Analysis of Parkinson’s Disease Using an Imbalanced-Speech Dataset by Employing Decision Tree Ensemble Methods
Abstract
1. Introduction
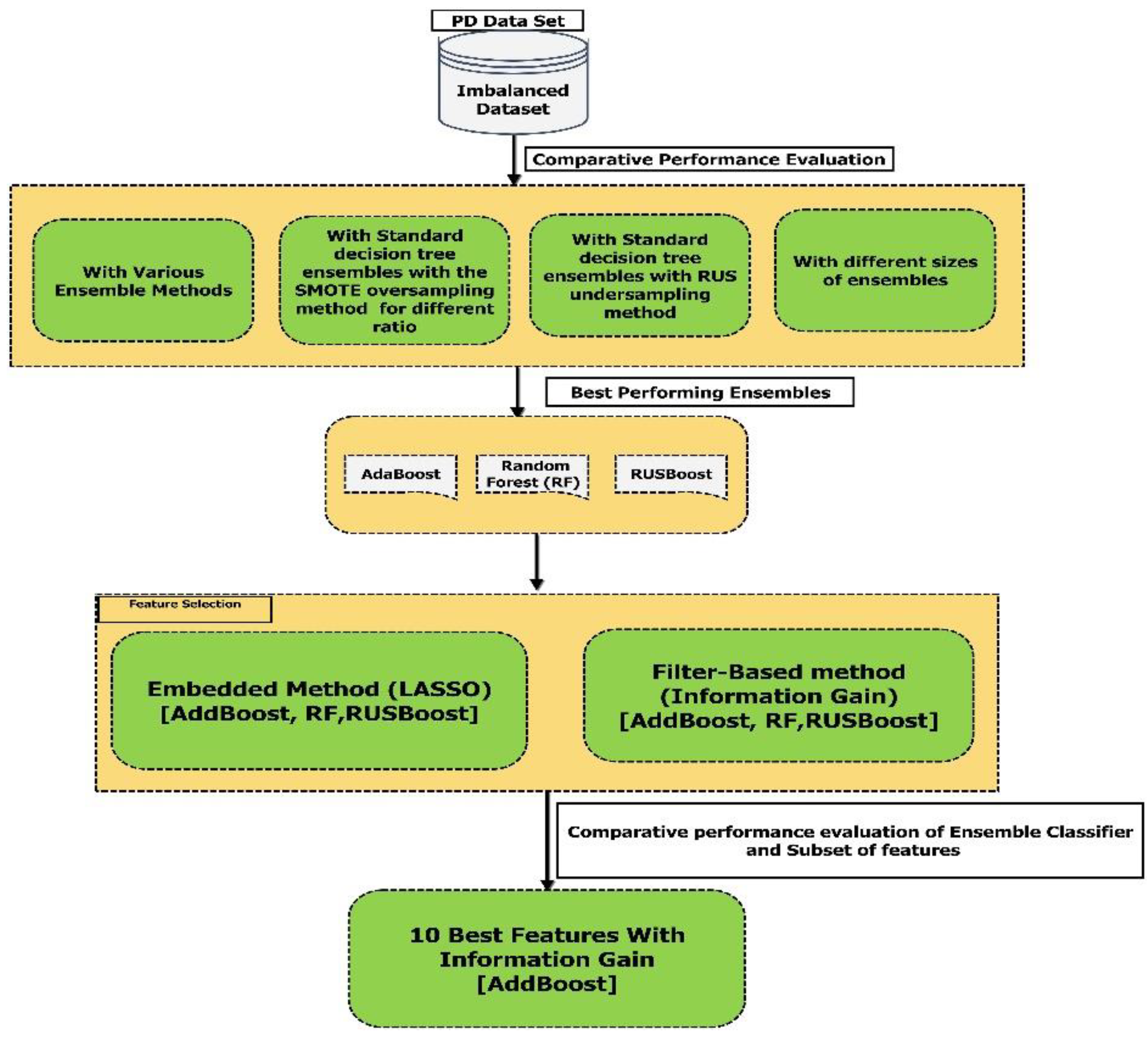
- We considered an imbalanced dataset and performed automatic classification between PD patients and healthy controls to evaluate the robustness of different ensemble methods for class imbalance.
- Decision tree ensembles have been shown to have excellent performance in different domains. In this study, we carried out extensive performance evaluations of different types of decision tree ensembles such as RUSBoost, isolation forset, RUSBagging, balanced bagging etc.; developed for imbalanced data. To the best of our knowledge, this has never been used by other researcher in this area.
- We carried out the feature selection using lasso and the information gain method, to achieve the best set of features.
2. Related Work
Related Literature Which Addresses the Imbalance Problem
3. Materials and Methods
3.1. Parkinson’s Diseases Speech Vocal Dataset
3.2. Decision Tree Classifier
3.3. Decision Tree Ensembles
3.4. Decision Tree Ensembles for Imbalanced Datasets
3.5. Feature Selection Methods
3.5.1. Feature Selection Using Information Gain (IG)
3.5.2. Least Absolute Shrinkage and Selection Operator (Lasso) or L1 Regularization
3.6. Evaluation Metrics
3.6.1. The Area under the Receiver Operating Characteristic (ROC) Curve
3.6.2. Area under the Precision-Recall (PR) Curve (AUPRC)
3.6.3. Geometric Mean (G-Mean)
3.6.4. Sensitivity
3.6.5. Specificity
4. Experimental Setup, Results, and Discussion
4.1. Experimental Setup and Software Packages
4.2. Comparative Study of Various Decision Tree Ensemble Models Built Using the Imbalanced Dataset
4.3. Decision Tree Ensembles Using Various Sampling Techniques
4.4. Ensemble Size Effect in Imbalanced Datasets
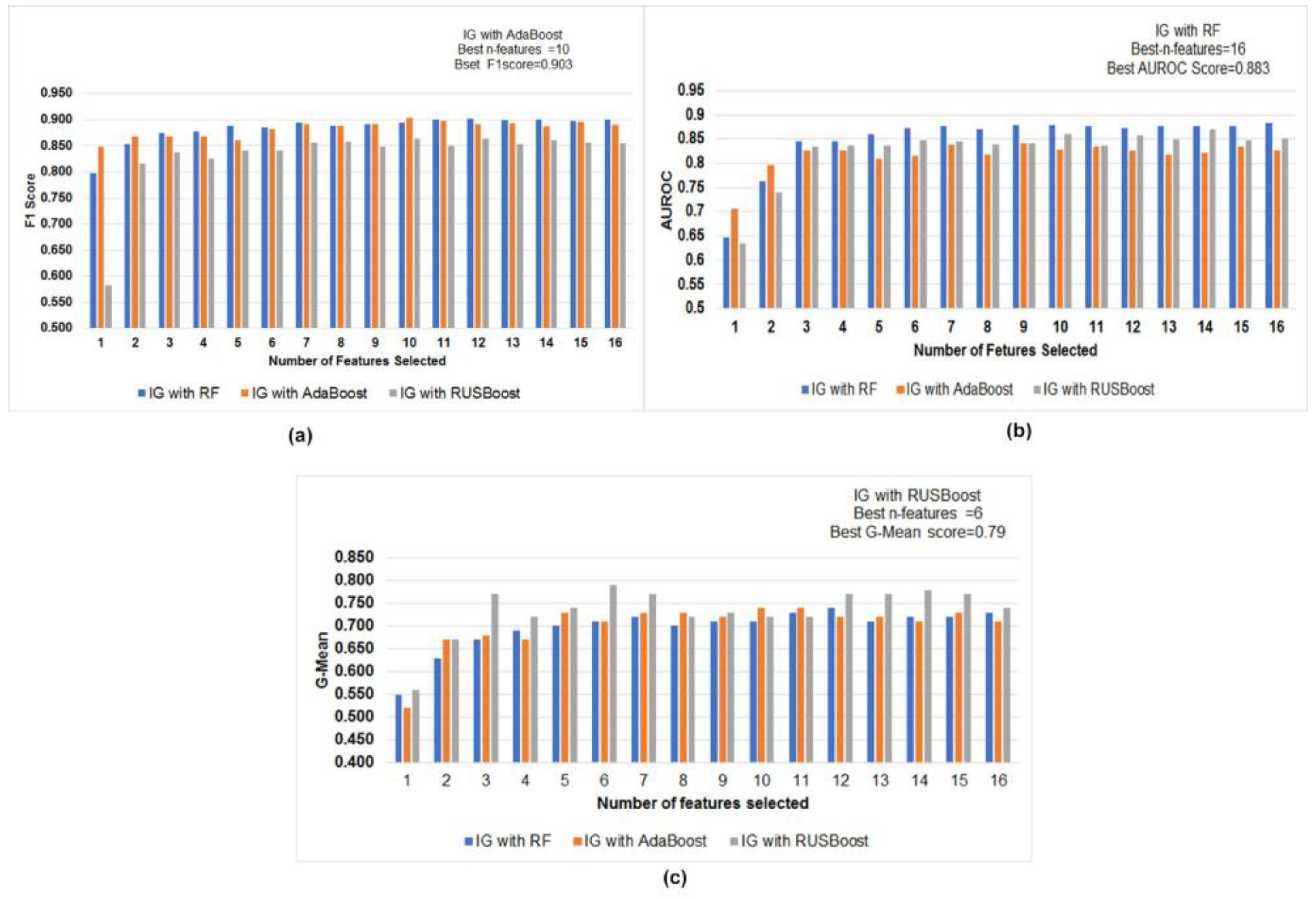
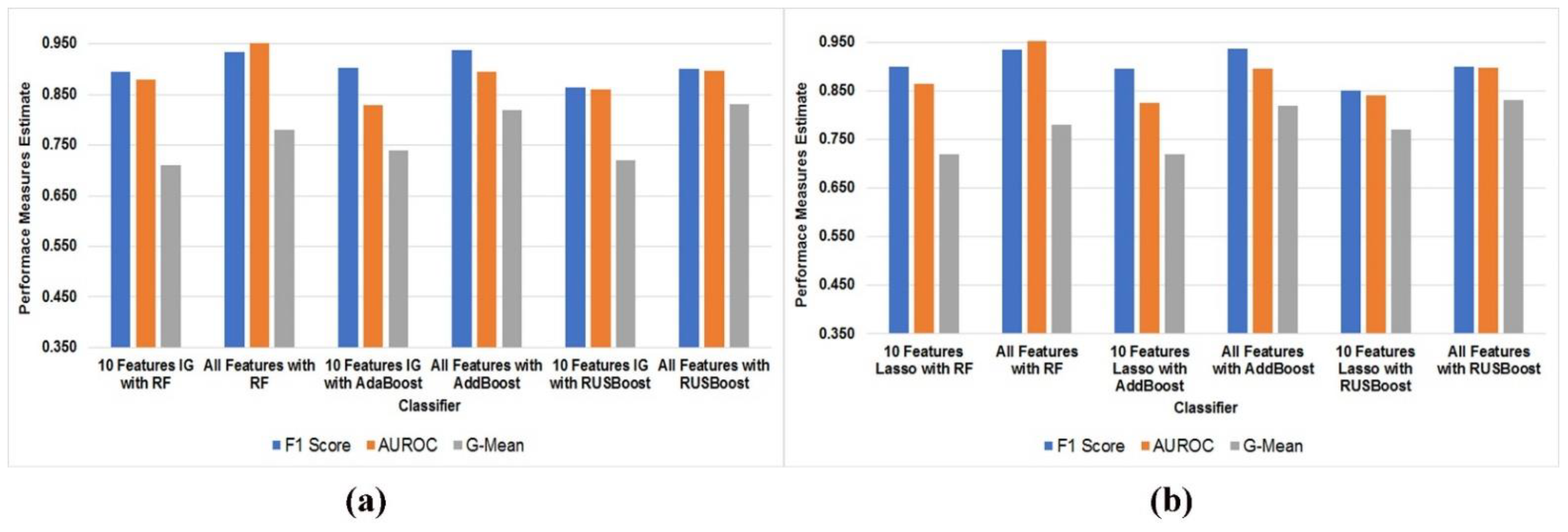
4.5. Feature Selection and Comparative Performance Evaluation of Best Ensemble Classifiers with a Different Subset of Features
5. Conclusions and Future Scope
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balestrino, R.; Schapira, A.H.V. Parkinson disease. Eur. J. Neurol. 2020, 27, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.X.; Chen, L. Economic Burden Analysis of ’Parkinson’s Disease Patients in China. Park. Dis. 2017, 2017, 8762939. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.J.; Diener, M.D.; Kaltenboeck, A.; Birnbaum, H.G.; Siderowf, A.D. An economic model of Parkinson’s disease: Implications for slowing progression in the United States. Mov. Disord. 2013, 28, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Kowal, S.L.; Dall, T.M.; Chakrabarti, R.; Storm, M.V.; Jain, A. The current and projected economic burden of Parkinson’s disease in the United States. Mov. Disord. 2013, 28, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Zesiewicz, T.A.; Bezchlibnyk, Y.; Dohse, N.; Ghanekar, S.D. Management of Early Parkinson Disease. Clin. Geriatr. Med. 2020, 36, 35–41. [Google Scholar] [CrossRef]
- Berus, L.; Klancnik, S.; Brezocnik, M.; Ficko, M. Classifying parkinson’s disease based on acoustic measures using artificial neural networks. Sensors 2019, 19, 16. [Google Scholar] [CrossRef]
- Arena, J.E.; Stoessl, A.J. Optimizing diagnosis in Parkinson’s disease: Radionuclide imaging. Park. Relat. Disord. 2016, 22, S47–S51. [Google Scholar] [CrossRef]
- Naseer, A.; Rani, M.; Naz, S.; Razzak, M.I.; Imran, M.; Xu, G. Refining Parkinson’s neurological disorder identification through deep transfer learning. Neural Comput. Appl. 2020, 32, 839–854. [Google Scholar] [CrossRef]
- Almeida, J.S.; Filho, P.P.R.; Carneiro, T.; Wei, W.; Damaševičius, R.; Maskeliūnas, R.; de Albuquerque, V.H.C. Detecting Parkinson’s disease with sustained phonation and speech signals using machine learning techniques. Pattern Recognit. Lett. 2019, 125, 55–62. [Google Scholar] [CrossRef]
- Bernardo, L.S.; Quezada, A.; Munoz, R.; Maia, F.M.; Pereira, C.R.; Wu, W.; de Albuquerque, V.H.C. Handwritten pattern recognition for early Parkinson’s disease diagnosis. Pattern Recognit. Lett. 2019, 125, 78–84. [Google Scholar] [CrossRef]
- De Souza, J.W.M.; Alves, S.S.A.; Rebouças, E.D.S.; Almeida, J.S.; Filho, P.P.R. A New Approach to Diagnose Parkinson’s Disease Using a Structural Cooccurrence Matrix for a Similarity Analysis. Comput. Intell. Neurosci. 2018, 2018, 7613282. [Google Scholar] [CrossRef]
- Pereira, C.R.; Pereira, D.R.; Silva, F.A.; Masieiro, J.P.; Weber, S.A.; Hook, C.; Papa, J.P. A new computer vision-based approach to aid the diagnosis of Parkinson’s disease. Comput. Methods Programs Biomed. 2016, 136, 79–88. [Google Scholar] [CrossRef]
- Lauraitis, A.; Maskeliūnas, R.; Damaševičius, R. ANN and Fuzzy Logic Based Model to Evaluate Huntington Disease Symptoms. J. Health Eng. 2018, 2018, 4581272. [Google Scholar] [CrossRef]
- Peixoto, E., Jr.; Delmiro, I.L.D.; Magaia, N.; Maia, F.M.; Hassan, M.M.; Albuquerque, V.H.C.; Fortino, G. Intelligent Sensory Pen for Aiding in the Diagnosis of Parkinson’s Disease from Dynamic Handwriting Analysis. Sensors 2020, 20, 5840. [Google Scholar] [CrossRef]
- Fernandez, M.L.; Vergara-Jimenez, M.; Missimer, A.; DiMarco, D.M.; Andersen, C.J.; Murillo, A.G. Evaluation of Family History, Antioxidant Intake and Activity Level as Indicators for Chronic Disease in a Healthy Young Population. EC Nutr. 2015, 1, 164–173. [Google Scholar] [CrossRef]
- New, A.B.; Robin, D.A.; Parkinson, A.L.; Eickhoff, C.R.; Reetz, K.; Hoffstaedter, F.; Mathys, C.; Sudmeyer, M.; Michely, J.; Caspers, J.; et al. The intrinsic resting state voice network in Parkinson’s disease. Hum. Brain Mapp. 2015, 36, 1951–1962. [Google Scholar] [CrossRef]
- Pawlukowska, W.; Gołąb-Janowska, M.; Safranow, K.; Rotter, I.; Amernik, K.; Honczarenko, K.; Nowacki, P. Articulation disorders and duration, severity and l-dopa dosage in idiopathic Parkinson’s disease. Neurol. Neurochir. Pol. 2015, 49, 302–306. [Google Scholar] [CrossRef]
- Drotar, P.; Mekyska, J.; Rektorova, I.; Masarova, L.; Smekal, Z.; Faundez-Zanuy, M. Decision Support Framework for Parkinson’s Disease Based on Novel Handwriting Markers. IEEE Trans. Neural Syst. Rehabil. Eng. 2015, 23, 508–516. [Google Scholar] [CrossRef]
- Pereira, C.R.; Pereira, D.R.; Rosa, G.H.; Albuquerque, V.H.; Weber, S.A.; Hook, C.; Papa, J.P. Handwritten dynamics assessment through convolutional neural networks: An application to Parkinson’s disease identification. Artif. Intell. Med. 2018, 87, 67–77. [Google Scholar] [CrossRef]
- Yang, M.; Zheng, H.; Wang, H.; McClean, S. Feature selection and construction for the discrimination of neurodegenerative diseases based on gait analysis. In Proceedings of the 2009 3rd International Conference on Pervasive Computing Technologies for Healthcare, London, UK, 1–3 April 2009. [Google Scholar]
- Wahid, F.; Begg, R.K.; Hass, C.J.; Halgamuge, S.; Ackland, D.C. Classification of Parkinson’s disease gait using spatial-temporal gait features. IEEE J. Biomed. Health Inform. 2015, 19, 1794–1802. [Google Scholar] [CrossRef]
- Pham, T.D.; Yan, H. Tensor decomposition of gait dynamics in Parkinson’s disease. IEEE Trans. Biomed. Eng. 2018, 65, 1820–1827. [Google Scholar]
- Cherubini, A.; Morelli, M.; Nisticó, R.; Salsone, M.; Arabia, G.; Vasta, R.; Augimeri, A.; Msc, M.E.C.; Quattrone, A. Magnetic resonance support vector machine discriminates between Parkinson disease and progressive supranuclear palsy. Mov. Disord. 2014, 29, 266–269. [Google Scholar] [CrossRef]
- Choi, H.; Ha, S.; Im, H.J.; Paek, S.H.; Lee, D.S. Refining diagnosis of Parkinson’s disease with deep learning-based interpretation of dopamine transporter imaging. NeuroImage Clin. 2017, 16, 586–594. [Google Scholar] [CrossRef]
- Segovia, F.; Górriz, J.M.; Ramírez, J.; Martínez-Murcia, F.J.; Castillo-Barnes, D. Assisted diagnosis of parkinsonism based on the striatal morphology. Int. J. Neural Syst. 2019, 29, 1950011. [Google Scholar] [CrossRef]
- Sakar, B.E.; Isenkul, M.E.; Sakar, C.O.; Sertbas, A.; Gurgen, F.; Delil, S.; Apaydin, H.; Kursun, O. Collection and Analysis of a Parkinson Speech Dataset with Multiple Types of Sound Recordings. IEEE J. Biomed. Health Inform. 2013, 17, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Ouyang, J.; Chen, H.-L.; Zhao, X.-H. An Efficient Diagnosis System for Parkinson’s Disease Using Kernel-Based Extreme Learning Machine with Subtractive Clustering Features Weighting Approach. Comput. Math. Methods Med. 2014, 2014, 985789. [Google Scholar] [CrossRef] [PubMed]
- Lewitt, P.A.; Li, J.; Lu, M.; Beach, T.G.; Adler, C.H.; Guo, L. 3-hydroxykynurenine and other Parkinson’s disease biomarkers discovered by metabolomic analysis. Mov. Disord. 2013, 28, 1653–1660. [Google Scholar] [CrossRef]
- Maass, F.; Michalke, B.; Willkommen, D.; Leha, A.; Schulte, C.; Tönges, L.; Mollenhauer, B.; Trenkwalder, C.; Rückamp, D.; Börger, M.; et al. Elemental fingerprint: Reassessment of a cerebrospinal fluid biomarker for Parkinson’s disease. Neurobiol. Dis. 2020, 134, 104677. [Google Scholar] [CrossRef]
- Nuvoli, S.; Spanu, A.; Fravolini, M.L.; Bianconi, F.; Cascianelli, S.; Madeddu, G.; Palumbo, B. [123I]Metaiodobenzylguanidine (MIBG) Cardiac Scintigraphy and Automated Classification Techniques in Parkinsonian Disorders. Mol. Imaging Biol. 2019, 22, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Váradi, C.; Nehéz, K.; Hornyák, O.; Viskolcz, B.; Bones, J. Serum N-Glycosylation in Parkinson’s Disease: A Novel Approach for Potential Alterations. Molecules 2019, 24, 2220. [Google Scholar] [CrossRef] [PubMed]
- Nunes, A.; Silva, G.; Duque, C.; Januário, C.; Santana, I.; Ambrósio, A.F.; Castelo-Branco, M.; Bernardes, R. Retinal texture biomarkers may help to discriminate between Alzheimer’s, Parkinson’s, and healthy controls. PLoS ONE 2019, 14, e0218826. [Google Scholar] [CrossRef]
- Cherubini, A.; Nisticó, R.; Novellino, F.; Salsone, M.; Nigro, S.; Donzuso, G.; Quattrone, A. Magnetic resonance support vector machine discriminates essential tremor with rest tremor from tremor-dominant Parkinson disease. Mov. Disord. 2014, 29, 1216–1219. [Google Scholar] [CrossRef]
- Ramig, L.; Halpern, A.; Spielman, J.; Fox, C.; Freeman, K. Speech treatment in Parkinson’s disease: Randomized controlled trial (RCT). Mov. Disord. 2018, 33, 1777–1791. [Google Scholar] [CrossRef]
- Sakar, C.O.; Serbes, G.; Gunduz, A.; Tunc, H.C.; Nizam, H.; Sakar, B.E.; Tutuncu, M.; Aydin, T.; Isenkul, M.E.; Apaydin, H. A comparative analysis of speech signal processing algorithms for Parkinson’s disease classification and the use of the tunable Q-factor wavelet transform. Appl. Soft Comput. 2019, 74, 255–263. [Google Scholar] [CrossRef]
- Polat, K. A hybrid approach to Parkinson disease classification using speech signal: The combination of SMOTE and random forests. In Proceedings of the 2019 Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT), Istanbul, Turkey, 24–26 April 2019; pp. 2019–2021. [Google Scholar]
- Hoq, M.; Uddin, M.N.; Park, S.B. Vocal feature extraction-based artificial intelligent model for Parkinson’s disease detection. Diagnostics 2021, 11, 1076. [Google Scholar] [CrossRef]
- Pramanik, M.; Pradhan, R.; Nandy, P.; Bhoi, A.K.; Barsocchi, P. Machine Learning Methods with Decision Forests for Parkinson’s Detection. Appl. Sci. 2021, 11, 581. [Google Scholar] [CrossRef]
- Gunduz, H. Deep Learning-Based Parkinson’s Disease Classification Using Vocal Feature Sets. IEEE Access 2019, 7, 115540–115551. [Google Scholar] [CrossRef]
- Salzberg, S.L. C4.5: Programs for Machine Learning by J. Ross Quinlan; Morgan Kaufmann Publishers, Inc.: Burlington, MA, USA, 1993. [Google Scholar]
- Karaman, O.; Çakın, H.; Alhudhaif, A.; Polat, K. Robust automated Parkinson disease detection based on voice signals with transfer learning. Expert Syst. Appl. 2021, 178, 115013. [Google Scholar] [CrossRef]
- Mohammadi, A.G.; Mehralian, P.; Naseri, A.; Sajedi, H. Parkinson’s disease diagnosis: The effect of autoencoders on extracting features from vocal characteristics. Array 2021, 11, 100079. [Google Scholar] [CrossRef]
- Nissar, I.; Rizvi, D.R.; Masood, S.; Mir, A. Voice-Based Detection of Parkinson’s Disease through Ensemble Machine Learning Approach: A Performance Study. EAI Endorsed Trans. Pervasive Health Technol. 2019, 5, e2. [Google Scholar] [CrossRef]
- Yücelbaş, Ş. Simple Logistic Hybrid System Based on Greedy Stepwise Algorithm for Feature Analysis to Diagnose Parkinson’s Disease According to Gender. Arab. J. Sci. Eng. 2020, 45, 2001–2016. [Google Scholar] [CrossRef]
- Solana-Lavalle, G.; Rosas-Romero, R. Analysis of voice as an assisting tool for detection of Parkinson’s disease and its subsequent clinical interpretation. Biomed. Signal Process. Control 2021, 66, 102415. [Google Scholar] [CrossRef]
- Gunduz, H. An efficient dimensionality reduction method using filter-based feature selection and variational autoencoders on Parkinson’s disease classification. Biomed. Signal Process. Control 2021, 66, 102452. [Google Scholar] [CrossRef]
- Ashour, A.S.; Nour, M.K.A.; Polat, K.; Guo, Y.; Alsaggaf, W.; El-Attar, A. A Novel Framework of Two Successive Feature Selection Levels Using Weight-Based Procedure for Voice-Loss Detection in Parkinson’s Disease. IEEE Access 2020, 8, 76193–76203. [Google Scholar] [CrossRef]
- Yücelbaş, C. A new approach: Information gain algorithm-based k-nearest neighbors hybrid diagnostic system for Parkinson’s disease. Phys. Eng. Sci. Med. 2021, 44, 511–524. [Google Scholar] [CrossRef]
- Bchir, O. Parkinson’s Disease Classification using Gaussian Mixture Models with Relevance Feature Weights on Vocal Feature Sets. Int. J. Adv. Comput. Sci. Appl. 2020, 11, 413–419. [Google Scholar] [CrossRef]
- Sharma, S.R.; Singh, B.; Kaur, M. Classification of Parkinson disease using binary Rao optimization algorithms. Expert Syst. 2021, 38, e12674. [Google Scholar] [CrossRef]
- Thanoun, M.Y.; Yaseen, M.T. A Comparative Study of Parkinson Disease Diagnosis in Machine Learning. In Proceedings of the the 4th International Conference on Advances in Artificial Intelligence, London, UK, 9–11 October 2020; pp. 23–28. [Google Scholar]
- Gemci, F.; Ibrikci, T. Using Deep Learning Algorithm to Diagnose Parkinson Disease with High Accuracy. Kahramanmaraş Sütçü İmam Üniversitesi Mühendislik Bilim. Derg. 2019, 22, 19–25. [Google Scholar]
- Prasad, G.; Munasinghe, T.; Seneviratne, O. A two-step framework for Parkinson’s disease classification: Using multiple one-way ANOVA on speech features and decision trees. In Proceedings of the CEUR Workshop, Galway, Ireland, 19–23 October 2020; p. 2884. [Google Scholar]
- Xiong, Y.; Lu, Y. Deep Feature Extraction from the Vocal Vectors Using Sparse Autoencoders for Parkinson’s Classification. IEEE Access 2020, 8, 27821–27830. [Google Scholar] [CrossRef]
- Schellhas, D.; Neupane, B.; Thammineni, D.; Kanumuri, B.; Green, R.C. Distance Correlation Sure Independence Screening for Accelerated Feature Selection in Parkinson’s Disease Vocal Data. In Proceedings of the 2020 International Conference on Computational Science and Computational Intelligence, Las Vegas, NV, USA, 16–18 December 2020; pp. 1433–1438. [Google Scholar]
- Jain, D.; Mishra, A.K.; Das, S.K. Machine Learning Based Automatic Prediction of Parkinson’s Disease Using Speech Features BT. In Advances in Intelligent Systems and Computing; Springer: Berlin/Heidelberg, Germany, 2021; pp. 351–362. [Google Scholar]
- Lamba, R.; Gulati, T.; Alharbi, H.F.; Jain, A. A hybrid system for Parkinson’s disease diagnosis using machine learning techniques. Int. J. Speech Technol. 2021, 25, 583–593. [Google Scholar] [CrossRef]
- Wu, J.; Chen, S.; Zhou, W.; Wang, N.; Fan, Z. Evaluation of Feature Selection Methods Using Bagging and Boosting Ensemble Techniques on High Throughput Biological Data. In Proceedings of the 2020 10th International Conference on Biomedical Engineering and Technology, Tokyo, Japan, 15–18 September 2020. [Google Scholar]
- Freund, Y.; Schapire, R.E. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Richman, R.; Wüthrich, M.V. Bagging predictors. Risks 2020, 8, 83. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the KDD ’16: The 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Sun, Y.; Wong, A.K.C.; Kamel, M.S. Classification of imbalanced data: A review. Int. J. Pattern Recognit. Artif. Intell. 2009, 23, 687–719. [Google Scholar] [CrossRef]
- Guo, H.; Li, Y.; Shang, J.; Gu, M.; Huang, Y.; Gong, B. Learning from class-imbalanced data: Review of methods and applications. Expert Syst. Appl. 2017, 73, 220–239. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Seiffert, C.; Khoshgoftaar, T.M.; van Hulse, J.; Napolitano, A. RUSBoost: A Hybrid Approach to Alleviating Class Imbalance. IEEE Trans. Syst. Man Cybern. Part A Syst. Hum. 2010, 40, 185–197. [Google Scholar] [CrossRef]
- PYPI, Imbalanced-Learn 0.7.0. Available online: https://pypi.org/project/imbalanced-learn/ (accessed on 30 July 2021).
- Chawla, N.V.; Lazarevic, A.; Hall, L.O.; Bowyer, K.W. SMOTEBoost: Improving Prediction of the Minority Class in Boosting. In Knowledge Discovery in Databases: PKDD 2003; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2003; pp. 107–119. [Google Scholar]
- Wang, S.; Yao, X. Diversity analysis on imbalanced data sets by using ensemble models. In Proceedings of the 2009 IEEE Symposium on Computational Intelligence and Data Mining, Nashville, TN, USA, 30 March–2 April 2009; pp. 324–331. [Google Scholar]
- Chen, C.; Liaw, A.; Breiman, L. Using Random Forest to Learn Imbalanced Data; University of California, Berkeley: Berkeley, CA, USA, 1999; pp. 1–12. [Google Scholar]
- Frank, E.; Hall, M.; Holmes, G.; Kirkby, R.; Pfahringer, B.; Witten, I.H.; Trigg, L. Weka—A Machine Learning Workbench for Data Mining. In Data Mining and Knowledge Discovery Handbook; Maimon, O., Rokach, L., Eds.; Springer: Boston, MA, USA, 2010; pp. 1269–1277. [Google Scholar]
- Xgboost Python Package. 2020. Available online: https://xgboost.readthedocs.io/en/latest/python/pythonintro.html (accessed on 30 April 2022).
- Rehman, R.Z.U.; del Din, S.; Guan, Y.; Yarnall, A.J.; Shi, J.Q.; Rochester, L. Selecting Clinically Relevant Gait Characteristics for Classification of Early Parkinson’s Disease: A Comprehensive Machine Learning Approach. Sci. Rep. 2019, 9, 17269. [Google Scholar] [CrossRef]
- Fonti, V.; Belitser, E. Paper in Business Analytics Feature Selection Using LASSO; Vrije Universiteit Amsterdam: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Demir-Kavuk, O.; Kamada, M.; Akutsu, T.; Knapp, E.-W. Prediction using step-wise L1, L2 regularization and feature selection for small data sets with large number of features. BMC Bioinform. 2011, 12, 412. [Google Scholar] [CrossRef]
- Omuya, E.O.; Okeyo, G.O.; Kimwele, M.W. Feature Selection for Classification using Principal Component Analysis and Information Gain. Expert Syst. Appl. 2021, 174, 114765. [Google Scholar] [CrossRef]
- Gu, Q.; Zhu, L.; Cai, Z. Evaluation Measures of the Classification Performance of Imbalanced Data Sets. In Proceedings of the International Symposium on Intelligence Computation and Applications, Huangshi, China, 23–25 October 2009; Springer: Berlin/Heidelberg, Germany, 2009; pp. 461–471. [Google Scholar]
- Kuncheva, L.I.; Arnaiz-González, Á.; Díez-Pastor, J.-F.; Gunn, I.A.D. Instance selection improves geometric mean accuracy: A study on imbalanced data classification. Prog. Artif. Intell. 2019, 8, 215–228. [Google Scholar] [CrossRef]
- Dinga, R.; Penninx, B.W.J.H.; Veltman, D.J.; Schmaal, L.; Marquand, A.F. Beyond accuracy: Measures for assessing machine learning models, pitfalls and guidelines. bioRxiv 2019, 743138. [Google Scholar]
- Saito, T.; Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Martin, P.R.; Botha, H.; Ahlskog, J.E.; Bower, J.H.; Masumoto, J.Y.; Maraganore, D.; Hassan, A.; Eggers, S.; Boeve, B.F.; et al. Sensitivity and Specificity of Diagnostic Criteria for Progressive Supranuclear Palsy. Mov. Disord. 2019, 34, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Rushdi, R.A.; Rushdi, A.M. Karnaugh-Map Utility in Medical Studies: The Case of Fetal Malnutrition. Int. J. Math. Eng. Manag. Sci. 2018, 3, 220–244. [Google Scholar] [CrossRef]
- Rushdi, A.M.A.; Serag, H.A.M. Solutions of ternary problems of conditional probability with applications to mathematical epidemiology and the COVID-19 pandemic. Int. J. Math. Eng. Manag. Sci. 2020, 5, 787–811. [Google Scholar] [CrossRef]
- Rushdi, A.M.; Alghamdi, S.M. Measures, metrics, and indicators derived from the ubiquitous two-by-two contingency table, Part I: Background. Asian J. Med. Princ. Clin. Pract. 2021, 4, 51–65. [Google Scholar]
- Liang, X.; Jiang, A.; Li, T.; Xue, Y.; Wang, G. LR-SMOTE—An improved unbalanced data set oversampling based on K-means and SVM. Knowl. Based Syst. 2020, 196, 105845. [Google Scholar] [CrossRef]
- Fumera, G.; Roli, F.; Serrau, A. A Theoretical Analysis of Bagging as a Linear Combination of Classifiers. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 30, 1293–1299. [Google Scholar] [CrossRef]

| Features | Measure | Number of Features |
|---|---|---|
| Baseline features | Jitter variants | 5 |
| Shimmer variants | 6 | |
| Fundamental frequency parameters | 5 | |
| Harmonicity parameters | 2 | |
| Recurrence Period Density Entropy (RPDE) | 1 | |
| Detrended Fluctuation Analysis (DFA) | 1 | |
| Pitch Period Entropy (PPE) | 1 | |
| Time-Frequency Features | Intensity Parameters | 3 |
| Formant Frequencies | 4 | |
| Bandwidth | 4 | |
| Tunable Q-factor Wavelet Transform (TQWT) | TQWT features related to F0 | 432 |
| Wavelet Transform based Features | Wavelet Transform (WT) features related to F0 | 182 |
| Vocal fold features | Glottis Quotient (GQ) | 3 |
| Glottal to Noise | 6 | |
| Vocal Fold Excitation Ratio (VFER) | 7 | |
| Empirical Mode Decomposition (EMD) | 6 | |
| Mel Frequency Cepstral Coefficients (MFCCs) | MFCCs | 84 |
| Classifier (Decision Tree Ensembles) | Software Package |
|---|---|
| Bagging | Weka tool |
| C4.5 Decision tree | Weka tool |
| AdaBoost | Weka tool |
| Random forest (RF) | Weka tool |
| Balanced random forset | Imblearn (Python) |
| XGBoost | Python |
| Balanced Bagging | Imblearn (Python) |
| RUSBoost | Isolation Forset |
| Isolation Forset | Weka tool |
| Random under sampling with bagging (RUSBagging) | Filter (SpreadsubSample) (Weka), Weka tool |
| Random under sampling with Random Forest (RUS random forest) | Filter (SpreadsubSample) (Weka), Weka tool |
| Random under sampling with XGBoost (RUS XGBoost) | Imblearn (Python), XGBoost |
| Random under sampling with AdaBoost (RUS AdaBoost) | Filter (SpreadsubSample) (Weka), Weka tool |
| Oversampling with Random Froest (SMOTE RF) | SMOTE, Weka Tool |
| Oversampling with Bagging (SMOTE Bagging) | SMOTE, Weka Tool SMOTE, Weka Tool |
| Oversampling with XGBoost (SMOTE XGBoost) | Imblearn (Python), XGBoost (Python) |
| Oversampling with AdaBoost (SMOTE AdaBoost) | SMOTE, Weka Tool |
| Ensemble | Accuracy | AUROC | SN | SP |
|---|---|---|---|---|
| Single decision tree [J48] | 0.855 | 0.848 | 0.896 | 0.730 |
| Bagging | 0.882 | 0.940 | 0.948 | 0.676 |
| Random forest [RF] | 0.895 | 0.952 | 0.983 | 0.622 |
| XGBoost | 0.800 | 0.928 | 0.972 | 0.625 |
| AdaBoost | 0.901 | 0.895 | 0.965 | 0.703 |
| Ensembles for imbalanced datasets | ||||
| Balanced random forest | 0.820 | 0.897 | 0.844 | 0.792 |
| BalancedBagging | 0.780 | 0.883 | 0.794 | 0.750 |
| RUSBoost | 0.830 | 0.897 | 0.979 | 0.792 |
| Isolation forest | 0.750 | 0.567 | 0.974 | 0.541 |
| Ensemble | Original | Ratio = 1 | Ratio = 0.75 | Ratio = 0.50 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SN | SP | G-Mean | SN | SP | G-Mean | SN | SP | G-Mean | SN | SP | G-Mean | |
| Single decision tree | 0.896 | 0.730 | 0.810 | 0.861 | 0.568 | 0.690 | 0.878 | 0.541 | 0.690 | 0.896 | 0.649 | 0.760 |
| Bagging | 0.948 | 0.675 | 0.800 | 0.930 | 0.784 | 0.850 | 0.922 | 0.703 | 0.800 | 0.913 | 0.676 | 0.790 |
| Random forest | 0.983 | 0.622 | 0.780 | 0.913 | 0.730 | 0.820 | 0.939 | 0.703 | 0.810 | 0.939 | 0.784 | 0.860 |
| XGBoost | 0.972 | 0.625 | 0.780 | 0.915 | 0.667 | 0.780 | 0.950 | 0.667 | 0.830 | 0.957 | 0.604 | 0.760 |
| AdaBoost | 0.965 | 0.703 | 0.82 | 0.948 | 0.703 | 0.820 | 0.957 | 0.784 | 0.870 | 0.948 | 0.757 | 0.850 |
| Ensemble | Original | Ratio = 1 | Ratio = 0.75 | Ratio = 0.50 | ||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| Single decision tree | 0.848 | 0.914 | 0.752 | 0.871 | 0.727 | 0.848 | 0.760 | 0.875 |
| Bagging | 0.940 | 0.978 | 0.953 | 0.986 | 0.943 | 0.983 | 0.946 | 0.984 |
| Random forest | 0.952 | 0.984 | 0.949 | 0.982 | 0.953 | 0.984 | 0.963 | 0.988 |
| XGBoost | 0.928 | 0.974 | 0.940 | 0.980 | 0.930 | 0.976 | 0.927 | 0.974 |
| AdaBoost | 0.895 | 0.941 | 0.951 | 0.981 | 0.940 | 0.967 | 0.925 | 0.959 |
| Ensemble | Original | Ratio = 1 | Ratio = 0.75 | Ratio = 0.50 | ||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| Single decision tree | 0.848 | 0.914 | 0.780 | 0.888 | 0.869 | 0.938 | 0.772 | 0.884 |
| Bagging | 0.901 | 0.960 | 0.944 | 0.983 | 0.945 | 0.983 | 0.919 | 0.974 |
| Random forest | 0.967 | 0.990 | 0.928 | 0.977 | 0.943 | 0.981 | 0.945 | 0.981 |
| XGBoost | 0.928 | 0.974 | 0.893 | 0.963 | 0.907 | 0.969 | 0.917 | 0.972 |
| AdaBoost | 0.937 | 0.981 | 0.947 | 0.983 | 0.954 | 0.986 | 0.922 | 0.962 |
| Ensemble | Original | Ratio = 1 | Ratio = 0.75 | Ratio = 0.50 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SN | SP | G-Mean | SN | SP | G-Mean | SN | SP | G-Mean | SN | SP | G-Mean | |
| Single decision tree | 0.896 | 0.730 | 0.810 | 0.739 | 0.811 | 0.770 | 0.809 | 0.784 | 0.800 | 0.861 | 0.622 | 0.730 |
| Bagging | 0.948 | 0.676 | 0.800 | 0.844 | 0.892 | 0.870 | 0.878 | 0.838 | 0.800 | 0.887 | 0.703 | 0.790 |
| Random forest | 0.983 | 0.622 | 0.780 | 0.844 | 0.811 | 0.830 | 0.870 | 0.784 | 0.830 | 0.939 | 0.622 | 0.760 |
| XGBoost | 0.972 | 0.625 | 0.780 | 0.804 | 0.744 | 0.770 | 0.902 | 0.769 | 0.830 | 0.920 | 0.744 | 0.830 |
| AdaBoost | 0.965 | 0.703 | 0.820 | 0.852 | 0.865 | 0.860 | 0.878 | 0.838 | 0.860 | 0.922 | 0.784 | 0.850 |
| Ensemble | 20 | 50 | 100 | 200 |
|---|---|---|---|---|
| Bagging | 0.941 | 0.940 | 0.941 | 0.941 |
| Random forest | 0.940 | 0.952 | 0.967 | 0.969 |
| AdaBoost | 0.940 | 0.895 | 0.850 | 0.857 |
| XGBoost | 0.919 | 0.928 | 0.929 | 0.927 |
| Balanced random forest | 0.872 | 0.897 | 0.913 | 0.910 |
| BalancedBagging | 0.892 | 0.883 | 0.863 | 0.872 |
| RUSBoost | 0.916 | 0.922 | 0.932 | 0.938 |
| Isolation forest | 0.566 | 0.567 | 0.571 | 0.550 |
| Ensemble | 20 | 50 | 100 | 200 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUPRC | SN | SP | G-Mean | AUPRC | SN | SP | G-Mean | AUPRC | SN | SP | G-Mean | AUPRC | SN | SP | G-Mean | |
| Bagging | 0.980 | 0.965 | 0.703 | 0.820 | 0.978 | 0.948 | 0.676 | 0.800 | 0.979 | 0.948 | 0.649 | 0.780 | 0.980 | 0.957 | 0.595 | 0.750 |
| Random forest | 0.979 | 0.966 | 0.622 | 0.770 | 0.984 | 0.983 | 0.622 | 0.780 | 0.990 | 0.974 | 0.595 | 0.760 | 0.990 | 0.965 | 0.541 | 0.770 |
| AdaBoost | 0.976 | 0.965 | 0.649 | 0.790 | 0.941 | 0.965 | 0.703 | 0.820 | 0.916 | 0.974 | 0.649 | 0.790 | 0.917 | 0.983 | 0.703 | 0.830 |
| XGBoost | 0.970 | 0.955 | 0.615 | 0.770 | 0.974 | 0.972 | 0.625 | 0.780 | 0.974 | 0.884 | 0.718 | 0.800 | 0.974 | 0.893 | 0.769 | 0.830 |
| Balanced random forest | 0.955 | 0.773 | 0.750 | 0.760 | 0.963 | 0.844 | 0.792 | 0.820 | 0.970 | 0.830 | 0.813 | 0.820 | 0.969 | 0.837 | 0.792 | 0.810 |
| BalancedBagging | 0.957 | 0.837 | 0.770 | 0.800 | 0.959 | 0.794 | 0.750 | 0.790 | 0.947 | 0.844 | 0.771 | 0.810 | 0.956 | 0.851 | 0.750 | 0.800 |
| RUSBoost | 0.966 | 0.872 | 0.770 | 0.820 | 0.970 | 0.979 | 0.792 | 0.830 | 0.973 | 0.943 | 0.771 | 0.850 | 0.976 | 0.972 | 0.771 | 0.870 |
| Isolation forest | 0.819 | 0.948 | 0.541 | 0.230 | 0.807 | 0.974 | 0.541 | 0.230 | 0.830 | 0.965 | 0.541 | 0.230 | 0.815 | 0.974 | 0.541 | 0.230 |
| Rank | 10 Best Feature Selected with Information Gain | Coefficient | 10 Best Feature Set with Lasso | Common Feature in Both Feature Selection |
|---|---|---|---|---|
| 0.1398 | std_6th_delta_delta | 3.181598 | std_6th_delta_delta | std_6th_delta_delta |
| 0.139 | std_delta_delta_log_energy | 2.406294 × 10−1 | std_delta_delta_log_energy | std_delta_delta_log_energy |
| 0.1371 | mean_MFCC_2nd_coef | 2.559498 × 10−2 | mean_MFCC_2nd_coef | mean_MFCC_2nd_coef |
| 0.1324 | std_delta_log_energy | 3.515898 × 10−7 | tqwt_entropy_log_dec_26 | |
| 0.1321 | tqwt_TKEO_mean_dec_12 | 2.720495 × 10−1 | tqwt_minValue_dec_12 | |
| 0.1311 | tqwt_entropy_log_dec_11 | 2.045678 | std_7th_delta_delta | |
| 0.1282 | tqwt_entropy_shannon_dec_11 | 3.281899 | std_9th_delta_delta | |
| 0.1258 | tqwt_stdValue_dec_11 | −1.039598 | tqwt_stdValue_dec_11 | tqwt_stdValue_dec_11 |
| 0.1239 | std_8th_delta_delta | −3.670655 × 10−4 | tqwt_kurtosisValue_dec_27 | |
| 0.1233 | tqwt_entropy_shannon_dec_12 | −2.894499 × 10−4 | tqwt_kurtosisValue_dec_26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barukab, O.; Ahmad, A.; Khan, T.; Thayyil Kunhumuhammed, M.R. Analysis of Parkinson’s Disease Using an Imbalanced-Speech Dataset by Employing Decision Tree Ensemble Methods. Diagnostics 2022, 12, 3000. https://doi.org/10.3390/diagnostics12123000
Barukab O, Ahmad A, Khan T, Thayyil Kunhumuhammed MR. Analysis of Parkinson’s Disease Using an Imbalanced-Speech Dataset by Employing Decision Tree Ensemble Methods. Diagnostics. 2022; 12(12):3000. https://doi.org/10.3390/diagnostics12123000
Chicago/Turabian StyleBarukab, Omar, Amir Ahmad, Tabrej Khan, and Mujeeb Rahiman Thayyil Kunhumuhammed. 2022. "Analysis of Parkinson’s Disease Using an Imbalanced-Speech Dataset by Employing Decision Tree Ensemble Methods" Diagnostics 12, no. 12: 3000. https://doi.org/10.3390/diagnostics12123000
APA StyleBarukab, O., Ahmad, A., Khan, T., & Thayyil Kunhumuhammed, M. R. (2022). Analysis of Parkinson’s Disease Using an Imbalanced-Speech Dataset by Employing Decision Tree Ensemble Methods. Diagnostics, 12(12), 3000. https://doi.org/10.3390/diagnostics12123000







