H.; Rajeev, V.; Fann, D.Y.-W.; Jo, D.-G.; Arumugam, T.V. Intermittent fasting increases adult hippocampal neurogenesis. Brain Behav. 2020, 10, e01444. [Google Scholar] [CrossRef]
- Marchese, E.; Di Maria, V.; Samengo, D.; Pani, G.; Michetti, F.; Geloso, M.C. Post-Natal Deletion of Neuronal cAMP Responsive-Element Binding (CREB)-1 Promotes Pro-inflammatory Changes in the Mouse Hippocampus. Neurochem. Res. 2017, 42, 2230–2245. [Google Scholar] [CrossRef]
- Hu, Y.; Fang, Z.; Yang, Y.; Rohlsen-Neal, D.; Cheng, F.; Wang, J. Analyzing the genes related to nicotine addiction or schizophrenia via a pathway and network based approach. Sci. Rep. 2018, 8, 2894. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, S.J.; Son, T.G.; Chan, S.L.; Mattson, M.P. Interferon-γ is up-regulated in the hippocampus in response to intermittent fasting and protects hippocampal neurons against excitotoxicity. J. Neurosci. Res. 2006, 83, 1552–1557. [Google Scholar] [CrossRef] [PubMed]
- Carlezon, W.A.; Kim, W.; Missig, G.; Finger, B.C.; Landino, S.M.; Alexander, A.J.; Mokler, E.L.; Robbins, J.O.; Li, Y.; Bolshakov, V.Y.; et al. Maternal and early postnatal immune activation produce sex-specific effects on autism-like behaviors and neuroimmune function in mice. Sci. Rep. 2019, 9, 16928. [Google Scholar] [CrossRef]
- Ren, X.; Zhou, L.; Terwilliger, R.; Newton, S.; De Araujo, I. Sweet taste signaling functions as a hypothalamic glucose sensor. Front. Integr. Neurosci. 2009, 3, 12. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.-i. Neural insights into sweet taste transduction and hunger-induced taste modification in mice. Biosci. Biotechnol. Biochem. 2022, 86, 1485–1489. [Google Scholar] [CrossRef]
- Hardikar, S.; Höchenberger, R.; Villringer, A.; Ohla, K. Higher sensitivity to sweet and salty taste in obese compared to lean individuals. Appetite 2017, 111, 158–165. [Google Scholar] [CrossRef]
- Gal-Oz, S.T.; Maier, B.; Yoshida, H.; Seddu, K.; Elbaz, N.; Czysz, C.; Zuk, O.; Stranger, B.E.; Ner-Gaon, H.; Shay, T. ImmGen report: Sexual dimorphism in the immune system transcriptome. Nat. Commun. 2019, 10, 4295. [Google Scholar] [CrossRef]
- Fung, S.J.; Fillman, S.G.; Webster, M.J.; Shannon Weickert, C. Schizophrenia and bipolar disorder show both common and distinct changes in cortical interneuron markers. Schizophr. Res. 2014, 155, 26–30. [Google Scholar] [CrossRef]
- Moreno, C.L.; Ehrlich, M.E.; Mobbs, C.V. Protection by dietary restriction in the YAC128 mouse model of Huntington’s disease: Relation to genes regulating histone acetylation and HTT. Neurobiol. Dis. 2016, 85, 25–34. [Google Scholar] [CrossRef][Green Version]
- Matsuura, S.; Nishimoto, Y.; Endo, A.; Segi-Nishida, E. Hippocampal Inflammation and Neuronal Changes in Peripheral Lipopolysaccharide Challenged Mice Showing Anxiety-Like Behaviors. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4025946 (accessed on 15 September 2022).
- Hayase, T.; Tachibana, S.; Yamakage, M. Determination of the Effects of Sevoflurane Anesthesia in Different Maturing Stages of the Mouse Hippocampus by Transcriptome Analysis. J. Anesth. Clin. Res. 2017, 8, 2. [Google Scholar] [CrossRef]
- Collison, K.S.; Inglis, A.; Shibin, S.; Saleh, S.; Andres, B.; Ubungen, R.; Thiam, J.; Mata, P.; Al-Mohanna, F.A. Effect of developmental NMDAR antagonism with CGP 39551 on aspartame-induced hypothalamic and adrenal gene expression. PLoS ONE 2018, 13, e0194416. [Google Scholar] [CrossRef]
- Luoni, A.; Riva, M.A. MicroRNAs and psychiatric disorders: From aetiology to treatment. Pharmacol. Ther. 2016, 167, 13–27. [Google Scholar] [CrossRef]
- Cacabelos, R.; Torrellas, C.; Fernandez-Novoa, L.; Aliev, G. Neuroimmune crosstalk in CNS disorders: The histamine connection. Curr. Pharm. Des. 2016, 22, 819–848. [Google Scholar] [CrossRef]
- Rosmond, R.; Bouchard, C.; Björntorp, P. Allelic variants in the GABAAα6 receptor subunit gene (GABRA6) is associated with abdominal obesity and cortisol secretion. Int. J. Obesity 2002, 26, 938–941. [Google Scholar] [CrossRef]
- Lee, W.H.; Sonntag, W.E.; Lee, Y.W. Aging attenuates radiation-induced expression of pro-inflammatory mediators in rat brain. Neurosci. Lett. 2010, 476, 89–93. [Google Scholar] [CrossRef]
- Semple, B.D.; Kossmann, T.; Morganti-Kossmann, M.C. Role of Chemokines in CNS Health and Pathology: A Focus on the CCL2/CCR2 and CXCL8/CXCR2 Networks. J. Cereb. Blood Flow Metab. 2010, 30, 459–473. [Google Scholar] [CrossRef]
- He, Y.; Kim, J.; Tacconi, C.; Moody, J.; Dieterich, L.C.; Anzengruber, F.; Maul, J.-T.; Gousopoulos, E.; Restivo, G.; Levesque, M.P.; et al. Mediators of Capillary-to-Venule Conversion in the Chronic Inflammatory Skin Disease Psoriasis. J. Investig. Dermatol. 2022, 142, 3313–3326.e13. [Google Scholar] [CrossRef]
- Megat, S.; Mora, N.; Sanogo, J.; Catanese, A.; Alami, N.O.; Freischmidt, A.; Mingaj, X.; de Calbiac, H.; Muratet, F.; Dirrig-Grosch, S.; et al. Loss of nucleoporin Nup50 is a risk factor for amyotrophic lateral sclerosis. medRxiv 2021. [Google Scholar] [CrossRef]
- Jong, Y.-J.I.; Harmon, S.K.; O’Malley, K.L. GPCR Signaling from Intracellular Membranes. GPCRs Ther. Targets 2022, 1, 216–298. [Google Scholar] [CrossRef]
- Grace, A.A. Dopamine system dysregulation by the hippocampus: Implications for the pathophysiology and treatment of schizophrenia. Neuropharmacology 2012, 62, 1342–1348. [Google Scholar] [CrossRef]
- Deslauriers, J.; Larouche, A.; Sarret, P.; Grignon, S. Combination of prenatal immune challenge and restraint stress affects prepulse inhibition and dopaminergic/GABAergic markers. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2013, 45, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Woods, R.M.; Lorusso, J.M.; Potter, H.G.; Neill, J.C.; Glazier, J.D.; Hager, R. Maternal immune activation in rodent models: A systematic review of neurodevelopmental changes in gene expression and epigenetic modulation in the offspring brain. Neurosci. Biobehav. Rev. 2021, 129, 389–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-s.; Jiang, Y.-l.; Lu, L.; Feng, B.; Ma, X.; Zhang, K.; Guan, S.-y.; Yang, L.; Fan, Q.-y.; Zhu, X.-c.; et al. Activation of GIPR Exerts Analgesic and Anxiolytic-Like Effects in the Anterior Cingulate Cortex of Mice. Front. Endocrinol. 2022, 13, 887238. [Google Scholar] [CrossRef]
- Yamamoto, M.; Ben-Shlomo, A.; Kameda, H.; Fukuoka, H.; Deng, N.; Ding, Y.; Melmed, S. Somatostatin receptor subtype 5 modifies hypothalamic-pituitary-adrenal axis stress function. JCI Insight 2018, 3, 122932. [Google Scholar] [CrossRef] [PubMed]
- Masser, D.R.; Bixler, G.V.; Brucklacher, R.M.; Yan, H.; Giles, C.B.; Wren, J.D.; Sonntag, W.E.; Freeman, W.M. Hippocampal Subregions Exhibit Both Distinct and Shared Transcriptomic Responses to Aging and Nonneurodegenerative Cognitive Decline. J. Gerontol. Ser. A 2014, 69, 1311–1324. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, J.L.; Grant, M.; Norman, M.; Wang, X.P.; Moldovan, S.; de Mayo, F.J.; Brunicardi, C.; Kumar, U. Deficiency of somatostatin (SST) receptor type 5 (SSTR5) is associated with sexually dimorphic changes in the expression of SST and SST receptors in brain and pancreas. Mol. Cell. Endocrinol. 2004, 221, 105–119. [Google Scholar] [CrossRef]
- Hoenicka, J.; Garrido, E.; Ponce, G.; Rodríguez-Jiménez, R.; Martínez, I.; Rubio, G.; Jiménez-Arriero, M.Á.; Palomo, T. Sexually dimorphic interaction between the DRD1 and COMT genes in schizophrenia. Am. J. Med. Genet. Part B: Neuropsychiatr. Genet. 2010, 153B, 948–954. [Google Scholar] [CrossRef]
- Brivio, E.; Lopez, J.P.; Chen, A. Sex differences: Transcriptional signatures of stress exposure in male and female brains. Genes Brain Behav. 2020, 19, e12643. [Google Scholar] [CrossRef]
- Tchessalova, D.; Tronson, N.C. Enduring and Sex-specific Changes in Hippocampal Gene Expression after a Subchronic Immune Challenge. Neuroscience 2020, 428, 76–89. [Google Scholar] [CrossRef]
- Vied, C.; Ray, S.; Badger, C.-D.; Bundy, J.L.; Arbeitman, M.N.; Nowakowski, R.S. Transcriptomic analysis of the hippocampus from six inbred strains of mice suggests a basis for sex-specific susceptibility and severity of neurological disorders. J. Comp. Neurol. 2016, 524, 2696–2710. [Google Scholar] [CrossRef]
- McGann, J.C.; Spinner, M.A.; Garg, S.K.; Mullendorff, K.A.; Woltjer, R.L.; Mandel, G. The Genome-Wide Binding Profile for Human RE1 Silencing Transcription Factor Unveils a Unique Genetic Circuitry in Hippocampus. J. Neurosci. 2021, 41, 6582–6595. [Google Scholar] [CrossRef]
- Garcia-Manteiga, J.M.; D’Alessandro, R.; Meldolesi, J. News about the Role of the Transcription Factor REST in Neurons: From Physiology to Pathology. Int. J. Mol. Sci. 2020, 21, 235. [Google Scholar] [CrossRef]
- Chen, Z.; Fan, R.; Liang, J.; Xiao, Z.; Dang, J.; Zhao, J.; Weng, R.; Zhu, C.; Zheng, S.G.; Jiang, Y. NFIL3 deficiency alleviates EAE through regulating different immune cell subsets. J. Adv. Res. 2022, 39, 225–235. [Google Scholar] [CrossRef]
- Heavner, W.E.; Smith, S.E.P. Resolving the Synaptic versus Developmental Dichotomy of Autism Risk Genes. Trends Neurosci. 2020, 43, 227–241. [Google Scholar] [CrossRef]
- Jurk, D.; Wilson, C.; Passos, J.F.; Oakley, F.; Correia-Melo, C.; Greaves, L.; Saretzki, G.; Fox, C.; Lawless, C.; Anderson, R.; et al. Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nat. Commun. 2014, 5, 4172. [Google Scholar] [CrossRef]
- Herman, J.P.; Nawreen, N.; Smail, M.A.; Cotella, E.M. Brain mechanisms of HPA axis regulation: Neurocircuitry and feedback in context Richard Kvetnansky lecture. Stress 2020, 23, 617–632. [Google Scholar] [CrossRef]

| Enrichment Ratio 3 | |||||||
|---|---|---|---|---|---|---|---|
| GO:Identity 1 | Biological Process Name 2 | MxPF | PFxS | MxS | M | PF | S |
| GO:0043950 | positive reg. cAMP-mediated sig. * | 23.6 | 23.4 | 56.9 | 19.9 | 50.1 | |
| GO:0051281 | positive reg. release of seq. Ca | 21.5 | 33.4 | ||||
| GO:0006911 | phagocytosis, engulfment | 31.2 | |||||
| GO:0002548 | monocyte chemotaxis | 15.1 | 21.5 | ||||
| GO:0045744 | negative reg. GPCR sig. pathway * | 20.8 | |||||
| GO:0042119 | neutrophil activation | 18.7 | 15.9 | ||||
| GO:0071347 | cellular resp. interleukin-1 | 12.5 | 16.9 | ||||
| GO:1903169 | reg. Ca transmembrane transport | 16.2 | |||||
| GO:1904064 | positive reg. cation transport | 16.1 | |||||
| GO:0048247 | lymphocyte chemotaxis | 11.8 | 12.5 | 16.1 | |||
| GO:0060078 | regulation of postsynaptic potential | 15.4 | |||||
| GO:0070098 | chemokine-mediated sig. pathway | 9.2 | 12.5 | 14.7 | |||
| GO:0051279 | reg. release of seq. Ca | 12.5 | 10.6 | ||||
| GO:2000351 | reg. endothelial cell apoptosis * | 12.0 | |||||
| GO:0050810 | reg. steroid biosynthetic process | 11.8 | |||||
| GO:0051928 | positive reg. Ca ion transport | 10.7 | |||||
| GO:0002224 | Toll-like receptor sig. pathway | 9.8 | |||||
| GO:0061844 | antimicrobial humoral immune resp. | 9.6 | |||||
| GO:1904894 | positive reg. STAT cascade | 9.2 | |||||
| GO:0046427 | positive reg. JAK-STAT cascade | 9.2 | |||||
| GO:0071346 | cellular resp.interferon-γ | 8.8 | |||||
| GO:0071356 | cellular resp. tumor necrosis factor | 8.7 | |||||
| GO:0071774 | response to fibroblast growth factor | 8.5 | |||||
| GO:0051607 | defense resp. virus | 6.9 | 8.4 | ||||
| GO:0120032 | reg. plasma membrane projection | 8.2 | |||||
| GO:0071222 | cellular resp. lipopolysaccharide | 8.0 | 8.1 | 7.7 | |||
| GO:0007189 | adenylate cyclase-activate GPCR sig. | 7.9 | 7.0 | 6.7 | |||
| GO:0030595 | leukocyte chemotaxis | 7.3 | |||||
| GO:0097529 | myeloid leukocyte migration | 6.9 | |||||
| GO:0043547 | positive reg. GTPase activity | 5.6 | |||||
| GO:0046879 | hormone secretion | 4.8 | |||||
| KEGG | Enrichment Score | ||||||
|---|---|---|---|---|---|---|---|
| Identity 1 | Pathway Name 2 | MxF 3 | MxP | PxS | M | p | S |
| ssc04668 | TNF signaling * | −2.5 | 2.6 | −2.1 | |||
| ssc04657 | IL-17 signaling | −2.2 | 2.5 | −1.9 | |||
| ssc04933 | AGE-RAGE signaling in diabetes | −1.8 | −1.8 | 2.4 | |||
| ssc04064 | NF-kappa B signaling | −2.1 | 2.4 | −2.0 | |||
| ssc04742 | Taste transduction * | 2.3 | |||||
| ssc04620 | Toll-like receptor signaling | −2.0 | 2.2 | −2.3 | |||
| ssc04630 | JAK-STAT signaling | −2.1 | −1.9 | 2.2 | |||
| ssc05142 | Chagas disease * | −1.7 | 2.2 | −1.7 | |||
| ssc05332 | Graft-versus-host disease | 2.2 | 1.8 | ||||
| ssc04664 | Fc epsilon RI signaling | 2.1 | |||||
| ssc05140 | Leishmaniasis | −1.8 | 2.0 | 2.0 | −2.1 | ||
| ssc05150 | Staphylococcus aureus infection * | 1.9 | 1.9 | −2.1 | |||
| ssc05033 | Nicotine addiction * | 2.1 | |||||
| ssc04917 | Prolactin signaling | −1.9 | −1.8 | 2.1 | |||
| ssc03050 | Proteasome | −2.1 | |||||
| ssc04672 | Intestinal immune network for IgA | 1.9 | −2.1 | ||||
| ssc04662 | B cell receptor signaling | 2.1 | −1.8 | ||||
| ssc05416 | Viral myocarditis | 2.0 | −2.0 | ||||
| ssc04622 | RIG-I-like receptor signaling | −2.0 | 2.0 | −1.9 | |||
| ssc04623 | Cytosolic DNA-sensing | −1.9 | 2.0 | ||||
| ssc04658 | Th1 and Th2 cell differentiation | −1.8 | 2.0 | −1.8 | |||
| ssc05160 | Hepatitis C | 2.0 | |||||
| ssc04918 | Thyroid hormone synthesis * | −1.9 | |||||
| ssc04512 | ECM-receptor interaction | −1.9 | 1.9 | ||||
| ssc05162 | Measles | −1.8 | 1.9 | −1.8 | |||
| ssc04659 | Th17 cell differentiation | 1.9 | −1.9 | ||||
| ssc04115 | p53 signaling * | 1.9 | |||||
| ssc04650 | Natural killer cell cytotoxicity | 1.8 | |||||
| ssc04744 | Phototransduction | −1.8 | |||||
| ssc04726 | Serotonergic synapse * | −1.7 | |||||
| Signaling Process | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| and Gene Symbol | M 1 | F | p | S | MxF | MxP | FxS | PxS | MxS |
| Neuronal | |||||||||
| CALB1 | 3.0 | 7.0 | 2.0 | 4.1 | 2.4 | ||||
| CBLN3 | 5.0 | 1.5 | |||||||
| GABRA6 | 2.0 | 8.6 | 2.1 | ||||||
| GPR88 | 5.3 | ||||||||
| NPSR1 | −3.0 | 3.1 | 3.7 | 3.4 | −3.0 | −4.4 | −7.5 | ||
| NTS | −2.8 | 5.3 | −2.2 | ||||||
| Inflammatory | |||||||||
| CXCL2 | 7.1 | −3.1 | |||||||
| CXCL8 | 6.7 | 2.7 | 2.3 | ||||||
| SELE | 8.3 | ||||||||
| Hormonal | |||||||||
| GH1 | −4.3 | 5.6 | 2.5 | −3.3 | −4.7 | −6.1 | −6.4 | −7.5 | |
| PRL | −5.1 | 5.7 | 3.0 | 7.1 | −3.5 | −4.7 | −5.4 | −6.1 | −8.5 |
| SIX3 | 5.9 | ||||||||
| cAMP | |||||||||
| ADORA2A | 3.8 | ||||||||
| DRD1 | 4.4 | ||||||||
| DRD2 | 4.2 | ||||||||
| GIPR | −1.5 | −1.7 | |||||||
| PPP1R1B | 3.1 |
| Transcription | Number of Target Genes | |||||
|---|---|---|---|---|---|---|
| Factor 1 | MIAxPF | PFxSex | MIAxSex | MIA | PF | S |
| REST | 79 | 15 | 55 | 37 | ||
| CTBP2 | 64 | 35 | ||||
| STAT1 | 11 | 43 | ||||
| SUZ12 | 38 | 32 | 32 | 22 | 29 | |
| NFKB1 | 29 | |||||
| NFIL3 | 28 | |||||
| IRF1 | 26 | |||||
| TCF12 | 3 | 23 | ||||
| EP300 | 19 | |||||
| CHD1 | 18 | |||||
| ETS2, NR3C1 | 18 | |||||
| NFIC | 18 | |||||
| STAT2 | 12 | 14 | 17 | |||
| RXRA | 15 | |||||
| TEAD4 | 14 | |||||
| SMARCA4 | 7 | 13 | ||||
| TP53 | 6 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez-Zas, S.L.; Southey, B.R.; Rymut, H.E.; Rund, L.A.; Johnson, R.W. Hippocampal Changes Elicited by Metabolic and Inflammatory Stressors following Prenatal Maternal Infection. Genes 2023, 14, 77. https://doi.org/10.3390/genes14010077
Rodriguez-Zas SL, Southey BR, Rymut HE, Rund LA, Johnson RW. Hippocampal Changes Elicited by Metabolic and Inflammatory Stressors following Prenatal Maternal Infection. Genes. 2023; 14(1):77. https://doi.org/10.3390/genes14010077
Chicago/Turabian StyleRodriguez-Zas, Sandra L., Bruce R. Southey, Haley E. Rymut, Laurie A. Rund, and Rodney W. Johnson. 2023. "Hippocampal Changes Elicited by Metabolic and Inflammatory Stressors following Prenatal Maternal Infection" Genes 14, no. 1: 77. https://doi.org/10.3390/genes14010077
APA StyleRodriguez-Zas, S. L., Southey, B. R., Rymut, H. E., Rund, L. A., & Johnson, R. W. (2023). Hippocampal Changes Elicited by Metabolic and Inflammatory Stressors following Prenatal Maternal Infection. Genes, 14(1), 77. https://doi.org/10.3390/genes14010077






