Ensemble Deep Learning for Cervix Image Selection toward Improving Reliability in Automated Cervical Precancer Screening
Abstract
1. Introduction
2. Image Data
2.1. MobileODT Dataset
2.2. Kaggle Dataset
2.3. COCO Dataset
2.4. SEVIA Dataset
3. Methods
3.1. RetinaNet
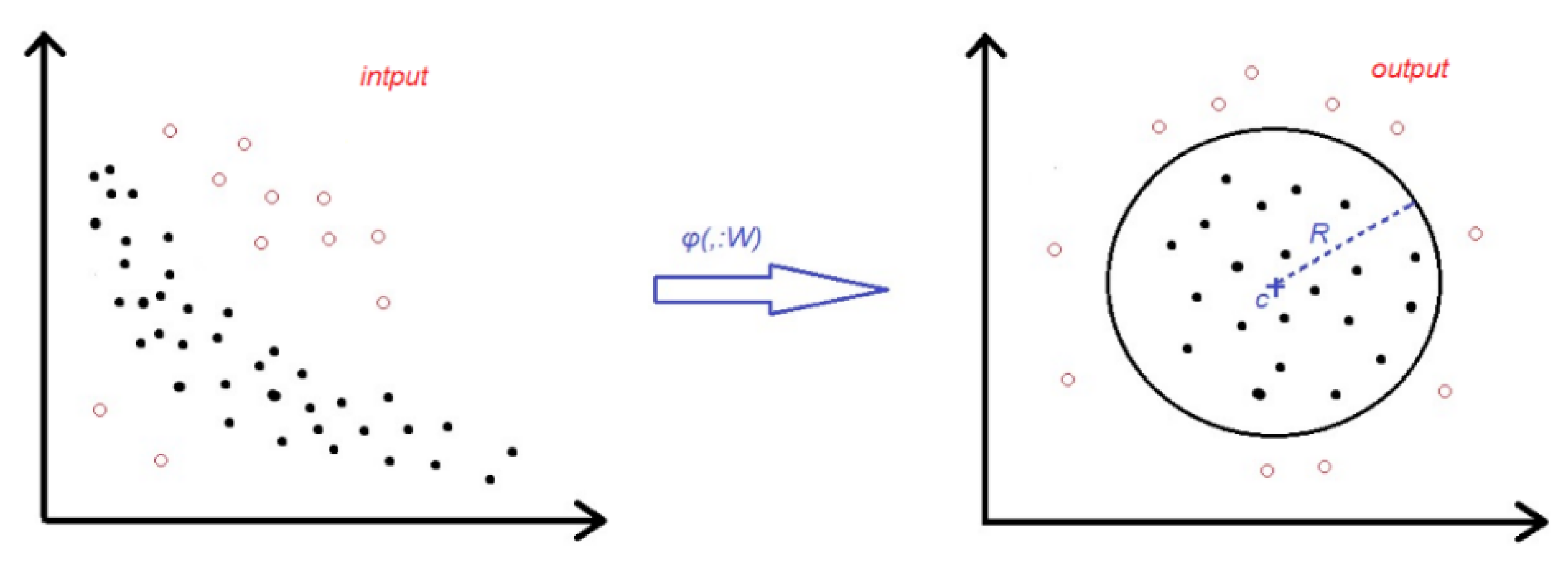
3.2. Deep SVDD
3.3. Customized CNN
3.4. Ensemble Method
4. Experiments
4.1. Data Preparation and Implementation Details
- RetinaNet: The batch size is 4, and a learning rate starting at 0.001. The weight decay is set as 0.0001 and the momentum is 0.9. The model uses ResNet50 [23] as the backbone and is initialized with pre-trained RetinaNet weights that were obtained in [10]. Augmentations of rotation, shearing, shifting and x/y flipping are used. Python 3.6 and Keras 2.2.4 are used.
- Deep SVDD: The batch size is 8 and the weight decay factor of λ = 0.5 × 10−5. Base learning rate is set as 0.001. Python version is 3.7 and PyTorch version is 0.5. Our Deep SVDD network is modified based on the PyTorch implementation in [24].
- Customized CNN: The batch size is 16. Learning rate is set as 10−4 and momentum is set as 0.9. “Categorical cross entropy” loss is used and updated via Stochastic Gradient Descent (SGD). Augmentations of scaling, horizontal/vertical shift and rotation are used. Python 3.6 and Keras 2.2.4 are used.
4.2. Results and Disccussion
4.2.1. Results of RetinaNet
4.2.2. Results of Deep SVDD
4.2.3. Results of Customized CNN
4.2.4. Results of Ensemble Method
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Human Papillomavirus (HPV) and Cervical Cancer. Available online: https://www.who.int/en/news-room/fact-sheets/detail/human-papillomavirus-(hpv)-and-cervical-cancer (accessed on 2 October 2019).
- Bhattacharyya, A.; Nath, J.; Deka, H. Comparative study between pap smear and visual inspection with acetic acid (via) in screening of CIN and early cervical cancer. J. Mid Life Health 2015, 6, 53–58. [Google Scholar]
- Egede, J.; Ajah, L.; Ibekwe, P.; Agwu, U.; Nwizu, E.; Iyare, F. Comparison of the accuracy of papanicolaou test cytology, Visual Inspection with Acetic acid, and Visual Inspection with Lugol Iodine in screening for cervical neoplasia in southeast Nigeria. J. Glob. Oncol. 2018, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sritipsukho, P.; Thaweekul, Y. Accuracy of visual inspection with acetic acid (VIA) for cervical cancer screening: A systematic review. J. Med. Assoc. Thai. 2010, 93, 254–261. [Google Scholar]
- Jeronimo, J.; Massad, L.S.; Castle, P.E.; Wacholder, S.; Schiffman, M.; National Institutes of Health (NIH)-American Society for Colposcopy and Cervical Pathology (ASCCP) Research Group. Interobserver agreement in the evaluation of digitized cervical images. Obstet. Gynecol. 2007, 110, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Bell, D.; Antani, S.; Xue, Z.; Yu, K.; Horning, M.P.; Gachuhi, N.; Wilson, B.; Jaiswal, M.S.; Befano, B.; et al. An observational study of deep learning and automated evaluation of cervical images for cancer screening. J. Natl. Cancer Inst. 2019, 111, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Bratti, M.C.; Rodriguez, A.C.; Schiffman, M.; Hildesheim, A.; Morales, J.; Alfaro, M.; Guillén, D.; Hutchinson, M.; Sherman, M.E.; Eklund, C.; et al. Description of a seven-year prospective study of human papillomavirus infection and cervical neoplasia among 10000 women in Guanacaste, Costa Rica. Rev. Panam. Salud Publica 2004, 152, 75–89. [Google Scholar]
- Herrero, R.; Schiffman, M.; Bratti, C.; Hildesheim, A.; Balmaceda, I.; Sherman, M.E.; Greenberg, M.; Cárdenas, F.; Gómez, V.; Helgesen, K.; et al. Design and methods of a population-based natural history study of cervical neoplasia in a rural province of Costa Rica: The Guanacaste Project. Rev. Panam. Salud Publica 1997, 15, 362–375. [Google Scholar] [CrossRef] [PubMed]
- AI Approach Outperformed Human Experts in Identifying Cervical Precancer. Available online: https://www.cancer.gov/news-events/press-releases/2019/deep-learning-cervical-cancer-screening (accessed on 15 May 2020).
- Guo, P.; Singh, S.; Xue, Z.; Long, L.R.; Antani, S. Deep learning for assessing image focus for automated cervical cancer screening. In Proceedings of the 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Chicago, IL, USA, 19–22 May 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Guo, P.; Xue, Z.; Long, L.R.; Antani, S. Deep learning cervix anatomical landmark segmentation and evaluation. SPIE Med. Imaging 2020. [Google Scholar] [CrossRef]
- Ruff, L.; Vandermeulen, R.A.; Geornitz, N.; Deecke, L.; Siddiqui, S.A.; Binder, A.; Müller, E.; Kloft, M. Deep one-class classification. In Proceedings of the 35th International Conference on Machine Learning, PMLR, Stockholm, Sweden, 10–15 July 2018; Volume 80, pp. 4393–4402. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal loss for dense object detection. In ICCV; IEEE: Piscataway, NJ, USA, 2017. [Google Scholar]
- Common Objects in Context (COCO) Dataset. Available online: http://cocodataset.org/#home (accessed on 20 May 2020).
- Intel & MobileODT Cervical Cancer Screening Competition, March 2017. Available online: https://www.kaggle.com/c/intel-mobileodt-cervical-cancer-screening (accessed on 13 May 2020).
- Fernandes, K.; Cardoso, J.S. Ordinal image segmentation using deep neural networks. In Proceedings of the International Joint Conference on Neural Networks, Rio de Janeiro, Brazil, 8–13 July 2018; pp. 1–7. [Google Scholar]
- Yeates, K.; Sleeth, J.; Hopman, W.; Ginsburg, O.; Heus, K.; Andrews, L.; Giattas, M.R.; Yuma, S.; Macheku, G.; Msuya, A.; et al. Evaluation of a Smartphone-Based Training Strategy among Health Care Workers Screening for Cervical Cancer in Northern Tanzania: The Kilimanjaro Method. J. Glob. Oncol. 2016, 2, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Moya, M.M.; Koch, M.W.; Hostetler, L.D. One-class classifier networks for target recognition applications. In Proceedings World Congress on Neural Networks; International Neural Network Society: Portland, OR, USA, 1993; pp. 797–801. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2019. [Google Scholar]
- Lin, T.Y.; Dollar, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 27. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In CVPR; IEEE: Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- A PyTorch Implementation of the Deep SVDD Anomaly Detection Method. Available online: https://github.com/lukasruff/Deep-SVDD-PyTorch (accessed on 23 April 2020).







| Dataset | Training | Validation | Testing | ||
|---|---|---|---|---|---|
| RetinaNet | Deep SVDD | Custom CNN | All Methods | All Methods | |
| MobileODT | 3118 | 3118 | 7170 | 814 | 0 |
| Kaggle | 1481 | 1481 | 1481 | 512 | 0 |
| COCO | 4599 | 0 | 8561 (contains the 4599 images used for training RetinaNet) | 1326 | 0 |
| SEVIA | 0 | 0 | 0 | 0 | 30,151 + 1816 (in 10 folds, last fold has 3016 cervix images) |
| Methods | Resized Dimension (w × h) |
|---|---|
| Deep SVDD | 128 × 128 |
| RetinaNet | 480 × 640 |
| Customized CNN | 256 × 256 |
| (a) | |||||
| Methods | Accuracy (Avg./Std.) | Sens. (Avg.) | Spec. (Avg./Std.) | F-1 Score (Avg.) | |
| RetinaNet | 0.911/0.010 | 0.918 | 0.908/0.011 | 0.877 | |
| (b) | |||||
| True Positive | True Negative | ||||
| Predicted Positive | 1667 | 279 | |||
| Predicted Negative | 149 | 2736 | |||
| (a) | |||||
| Methods | Accuracy (Avg./Std.) | Sens. (Avg.) | Spec. (Avg./Std.) | F-1 Score (Avg.) | |
| Deep SVDD | 0.889/0.010 | 0.922 | 0.876/0.010 | 0.863 | |
| (b) | |||||
| True Positive | True Negative | ||||
| Predicted Positive | 1674 | 394 | |||
| Predicted Negative | 142 | 2621 | |||
| (a) | |||||
| Methods | Accuracy (Avg./Std.) | Sens. (Avg.) | Spec. (Avg./Std.) | F-1 Score (Avg.) | |
| Customized CNN | 0.830/0.010 | 0.924 | 0.780/0.012 | 0.810 | |
| (b) | |||||
| True Positive | True Negative | ||||
| Predicted Positive | 1678 | 667 | |||
| Predicted Negative | 138 | 2348 | |||
| (a) | |||||
| Method | Accuracy (Avg./Std.) | Sens. (Avg.) | Spec. (Avg./Std.) | F-1 Score (Avg.) | |
| Ensemble | 0.916/0.004 | 0.935 | 0.900/0.008 | 0.890 | |
| (b) | |||||
| True Positive | True Negative | ||||
| Predicted Positive | 1698 | 301 | |||
| Predicted Negative | 118 | 2715 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, P.; Xue, Z.; Mtema, Z.; Yeates, K.; Ginsburg, O.; Demarco, M.; Long, L.R.; Schiffman, M.; Antani, S. Ensemble Deep Learning for Cervix Image Selection toward Improving Reliability in Automated Cervical Precancer Screening. Diagnostics 2020, 10, 451. https://doi.org/10.3390/diagnostics10070451
Guo P, Xue Z, Mtema Z, Yeates K, Ginsburg O, Demarco M, Long LR, Schiffman M, Antani S. Ensemble Deep Learning for Cervix Image Selection toward Improving Reliability in Automated Cervical Precancer Screening. Diagnostics. 2020; 10(7):451. https://doi.org/10.3390/diagnostics10070451
Chicago/Turabian StyleGuo, Peng, Zhiyun Xue, Zac Mtema, Karen Yeates, Ophira Ginsburg, Maria Demarco, L. Rodney Long, Mark Schiffman, and Sameer Antani. 2020. "Ensemble Deep Learning for Cervix Image Selection toward Improving Reliability in Automated Cervical Precancer Screening" Diagnostics 10, no. 7: 451. https://doi.org/10.3390/diagnostics10070451
APA StyleGuo, P., Xue, Z., Mtema, Z., Yeates, K., Ginsburg, O., Demarco, M., Long, L. R., Schiffman, M., & Antani, S. (2020). Ensemble Deep Learning for Cervix Image Selection toward Improving Reliability in Automated Cervical Precancer Screening. Diagnostics, 10(7), 451. https://doi.org/10.3390/diagnostics10070451







