The Genetic Basis of Anthocyanin Acylation in North American Grapes (Vitis spp.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Sampling
2.2. Phenotyping and Data Analysis
2.3. rhAmpSeq Genotyping and Quality Control
2.4. Genetic Map Construction
2.5. QTL Analysis
3. Results
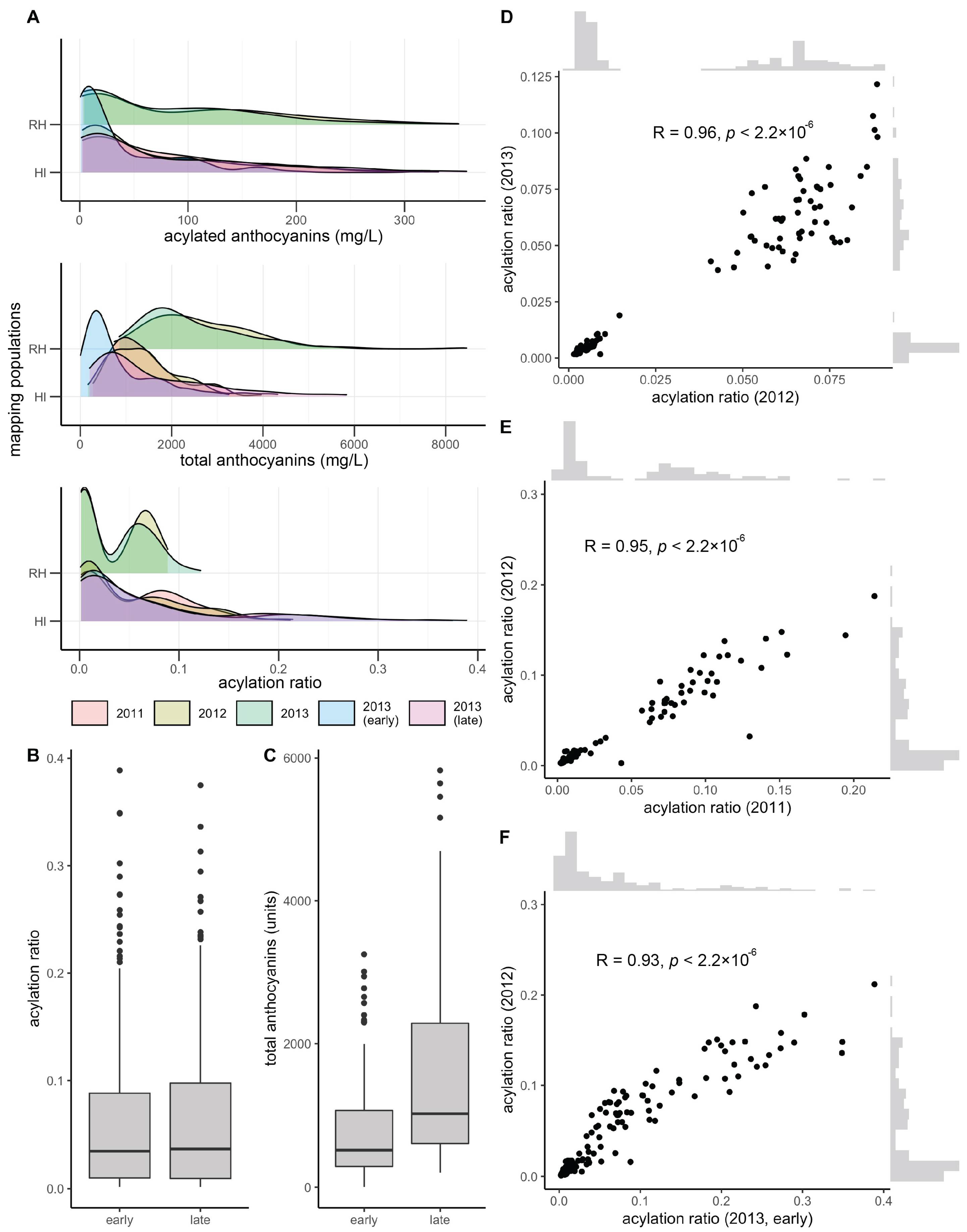
3.1. Mapping Population Showed Presence/Absence Patterns for Anthocyanin Acylation
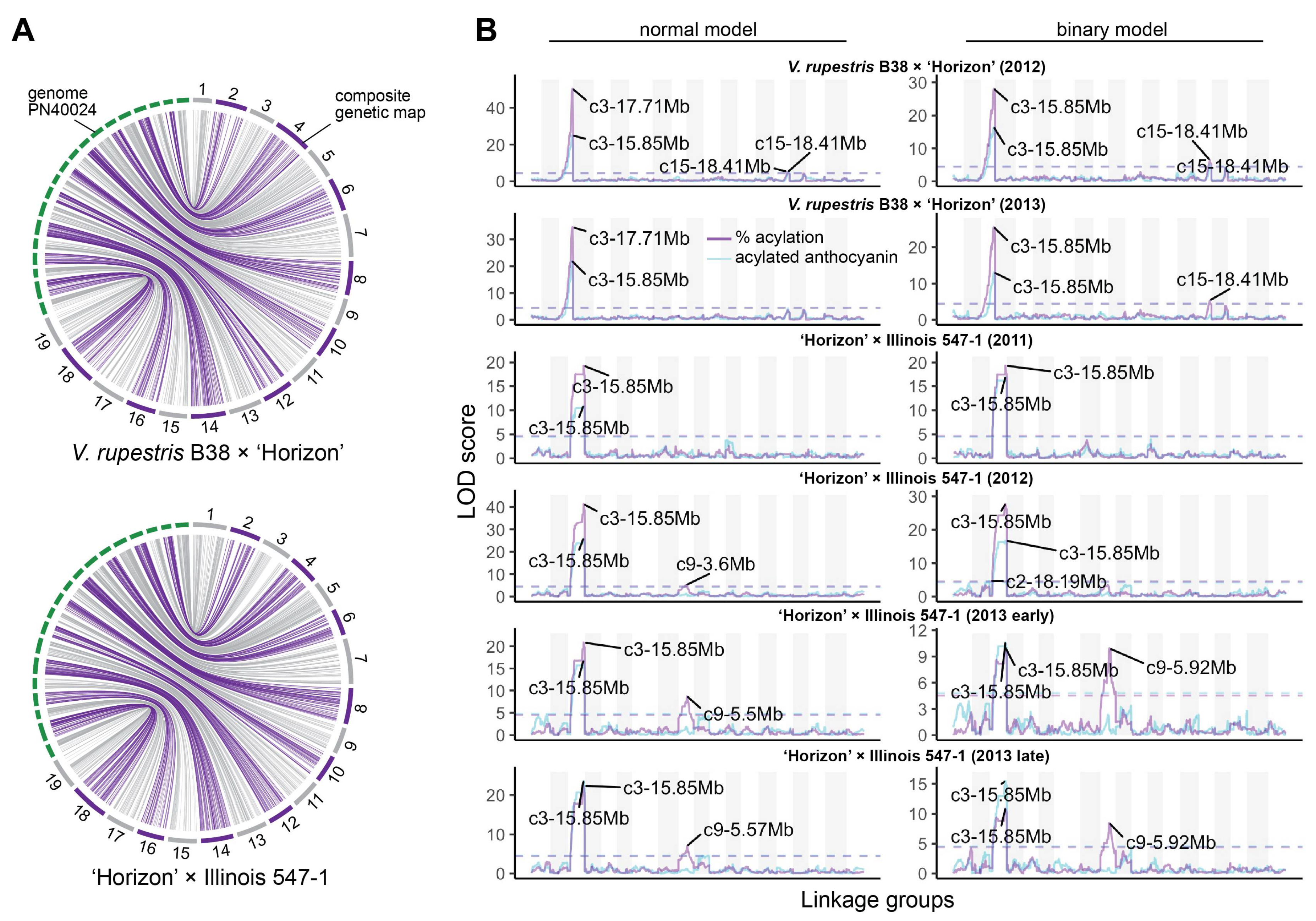
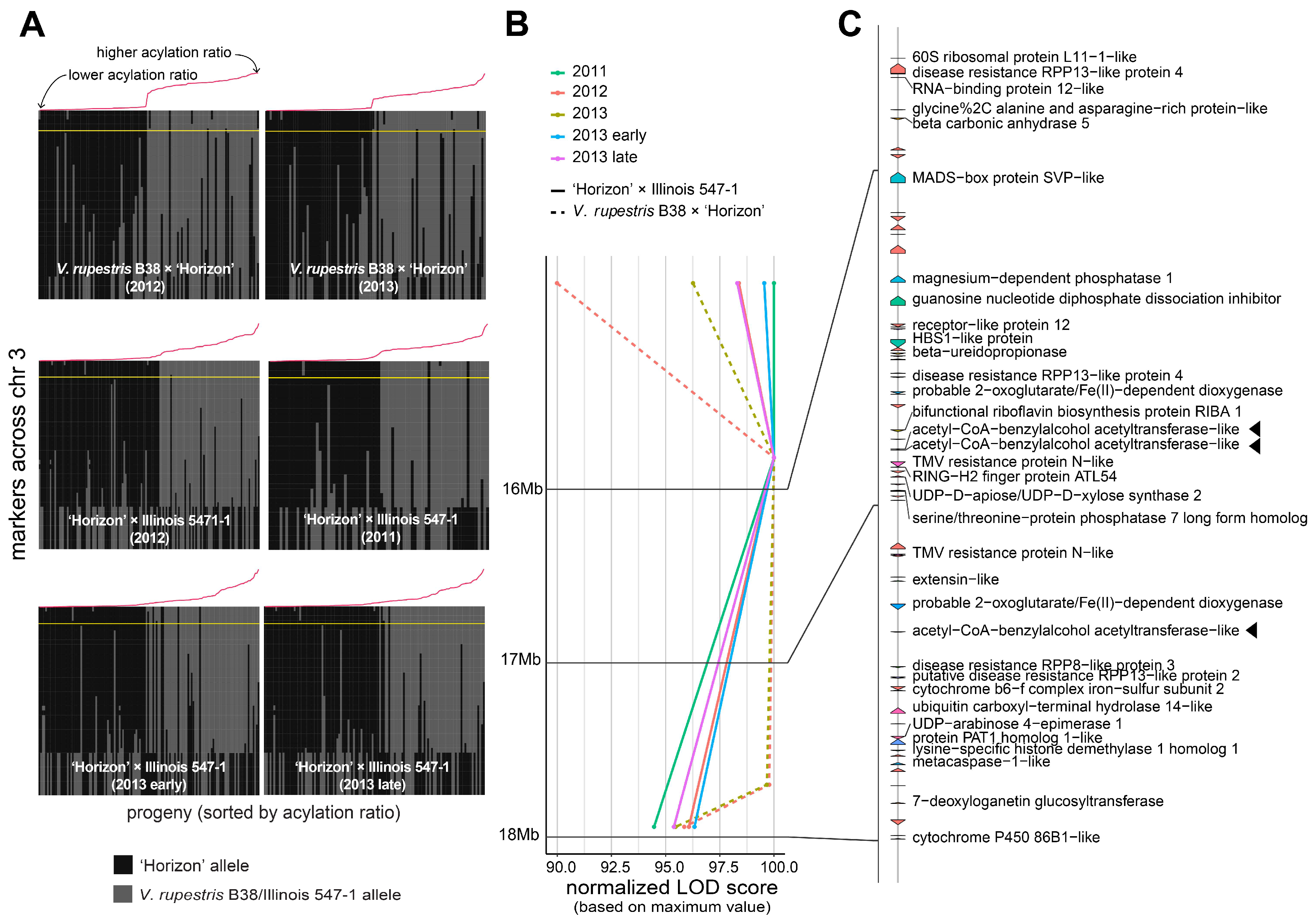
3.2. Stable QTL for Acylation Ratio across Genetic Backgrounds
3.3. Candidate Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waterhouse, A.L.; Sacks, G.L.; Jeffery, D.W. Anthocyanins. In Understanding Wine Chemistry; Waterhouse, A.L., Sacks, G.L., Jeffery, D.W., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2016; pp. 131–139. ISBN 9781118730720. [Google Scholar]
- Boss, P.K.; Davies, C. Molecular Biology of Anthocyanin Accumulation in Grape Berries. In Grapevine Molecular Physiology & Biotechnology; Roubelakis-Angelakis, K.A., Ed.; Springer: Dordrecht, The Netherlands, 2009; pp. 263–292. ISBN 9789048123056. [Google Scholar]
- He, F.; Liang, N.-N.; Mu, L.; Pan, Q.-H.; Wang, J.; Reeves, M.J.; Duan, C.-Q. Anthocyanins and Their Variation in Red Wines I. Monomeric Anthocyanins and Their Color Expression. Molecules 2012, 17, 1571–1601. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.-L.; Yu, Y.-Q.; Chen, Z.-J.; Wen, G.-S.; Wei, F.-G.; Zheng, Q.; Wang, C.-D.; Xiao, X.-L. Stability-Increasing Effects of Anthocyanin Glycosyl Acylation. Food Chem. 2017, 214, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Pomar, F.; Novo, M.; Masa, A. Varietal Differences among the Anthocyanin Profiles of 50 Red Table Grape Cultivars Studied by High Performance Liquid Chromatography. J. Chromatogr. A 2005, 1094, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Yang, Y.; Cheng, L.; Zhong, G.-Y. Polyphenolic Composition and Content in the Ripe Berries of Wild Vitis Species. Food Chem. 2012, 132, 730–738. [Google Scholar] [CrossRef]
- Charron, C.S.; Kurilich, A.C.; Clevidence, B.A.; Simon, P.W.; Harrison, D.J.; Britz, S.J.; Baer, D.J.; Novotny, J.A. Bioavailability of Anthocyanins from Purple Carrot Juice: Effects of Acylation and Plant Matrix. J. Agric. Food Chem. 2009, 57, 1226–1230. [Google Scholar] [CrossRef] [PubMed]
- Kurilich, A.C.; Clevidence, B.A.; Britz, S.J.; Simon, P.W.; Novotny, J.A. Plasma and Urine Responses Are Lower for Acylated vs Nonacylated Anthocyanins from Raw and Cooked Purple Carrots. J. Agric. Food Chem. 2005, 53, 6537–6542. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Suarez, J.M.; Giampieri, F.; Tulipani, S.; Casoli, T.; Di Stefano, G.; González-Paramás, A.M.; Santos-Buelga, C.; Busco, F.; Quiles, J.L.; Cordero, M.D.; et al. One-Month Strawberry-Rich Anthocyanin Supplementation Ameliorates Cardiovascular Risk, Oxidative Stress Markers and Platelet Activation in Humans. J. Nutr. Biochem. 2014, 25, 289–294. [Google Scholar] [CrossRef]
- Li, S.; Wu, B.; Fu, W.; Reddivari, L. The Anti-Inflammatory Effects of Dietary Anthocyanins against Ulcerative Colitis. Int. J. Mol. Sci. 2019, 20, 2588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morais, C.A.; de Rosso, V.V.; Estadella, D.; Pisani, L.P. Anthocyanins as Inflammatory Modulators and the Role of the Gut Microbiota. J. Nutr. Biochem. 2016, 33, 1–7. [Google Scholar] [CrossRef]
- Fournier-Level, A.; Le Cunff, L.; Gomez, C.; Doligez, A.; Ageorges, A.; Roux, C.; Bertrand, Y.; Souquet, J.-M.; Cheynier, V.; This, P. Quantitative Genetic Bases of Anthocyanin Variation in Grape (Vitis Vinifera L. Ssp. Sativa) Berry: A Quantitative Trait Locus to Quantitative Trait Nucleotide Integrated Study. Genetics 2009, 183, 1127–1139. [Google Scholar] [CrossRef] [Green Version]
- Rinaldo, A.R.; Cavallini, E.; Jia, Y.; Moss, S.M.A.; McDavid, D.A.J.; Hooper, L.C.; Robinson, S.P.; Tornielli, G.B.; Zenoni, S.; Ford, C.M.; et al. A Grapevine Anthocyanin Acyltransferase, Transcriptionally Regulated by VvMYBA, Can Produce Most Acylated Anthocyanins Present in Grape Skins. Plant Physiol. 2015, 169, 1897–1916. [Google Scholar] [CrossRef] [PubMed]
- Massonnet, M.; Fasoli, M.; Tornielli, G.B.; Altieri, M.; Sandri, M.; Zuccolotto, P.; Paci, P.; Gardiman, M.; Zenoni, S.; Pezzotti, M. Ripening Transcriptomic Program in Red and White Grapevine Varieties Correlates with Berry Skin Anthocyanin Accumulation. Plant Physiol. 2017, 174, 2376–2396. [Google Scholar] [CrossRef] [Green Version]
- Costantini, L.; Malacarne, G.; Lorenzi, S.; Troggio, M.; Mattivi, F.; Moser, C.; Grando, M.S. New Candidate Genes for the Fine Regulation of the Colour of Grapes. J. Exp. Bot. 2015, 66, 4427–4440. [Google Scholar] [CrossRef] [PubMed]
- Reisch, B.; Robinson, W.B.; Kimball, K.; Pool, R.; Watson, J. “Horizon” Grape. HortScience 1983, 18, 108–109. [Google Scholar]
- Manns, D.C.; Mansfield, A.K. A Core-Shell Column Approach to a Comprehensive High-Performance Liquid Chromatography Phenolic Analysis of Vitis vinifera L. and Interspecific Hybrid Grape Juices, Wines, and Other Matrices Following Either Solid Phase Extraction or Direct Injection. J. Chromatogr. A 2012, 1251, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Hyma, K.E.; Barba, P.; Wang, M.; Londo, J.P.; Acharya, C.B.; Mitchell, S.E.; Sun, Q.; Reisch, B.; Cadle-Davidson, L. Heterozygous Mapping Strategy (HetMappS) for High Resolution Genotyping-By-Sequencing Markers: A Case Study in Grapevine. PLoS ONE 2015, 10, e0134880. [Google Scholar] [CrossRef] [Green Version]
- Zou, C.; Karn, A.; Reisch, B.; Nguyen, A.; Sun, Y.; Bao, Y.; Campbell, M.S.; Church, D.; Williams, S.; Xu, X.; et al. Haplotyping the Vitis Collinear Core Genome with rhAmpSeq Improves Marker Transferability in a Diverse Genus. Nat. Commun. 2020, 11, 413. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Karn, A.; Cadle-Davidson, L.; Zou, C.; Underhill, A.; Atkins, P.; Treiber, E.; Voytas, D.; Clark, M. Fine Mapping of Leaf Trichome Density Revealed a 747-Kb Region on Chromosome 1 in Cold-Hardy Hybrid Wine Grape Populations. Front. Plant Sci. 2021, 12, 587640. [Google Scholar] [CrossRef]
- Reshef, N.; Karn, A.; Manns, D.C.; Mansfield, A.K.; Cadle-Davidson, L.; Reisch, B.; Sacks, G.L. Stable QTL for Malate Levels in Ripe Fruit and Their Transferability across Vitis Species. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Rastas, P. Lep-MAP3: Robust Linkage Mapping Even for Low-Coverage Whole Genome Sequencing Data. Bioinformatics 2017, 33, 3726–3732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canaguier, A.; Grimplet, J.; Di Gaspero, G.; Scalabrin, S.; Duchêne, E.; Choisne, N.; Mohellibi, N.; Guichard, C.; Rombauts, S.; Le Clainche, I.; et al. A New Version of the Grapevine Reference Genome Assembly (12X.v2) and of Its Annotation (VCost.v3). Genom. Data 2017, 14, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Broman, K.W.; Wu, H.; Sen, S.; Churchill, G.A. R/qtl: QTL Mapping in Experimental Crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Fresnedo-Ramírez, J.; Wang, M.; Cote, L.; Schweitzer, P.; Barba, P.; Takacs, E.M.; Clark, M.; Luby, J.; Manns, D.C.; et al. A Next-Generation Marker Genotyping Platform (AmpSeq) in Heterozygous Crops: A Case Study for Marker-Assisted Selection in Grapevine. Hortic. Res. 2016, 3, 16002. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Fan, X.; Zhang, Y.; Jiang, J.; Sun, H.; Liu, C. Transcriptome Analysis of Genes Involved in Anthocyanins Biosynthesis and Transport in Berries of Black and White Spine Grapes (Vitis davidii). Hereditas 2016, 153, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulsmeyer, M.N.; Brown, P.J.; Juvik, J.A. Discovery of Anthocyanin Acyltransferase1 (AAT1) in Maize Using Genotyping-by-Sequencing (GBS). G3 2018, 8, 3669–3678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White Grapes Arose through the Mutation of Two Similar and Adjacent Regulatory Genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Association of VvmybA1 Gene Expression with Anthocyanin Production in Grape (Vitis vinifera) Skin-Color Mutants. J. Jpn. Soc. Hotic. Sci. 2005, 74, 196–203. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Li, S.; Jiang, J.; Tang, X.; Fan, X.; Zhang, Y.; Liu, J.; Liu, C. New Quantitative Trait Locus (QTLs) and Candidate Genes Associated with the Grape Berry Color Trait Identified Based on a High-Density Genetic Map. BMC Plant Biol. 2020, 20, 302. [Google Scholar] [CrossRef] [PubMed]
- Cantu, D.; Walker, M.A. (Eds.) The Grape Genome; Compendium of Plant Genomes; Springer: Cham, Switzerland, 2019. [Google Scholar]
| Chr | V. rupestris B38 × ‘Horizon’ | ‘Horizon’ × Illinois 547-1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Markers | Size (cM) | Average Gap (cM) | Maximum Gap (cM) | Recomb. Rate (cM/Mb) | Markers | Size (cM) | Average Gap (cM) | Maximum Gap (cM) | Recomb. Rate (cM/Mb) | |
| 1 | 42 | 37.30 | 0.91 | 4.83 | 0.98 | 85 | 62.34 | 0.72 | 4.97 | 1.99 |
| 2 | 40 | 62.51 | 1.60 | 12.0 | 2.84 | 50 | 56.82 | 1.16 | 12.0 | 4.31 |
| 3 | 42 | 51.08 | 1.25 | 5.13 | 3.33 | 40 | 55.70 | 1.43 | 18.5 | 3.70 |
| 4 | 64 | 75.39 | 1.20 | 6.35 | 0.56 | 68 | 54.52 | 0.81 | 5.46 | 1.88 |
| 5 | 81 | 61.89 | 0.77 | 4.16 | 1.97 | 90 | 49.94 | 0.56 | 3.13 | 0.93 |
| 6 | 56 | 66.27 | 1.20 | 5.43 | 3.08 | 65 | 49.49 | 0.77 | 6.01 | 0.11 |
| 7 | 74 | 85.58 | 1.17 | 8.31 | 1.69 | 93 | 84.12 | 0.91 | 9.08 | 2.94 |
| 8 | 76 | 67.94 | 0.91 | 5.82 | 1.79 | 85 | 66.60 | 0.79 | 4.40 | 2.00 |
| 9 | 47 | 53.88 | 1.17 | 8.64 | 1.86 | 46 | 49.14 | 1.09 | 8.80 | 5.00 |
| 10 | 51 | 62.53 | 1.25 | 6.42 | 2.35 | 45 | 51.09 | 1.16 | 8.30 | 1.91 |
| 11 | 47 | 62.28 | 1.35 | 6.53 | 3.96 | 47 | 52.77 | 1.15 | 5.56 | 5.25 |
| 12 | 59 | 60.65 | 1.05 | 9.31 | 2.27 | 55 | 49.23 | 0.91 | 3.62 | 4.03 |
| 13 | 67 | 65.30 | 0.99 | 10.9 | 1.73 | 67 | 56.79 | 0.86 | 5.76 | 0.94 |
| 14 | 84 | 68.54 | 0.83 | 4.60 | 2.24 | 78 | 60.07 | 0.78 | 3.58 | 2.46 |
| 15 | 43 | 55.54 | 1.32 | 8.24 | 4.05 | 31 | 53.02 | 1.77 | 8.51 | 5.03 |
| 16 | 47 | 59.24 | 1.29 | 7.20 | 2.62 | 42 | 50.41 | 1.23 | 5.32 | 2.10 |
| 17 | 53 | 67.88 | 1.31 | 10.7 | 1.19 | 52 | 53.58 | 1.05 | 6.89 | 1.49 |
| 18 | 67 | 82.16 | 1.24 | 8.18 | 1.27 | 76 | 69.94 | 0.93 | 6.58 | 3.24 |
| 19 | 52 | 60.62 | 1.19 | 6.22 | 2.80 | 56 | 56.59 | 1.03 | 8.10 | 2.88 |
| Total | 1092 | 1206.58 | 1.16 | 7.31 | 2.24 | 1171 | 1082.16 | 1.01 | 7.08 | 2.75 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karn, A.; Diaz-Garcia, L.; Reshef, N.; Zou, C.; Manns, D.C.; Cadle-Davidson, L.; Mansfield, A.K.; Reisch, B.I.; Sacks, G.L. The Genetic Basis of Anthocyanin Acylation in North American Grapes (Vitis spp.). Genes 2021, 12, 1962. https://doi.org/10.3390/genes12121962
Karn A, Diaz-Garcia L, Reshef N, Zou C, Manns DC, Cadle-Davidson L, Mansfield AK, Reisch BI, Sacks GL. The Genetic Basis of Anthocyanin Acylation in North American Grapes (Vitis spp.). Genes. 2021; 12(12):1962. https://doi.org/10.3390/genes12121962
Chicago/Turabian StyleKarn, Avinash, Luis Diaz-Garcia, Noam Reshef, Cheng Zou, David C. Manns, Lance Cadle-Davidson, Anna Katharine Mansfield, Bruce I. Reisch, and Gavin L. Sacks. 2021. "The Genetic Basis of Anthocyanin Acylation in North American Grapes (Vitis spp.)" Genes 12, no. 12: 1962. https://doi.org/10.3390/genes12121962







