Biomarkers to Monitor Adherence to Gluten-Free Diet by Celiac Disease Patients: Gluten Immunogenic Peptides and Urinary miRNAs
Abstract
:1. Introduction
2. Monitoring Adherence to GFD
3. Production of GIPs by the Hydrolysis of Gluten
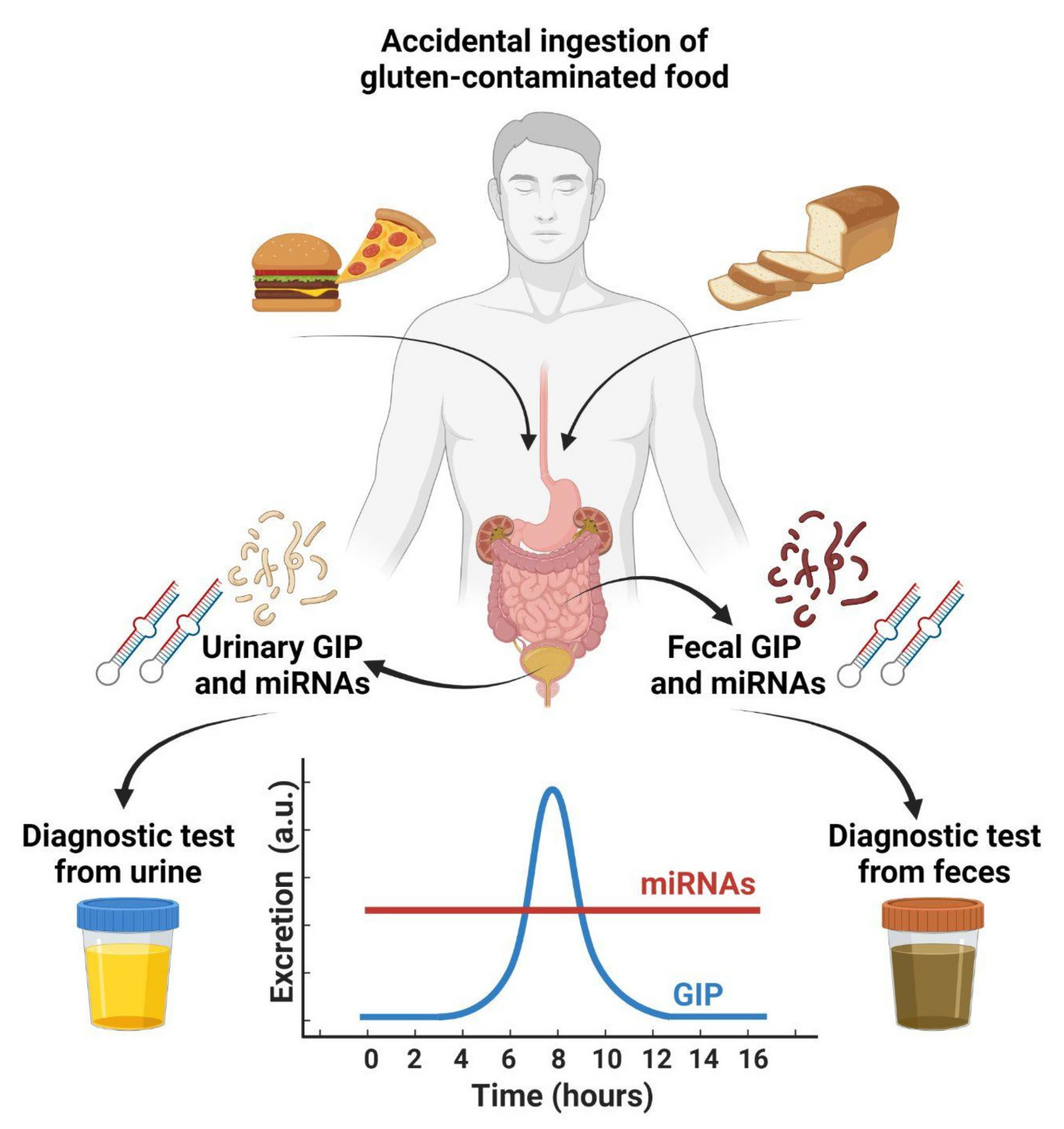
4. Detection of GIPs in Feces
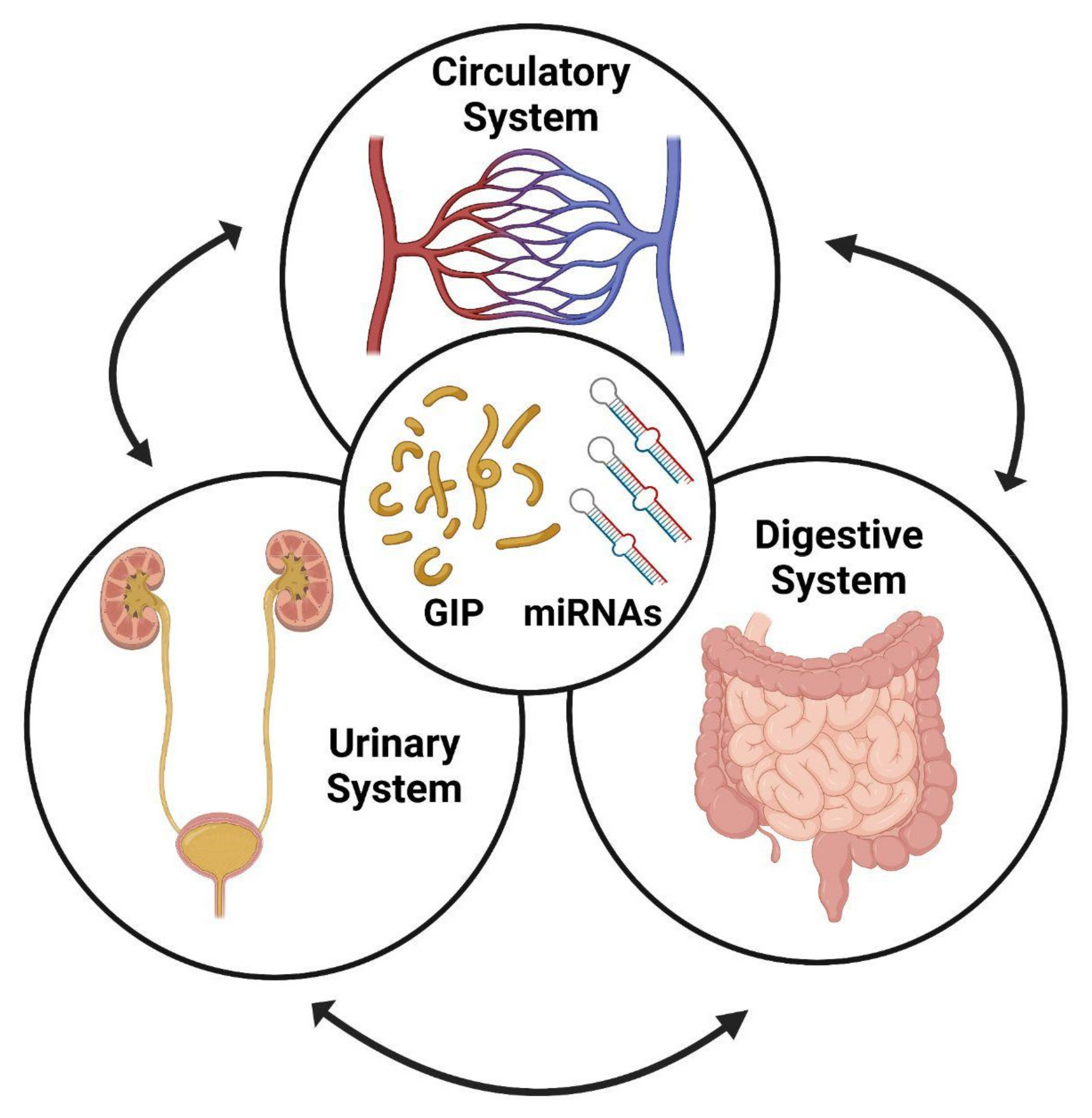
5. Detection of GIPs in Urines
6. Urinary miRNAs as Biomarkers of Kidney Diseases
7. Diagnosis of CD and Adherence to GFD by Circulating miRNAs
8. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gujral, N.; Freeman, H.J.; Thomson, A.B. Celiac disease: Prevalence, diagnosis, pathogenesis and treatment. World J. Gastroenterol. 2012, 18, 6036–6059. [Google Scholar] [CrossRef] [PubMed]
- Mariné, M.; Farre, C.; Alsina, M.; Vilar, P.; Cortijo, M.; Salas, A.; Fernández-Bañares, F.; Rosinach, M.; Santaolalla, R.; Loras, C.; et al. The prevalence of coeliac disease is significantly higher in children compared with adults. Aliment. Pharmacol. Ther. 2011, 33, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Lindfors, K.; Ciacci, C.; Kurppa, K.; Lundin, K.E.A.; Makharia, G.K.; Mearin, M.L.; Murray, J.A.; Verdu, E.F.; Kaukinen, K. Coeliac disease. Nat. Rev. Dis. Primers 2019, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Balakireva, A.V.; Zamyatnin, A.A. Properties of Gluten Intolerance: Gluten Structure, Evolution, Pathogenicity and Detoxification Capabilities. Nutrients 2016, 8, 644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, S.; Perez-Andres, J.; Martinez-Blanco, H.; Ferrero, M.A.; Vaquero, L.; Vivas, S.; Casqueiro, J.; Rodriguez-Aparicio, L.B. The human digestive tract has proteases capable of gluten hydrolysis. Mol. Metab. 2017, 6, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Urade, R.; Sato, N.; Sugiyama, M. Gliadins from wheat grain: An overview, from primary structure to nanostructures of aggregates. Biophys. Rev. 2018, 10, 435–443. [Google Scholar] [CrossRef] [Green Version]
- Arentz-Hansen, H.; Fleckenstein, B.; Molberg, O.; Scott, H.; Koning, F.; Jung, G.; Roepstorff, P.; Lundin, K.E.; Sollid, L.M. The molecular basis for oat intolerance in patients with celiac disease. PLoS Med. 2004, 1, e1. [Google Scholar] [CrossRef]
- Cebolla, A.; Moreno, M.L.; Coto, L.; Sousa, C. Gluten Immunogenic Peptides as Standard for the Evaluation of Potential Harmful Prolamin Content in Food and Human Specimen. Nutrients 2018, 10, 1927. [Google Scholar] [CrossRef] [Green Version]
- Janssen, F.; Pauly, A.; Rombouts, I.; Jansens, K.J.A.; Deleu, L.J.; Delcour, J.A. Proteins of Amaranth (Amaranthus spp.), Buckwheat (Fagopyrum spp.), and Quinoa (Chenopodium spp.): A Food Science and Technology Perspective. Compr. Rev. Food Sci. Food Saf. 2017, 16, 39–58. [Google Scholar] [CrossRef]
- Schuppan, D.; Junker, Y.; Barisani, D. Celiac disease: From pathogenesis to novel therapies. Gastroenterology 2009, 137, 1912–1933. [Google Scholar] [CrossRef]
- Kagnoff, M.F. Celiac disease: Pathogenesis of a model immunogenetic disease. J. Clin. Investig. 2007, 117, 41–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comino, I.; Real, A.; Vivas, S.; Siglez, M.A.; Caminero, A.; Nistal, E.; Casqueiro, J.; Rodriguez-Herrera, A.; Cebolla, A.; Sousa, C. Monitoring of gluten-free diet compliance in celiac patients by assessment of gliadin 33-mer equivalent epitopes in feces. Am. J. Clin. Nutr. 2012, 95, 670–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amarri, S.; Alvisi, P.; De Giorgio, R.; Gelli, M.C.; Cicola, R.; Tovoli, F.; Sassatelli, R.; Caio, G.; Volta, U. Antibodies to deamidated gliadin peptides: An accurate predictor of coeliac disease in infancy. J. Clin. Immunol. 2013, 33, 1027–1030. [Google Scholar] [CrossRef] [PubMed]
- Guandalini, S.; Assiri, A. Celiac disease: A review. JAMA Pediatr. 2014, 168, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Silvester, J.A.; Kurada, S.; Szwajcer, A.; Kelly, C.P.; Leffler, D.A.; Duerksen, D.R. Tests for Serum Transglutaminase and Endomysial Antibodies Do Not Detect Most Patients with Celiac Disease and Persistent Villous Atrophy on Gluten-Free Diets: A Meta-Analysis. Gastroenterology 2017, 153, 689–701.e1. [Google Scholar] [CrossRef]
- Husby, S.; Koletzko, S.; Korponay-Szabó, I.R.; Mearin, M.L.; Phillips, A.; Shamir, R.; Troncone, R.; Giersiepen, K.; Branski, D.; Catassi, C.; et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 136–160. [Google Scholar] [CrossRef]
- Husby, S.; Koletzko, S.; Korponay-Szabó, I.; Kurppa, K.; Mearin, M.L.; Ribes-Koninckx, C.; Shamir, R.; Troncone, R.; Auricchio, R.; Castillejo, G.; et al. European Society Paediatric Gastroenterology, Hepatology and Nutrition Guidelines for Diagnosing Coeliac Disease 2020. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 141–156. [Google Scholar] [CrossRef] [Green Version]
- Green, P.H.; Lebwohl, B.; Greywoode, R. Celiac disease. J. Allergy Clin. Immunol. 2015, 135, 1099–1106, quiz 1107. [Google Scholar] [CrossRef]
- Singh, A.; Verma, A.K.; Das, P.; Prakash, S.; Pramanik, R.; Nayak, B.; Datta Gupta, S.; Sreenivas, V.; Kumar, L.; Ahuja, V.; et al. Non-immunological biomarkers for assessment of villous abnormalities in patients with celiac disease. J. Gastroenterol. Hepatol. 2020, 35, 438–445. [Google Scholar] [CrossRef]
- Sugai, E.; Nachman, F.; Vaquez, H.; Gonzalez, A.; Andrenacci, P.; Czech, A.; Niveloni, S.; Mazure, R.; Smecuol, E.; Cabanne, A.; et al. Dynamics of celiac disease-specific serology after initiation of a gluten-free diet and use in the assessment of compliance with treatment. Dig. Liver Dis. 2010, 42, 352–358. [Google Scholar] [CrossRef]
- Fasano, A.; Catassi, C. Current approaches to diagnosis and treatment of celiac disease: An evolving spectrum. Gastroenterology 2001, 120, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, L.M.; Corbett, G.; Currie, E.; Lee, J.; Sweeney, N.; Woodward, J.M. Optimising delivery of care in coeliac disease-comparison of the benefits of repeat biopsy and serological follow-up. Aliment. Pharmacol. Ther. 2013, 38, 1278–1291. [Google Scholar] [CrossRef] [PubMed]
- Felli, C.; Baldassarre, A.; Uva, P.; Alisi, A.; Cangelosi, D.; Ancinelli, M.; Caruso, M.; Paolini, A.; Montano, A.; Silano, M.; et al. Circulating microRNAs as novel non-invasive biomarkers of paediatric celiac disease and adherence to gluten-free diet. EBioMedicine 2022, 76, 103851. [Google Scholar] [CrossRef] [PubMed]
- Bascunan, K.A.; Perez-Bravo, F.; Gaudioso, G.; Vaira, V.; Roncoroni, L.; Elli, L.; Monguzzi, E.; Araya, M. A miRNA-Based Blood and Mucosal Approach for Detecting and Monitoring Celiac Disease. Dig. Dis. Sci. 2020, 65, 1982–1991. [Google Scholar] [CrossRef] [PubMed]
- Buoli Comani, G.; Panceri, R.; Dinelli, M.; Biondi, A.; Mancuso, C.; Meneveri, R.; Barisani, D. miRNA-regulated gene expression differs in celiac disease patients according to the age of presentation. Genes Nutr. 2015, 10, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, I.L.; de Almeida, R.C.; Modderman, R.; Stachurska, A.; Dekens, J.; Barisani, D.; Meijer, C.R.; Roca, M.; Martinez-Ojinaga, E.; Shamir, R.; et al. Circulating miRNAs as Potential Biomarkers for Celiac Disease Development. Front. Immunol. 2021, 12, 734763. [Google Scholar] [CrossRef]
- Domsa, E.M.; Berindan-Neagoe, I.; Budisan, L.; Braicu, C.; Para, I.; Tantau, A.I.; Orasan, O.H.; Ciobanu, L.; Pop, T.A.; Filip, G.A.; et al. Expression of Selected Genes and Circulating microRNAs in Patients with Celiac Disease. Medicina 2022, 58, 180. [Google Scholar] [CrossRef]
- Wieser, H.; Ruiz-Carnicer, A.; Segura, V.; Comino, I.; Sousa, C. Challenges of Monitoring the Gluten-Free Diet Adherence in the Management and Follow-Up of Patients with Celiac Disease. Nutrients 2021, 13, 2274. [Google Scholar] [CrossRef]
- See, J.A.; Kaukinen, K.; Makharia, G.K.; Gibson, P.R.; Murray, J.A. Practical insights into gluten-free diets. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 580–591. [Google Scholar] [CrossRef]
- Silvester, J.A.; Comino, I.; Kelly, C.P.; Sousa, C.; Duerksen, D.R.; DOGGIE BAG Study Group. Most Patients with Celiac Disease on Gluten-Free Diets Consume Measurable Amounts of Gluten. Gastroenterology 2020, 158, 1497–1499.e1. [Google Scholar] [CrossRef]
- Syage, J.A.; Kelly, C.P.; Dickason, M.A.; Ramirez, A.C.; Leon, F.; Dominguez, R.; Sealey-Voyksner, J.A. Determination of gluten consumption in celiac disease patients on a gluten-free diet. Am. J. Clin. Nutr. 2018, 107, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Comino, I.; Fernández-Bañares, F.; Esteve, M.; Ortigosa, L.; Castillejo, G.; Fambuena, B.; Ribes-Koninckx, C.; Sierra, C.; Rodríguez-Herrera, A.; Salazar, J.C.; et al. Corrigendum: Fecal Gluten Peptides Reveal Limitations of Serological Tests and Food Questionnaires for Monitoring Gluten-Free Diet in Celiac Disease Patients. Am. J. Gastroenterol. 2017, 112, 1208. [Google Scholar] [CrossRef] [PubMed]
- Monzani, A.; Rapa, A.; Fonio, P.; Tognato, E.; Panigati, L.; Oderda, G. Use of deamidated gliadin peptide antibodies to monitor diet compliance in childhood celiac disease. J. Pediatr. Gastroenterol. Nutr. 2011, 53, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Volta, U.; Granito, A.; Parisi, C.; Fabbri, A.; Fiorini, E.; Piscaglia, M.; Tovoli, F.; Grasso, V.; Muratori, P.; Pappas, G.; et al. Deamidated gliadin peptide antibodies as a routine test for celiac disease: A prospective analysis. J. Clin. Gastroenterol. 2010, 44, 186–190. [Google Scholar] [CrossRef]
- Mills, J.R.; Murray, J.A. Contemporary celiac disease diagnosis: Is a biopsy avoidable? Curr. Opin. Gastroenterol. 2016, 32, 80–85. [Google Scholar] [CrossRef]
- Leffler, D.A.; Edwards-George, J.; Dennis, M.; Schuppan, D.; Cook, F.; Franko, D.L.; Blom-Hoffman, J.; Kelly, C.P. Factors that influence adherence to a gluten-free diet in adults with celiac disease. Dig. Dis. Sci. 2008, 53, 1573–1581. [Google Scholar] [CrossRef]
- Laurikka, P.; Salmi, T.; Collin, P.; Huhtala, H.; Maki, M.; Kaukinen, K.; Kurppa, K. Gastrointestinal Symptoms in Celiac Disease Patients on a Long-Term Gluten-Free Diet. Nutrients 2016, 8, 429. [Google Scholar] [CrossRef] [Green Version]
- Jabri, B.; Kasarda, D.D.; Green, P.H. Innate and adaptive immunity: The yin and yang of celiac disease. Immunol. Rev. 2005, 206, 219–231. [Google Scholar] [CrossRef]
- Shan, L.; Molberg, Ø.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef] [Green Version]
- Stefanolo, J.P.; Talamo, M.; Dodds, S.; de la Paz Temprano, M.; Costa, A.F.; Moreno, M.L.; Pinto-Sanchez, M.I.; Smecuol, E.; Vazquez, H.; Gonzalez, A.; et al. Real-World Gluten Exposure in Patients with Celiac Disease on Gluten-Free Diets, Determined From Gliadin Immunogenic Peptides in Urine and Fecal Samples. Clin. Gastroenterol. Hepatol. 2021, 19, 484–491.e1. [Google Scholar] [CrossRef]
- Coto, L.; Mendia, I.; Sousa, C.; Bai, J.C.; Cebolla, A. Determination of gluten immunogenic peptides for the management of the treatment adherence of celiac disease: A systematic review. World J. Gastroenterol. 2021, 27, 6306–6321. [Google Scholar] [CrossRef] [PubMed]
- Roca, M.; Donat, E.; Masip, E.; Crespo-Escobar, P.; Canada-Martinez, A.J.; Polo, B.; Ribes-Koninckx, C. Analysis of gluten immunogenic peptides in feces to assess adherence to the gluten-free diet in pediatric celiac patients. Eur. J. Nutr. 2021, 60, 2131–2140. [Google Scholar] [CrossRef] [PubMed]
- Porcelli, B.; Ferretti, F.; Biviano, I.; Santini, A.; Cinci, F.; Vascotto, M.; Grande, E.; Quagliarella, F.; Terzuoli, L.; Bizzaro, N.; et al. Testing for fecal gluten immunogenic peptides: A useful tool to evaluate compliance with gluten-free diet by celiacs. Ann. Gastroenterol. 2020, 33, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Roca, M.; Donat, E.; Masip, E.; Crespo Escobar, P.; Fornes-Ferrer, V.; Polo, B.; Ribes-Koninckx, C. Detection and quantification of gluten immunogenic peptides in feces of infants and their relationship with diet. Rev. Esp. Enferm. Dig. 2019, 111, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Skodje, G.I.; van Megen, F.; Stendahl, M.; Henriksen, C.; Lundin, K.E.A.; Veierod, M.B. Detection of gluten immunogenic peptides and the Celiac Disease Adherence Test to monitor gluten-free diet: A pilot study. Eur. J. Clin. Nutr. 2022. [Google Scholar] [CrossRef]
- Laserna-Mendieta, E.J.; Casanova, M.J.; Arias, A.; Arias-Gonzalez, L.; Majano, P.; Mate, L.A.; Gordillo-Velez, C.H.; Jimenez, M.; Angueira, T.; Tebar-Romero, E.; et al. Poor Sensitivity of Fecal Gluten Immunogenic Peptides and Serum Antibodies to Detect Duodenal Mucosal Damage in Celiac Disease Monitoring. Nutrients 2020, 13, 98. [Google Scholar] [CrossRef]
- Coto, L.; Sousa, C.; Cebolla, A. Individual variability in patterns and dynamics of fecal gluten immunogenic peptides excretion after low gluten intake. Eur. J. Nutr. 2022, 7, 1–17. [Google Scholar] [CrossRef]
- Comino, I.; Segura, V.; Ortigosa, L.; Espin, B.; Castillejo, G.; Garrote, J.A.; Sierra, C.; Millan, A.; Ribes-Koninckx, C.; Roman, E.; et al. Prospective longitudinal study: Use of faecal gluten immunogenic peptides to monitor children diagnosed with coeliac disease during transition to a gluten-free diet. Aliment. Pharmacol. Ther. 2019, 49, 1484–1492. [Google Scholar] [CrossRef] [Green Version]
- Costa, A.F.; Sugai, E.; Temprano, M.P.; Niveloni, S.I.; Vazquez, H.; Moreno, M.L.; Dominguez-Flores, M.R.; Munoz-Suano, A.; Smecuol, E.; Stefanolo, J.P.; et al. Gluten immunogenic peptide excretion detects dietary transgressions in treated celiac disease patients. World J. Gastroenterol. 2019, 25, 1409–1420. [Google Scholar] [CrossRef]
- Monachesi, C.; Verma, A.K.; Catassi, G.N.; Franceschini, E.; Gatti, S.; Gesuita, R.; Lionetti, E.; Catassi, C. Determination of Urinary Gluten Immunogenic Peptides to Assess Adherence to the Gluten-Free Diet: A Randomized, Double-Blind, Controlled Study. Clin. Transl. Gastroenterol. 2021, 12, e00411. [Google Scholar] [CrossRef]
- Moreno, M.L.; Sanchez-Munoz, D.; Sanders, D.; Rodriguez-Herrera, A.; Sousa, C. Verifying Diagnosis of Refractory Celiac Disease with Urine Gluten Immunogenic Peptides as Biomarker. Front. Med. 2021, 7, 601854. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Carnicer, Á.; Garzón-Benavides, M.; Fombuena, B.; Segura, V.; García-Fernández, F.; Sobrino-Rodríguez, S.; Gómez-Izquierdo, L.; Montes-Cano, M.A.; Rodríguez-Herrera, A.; Millán, R.; et al. Negative predictive value of the repeated absence of gluten immunogenic peptides in the urine of treated celiac patients in predicting mucosal healing: New proposals for follow-up in celiac disease. Am. J. Clin. Nutr. 2020, 112, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.L.; Cebolla, A.; Munoz-Suano, A.; Carrillo-Carrion, C.; Comino, I.; Pizarro, A.; Leon, F.; Rodriguez-Herrera, A.; Sousa, C. Detection of gluten immunogenic peptides in the urine of patients with coeliac disease reveals transgressions in the gluten-free diet and incomplete mucosal healing. Gut 2017, 66, 250–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandini, E. Urinary microRNA and mRNA in Tumors. Methods Mol. Biol. 2021, 2292, 57–72. [Google Scholar] [PubMed]
- Lehto, M.; Groop, P.H. The Gut-Kidney Axis: Putative Interconnections between Gastrointestinal and Renal Disorders. Front. Endocrinol. 2018, 9, 553. [Google Scholar] [CrossRef] [PubMed]
- Sun, I.O.; Lerman, L.O. Urinary microRNA in kidney disease: Utility and roles. Am. J. Physiol. Renal Physiol. 2019, 316, F785–F793. [Google Scholar] [CrossRef]
- Kylokas, A.; Kaukinen, K.; Huhtala, H.; Collin, P.; Maki, M.; Kurppa, K. Type 1 and type 2 diabetes in celiac disease: Prevalence and effect on clinical and histological presentation. BMC Gastroenterol. 2016, 16, 76. [Google Scholar] [CrossRef] [Green Version]
- Chan, G.; Tang, S.C. Current practices in the management of diabetic nephropathy. J. R. Coll. Physicians. Edinb. 2013, 43, 330–332, quiz 333. [Google Scholar] [CrossRef] [Green Version]
- Fried, L.F.; Lewis, J. Rebuttal of the Pro View: Albuminuria Is an Appropriate Therapeutic Target in Patients with CKD. Clin. J. Am. Soc. Nephrol. 2015, 10, 1095–1098. [Google Scholar] [CrossRef] [Green Version]
- Delic, D.; Eisele, C.; Schmid, R.; Baum, P.; Wiech, F.; Gerl, M.; Zimdahl, H.; Pullen, S.S.; Urquhart, R. Urinary Exosomal miRNA Signature in Type II Diabetic Nephropathy Patients. PLoS ONE 2016, 11, e0150154. [Google Scholar] [CrossRef]
- Eissa, S.; Matboli, M.; Bekhet, M.M. Clinical verification of a novel urinary microRNA panal: 133b, -342 and -30 as biomarkers for diabetic nephropathy identified by bioinformatics analysis. Biomed. Pharmacother. 2016, 83, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Cardenas-Gonzalez, M.; Srivastava, A.; Pavkovic, M.; Bijol, V.; Rennke, H.G.; Stillman, I.E.; Zhang, X.; Parikh, S.; Rovin, B.H.; Afkarian, M.; et al. Identification, Confirmation, and Replication of Novel Urinary MicroRNA Biomarkers in Lupus Nephritis and Diabetic Nephropathy. Clin. Chem. 2017, 63, 1515–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sole, C.; Moline, T.; Vidal, M.; Ordi-Ros, J.; Cortes-Hernandez, J. An Exosomal Urinary miRNA Signature for Early Diagnosis of Renal Fibrosis in Lupus Nephritis. Cells 2019, 8, 773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argyropoulos, C.; Wang, K.; McClarty, S.; Huang, D.; Bernardo, J.; Ellis, D.; Orchard, T.; Galas, D.; Johnson, J. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS ONE 2013, 8, e54662. [Google Scholar] [CrossRef]
- Argyropoulos, C.; Wang, K.; Bernardo, J.; Ellis, D.; Orchard, T.; Galas, D.; Johnson, J.P. Urinary MicroRNA Profiling Predicts the Development of Microalbuminuria in Patients with Type 1 Diabetes. J. Clin. Med. 2015, 4, 1498–1517. [Google Scholar] [CrossRef] [Green Version]
- Conserva, F.; Barozzino, M.; Pesce, F.; Divella, C.; Oranger, A.; Papale, M.; Sallustio, F.; Simone, S.; Laviola, L.; Giorgino, F.; et al. Urinary miRNA-27b-3p and miRNA-1228-3p correlate with the progression of Kidney Fibrosis in Diabetic Nephropathy. Sci. Rep. 2019, 9, 11357. [Google Scholar] [CrossRef]
- El-Samahy, M.H.; Adly, A.A.; Elhenawy, Y.I.; Ismail, E.A.; Pessar, S.A.; Mowafy, M.E.; Saad, M.S.; Mohammed, H.H. Urinary miRNA-377 and miRNA-216a as biomarkers of nephropathy and subclinical atherosclerotic risk in pediatric patients with type 1 diabetes. J. Diabetes Complicat. 2018, 32, 185–192. [Google Scholar] [CrossRef]
- Peng, H.; Zhong, M.; Zhao, W.; Wang, C.; Zhang, J.; Liu, X.; Li, Y.; Paudel, S.D.; Wang, Q.; Lou, T. Urinary miR-29 correlates with albuminuria and carotid intima-media thickness in type 2 diabetes patients. PLoS ONE 2013, 8, e82607. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Wang, C.; Yu, H.; Ding, M.; Zhang, C.; Lu, X.; Zhang, C.Y.; Zhang, C. Increased urinary exosomal microRNAs in children with idiopathic nephrotic syndrome. EBioMedicine 2019, 39, 552–561. [Google Scholar] [CrossRef] [Green Version]
- Habura, I.; Fiedorowicz, K.; Wozniak, A.; Idasiak-Piechocka, I.; Kosikowski, P.; Oko, A. IgA nephropathy associated with coeliac disease. Cent. Eur. J. Immunol. 2019, 44, 106–108. [Google Scholar] [CrossRef]
- Gans, R.O.; Ueda, Y.; Ito, S.; Kohli, R.; Min, I.; Shafi, M.; Brentjens, J.R. The occurrence of IgA-nephropathy in patients with diabetes mellitus may not be coincidental: A report of five cases. Am. J. Kidney Dis. 1992, 20, 255–260. [Google Scholar] [CrossRef]
- Szeto, C.C.; Wang, G.; Ng, J.K.; Kwan, B.C.; Mac-Moune Lai, F.; Chow, K.M.; Luk, C.C.; Lai, K.B.; Li, P.K. Urinary miRNA profile for the diagnosis of IgA nephropathy. BMC Nephrol. 2019, 20, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, T.F.; Bekele, S.; O’Dwyer, M.J.; Prowle, J.R. MicroRNAs in Acute Kidney Injury. Nephron 2018, 140, 124–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, Y.F.; Wen, D.; Zhao, Q.; Shen, P.Y.; Shi, H.; Zhao, Q.; Chen, Y.X.; Zhang, W. Urinary MicroRNA-30c-5p and MicroRNA-192-5p as potential biomarkers of ischemia-reperfusion-induced kidney injury. Exp. Biol. Med. 2017, 242, 657–667. [Google Scholar] [CrossRef] [Green Version]
- Ramezani, A.; Devaney, J.M.; Cohen, S.; Wing, M.R.; Scott, R.; Knoblach, S.; Singhal, R.; Howard, L.; Kopp, J.B.; Raj, D.S. Circulating and urinary microRNA profile in focal segmental glomerulosclerosis: A pilot study. Eur. J. Clin. Investig. 2015, 45, 394–404. [Google Scholar] [CrossRef] [Green Version]
- Baldassarre, A.; Felli, C.; Prantera, G.; Masotti, A. Circulating microRNAs and Bioinformatics Tools to Discover Novel Diagnostic Biomarkers of Pediatric Diseases. Genes 2017, 8, 234. [Google Scholar] [CrossRef] [Green Version]
- Masotti, A.; Baldassarre, A.; Fabrizi, M.; Olivero, G.; Loreti, M.C.; Giammaria, P.; Veronelli, P.; Graziani, M.P.; Manco, M. Oral glucose tolerance test unravels circulating miRNAs associated with insulin resistance in obese preschoolers. Pediatr. Obes. 2017, 12, 229–238. [Google Scholar] [CrossRef]
- Masotti, A.; Baldassarre, A.; Guzzo, M.P.; Iannuccelli, C.; Barbato, C.; Di Franco, M. Circulating microRNA Profiles as Liquid Biopsies for the Characterization and Diagnosis of Fibromyalgia Syndrome. Mol. Neurobiol. 2017, 54, 7129–7136. [Google Scholar] [CrossRef]
- Felli, C.; Baldassarre, A.; Masotti, A. Intestinal and Circulating MicroRNAs in Coeliac Disease. Int. J. Mol. Sci. 2017, 18, 1907. [Google Scholar] [CrossRef] [Green Version]
- Sarshar, M.; Scribano, D.; Ambrosi, C.; Palamara, A.T.; Masotti, A. Fecal microRNAs as Innovative Biomarkers of Intestinal Diseases and Effective Players in Host-Microbiome Interactions. Cancers 2020, 12, 2174. [Google Scholar] [CrossRef]
- Liu, S.; da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell. Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Weiner, H.L. Control of the gut microbiome by fecal microRNA. Microb. Cell. 2016, 3, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Bañares, F.; Beltrán, B.; Salas, A.; Comino, I.; Ballester-Clau, R.; Ferrer, C.; Molina-Infante, J.; Rosinach, M.; Modolell, I.; Rodríguez-Moranta, F.; et al. Persistent Villous Atrophy in De Novo Adult Patients with Celiac Disease and Strict Control of Gluten-Free Diet Adherence: A Multicenter Prospective Study (CADER Study). Am. J. Gastroenterol. 2021, 116, 1036–1043. [Google Scholar] [CrossRef]
- Coto, L.; Sousa, C.; Cebolla, A. Dynamics and Considerations in the Determination of the Excretion of Gluten Immunogenic Peptides in Urine: Individual Variability at Low Gluten Intake. Nutrients 2021, 13, 2624. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paolini, A.; Sarshar, M.; Felli, C.; Bruno, S.P.; Rostami-Nejad, M.; Ferretti, F.; Masotti, A.; Baldassarre, A. Biomarkers to Monitor Adherence to Gluten-Free Diet by Celiac Disease Patients: Gluten Immunogenic Peptides and Urinary miRNAs. Foods 2022, 11, 1380. https://doi.org/10.3390/foods11101380
Paolini A, Sarshar M, Felli C, Bruno SP, Rostami-Nejad M, Ferretti F, Masotti A, Baldassarre A. Biomarkers to Monitor Adherence to Gluten-Free Diet by Celiac Disease Patients: Gluten Immunogenic Peptides and Urinary miRNAs. Foods. 2022; 11(10):1380. https://doi.org/10.3390/foods11101380
Chicago/Turabian StylePaolini, Alessandro, Meysam Sarshar, Cristina Felli, Stefania Paola Bruno, Mohammad Rostami-Nejad, Francesca Ferretti, Andrea Masotti, and Antonella Baldassarre. 2022. "Biomarkers to Monitor Adherence to Gluten-Free Diet by Celiac Disease Patients: Gluten Immunogenic Peptides and Urinary miRNAs" Foods 11, no. 10: 1380. https://doi.org/10.3390/foods11101380






