NADPH Oxidases: The Vital Performers and Center Hubs during Plant Growth and Signaling
Abstract
:1. Introduction
2. Protein Structure and Evolution of Plant NOXs/RBOHs
2.1. Highly Conservative Structure
2.2. Complex Evolution History
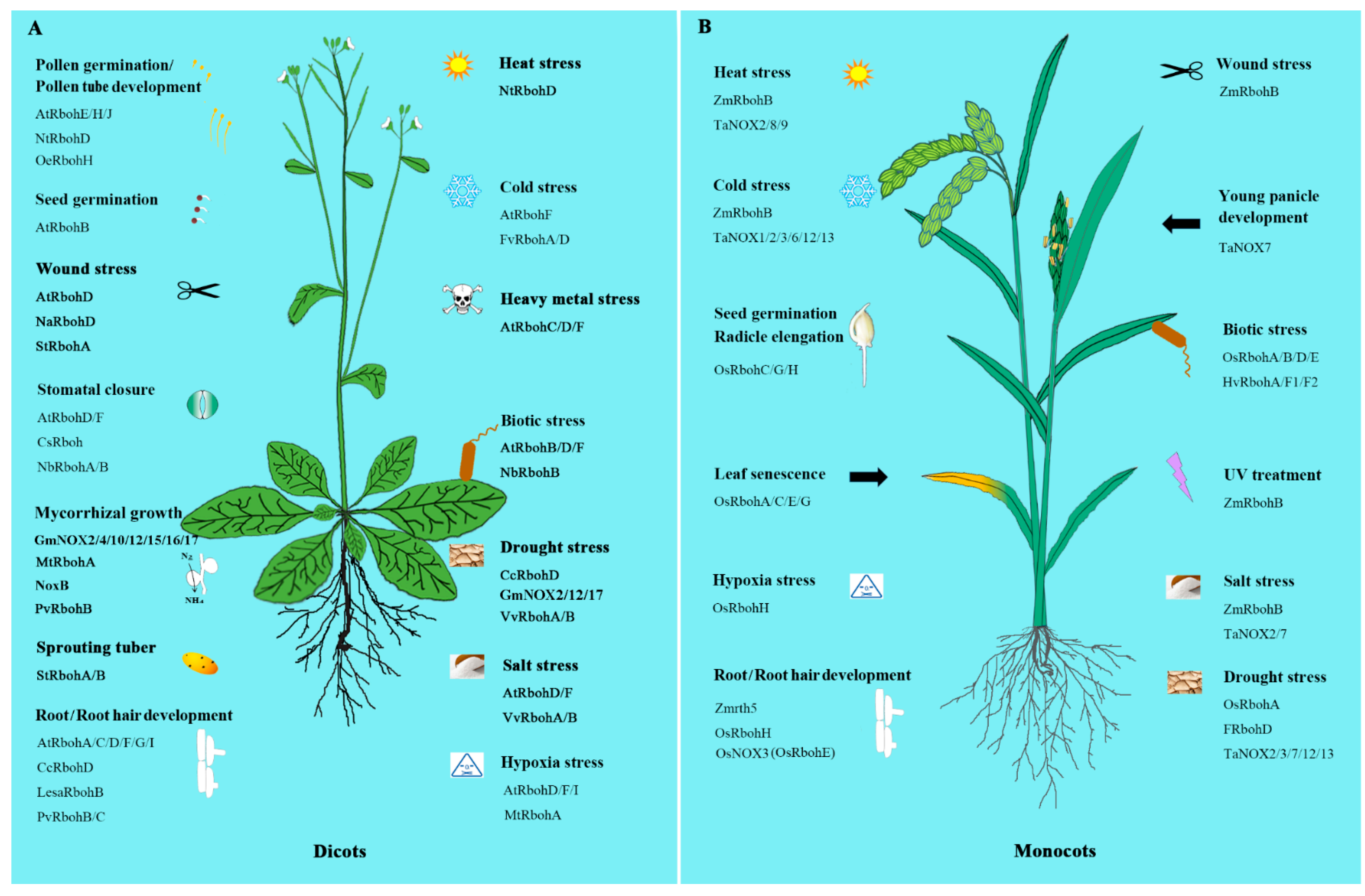
3. NOXs/RBOHs in Plant Development
3.1. Pollen Germination and Pollen Tube Growth
3.2. Root and Root Hair Development
3.3. Seed Germination
3.4. Plant-Microorganism Ineractions
3.5. Fungal Development
3.6. Other Aspects
4. NOXs/RBOHs in Stress Tolerance
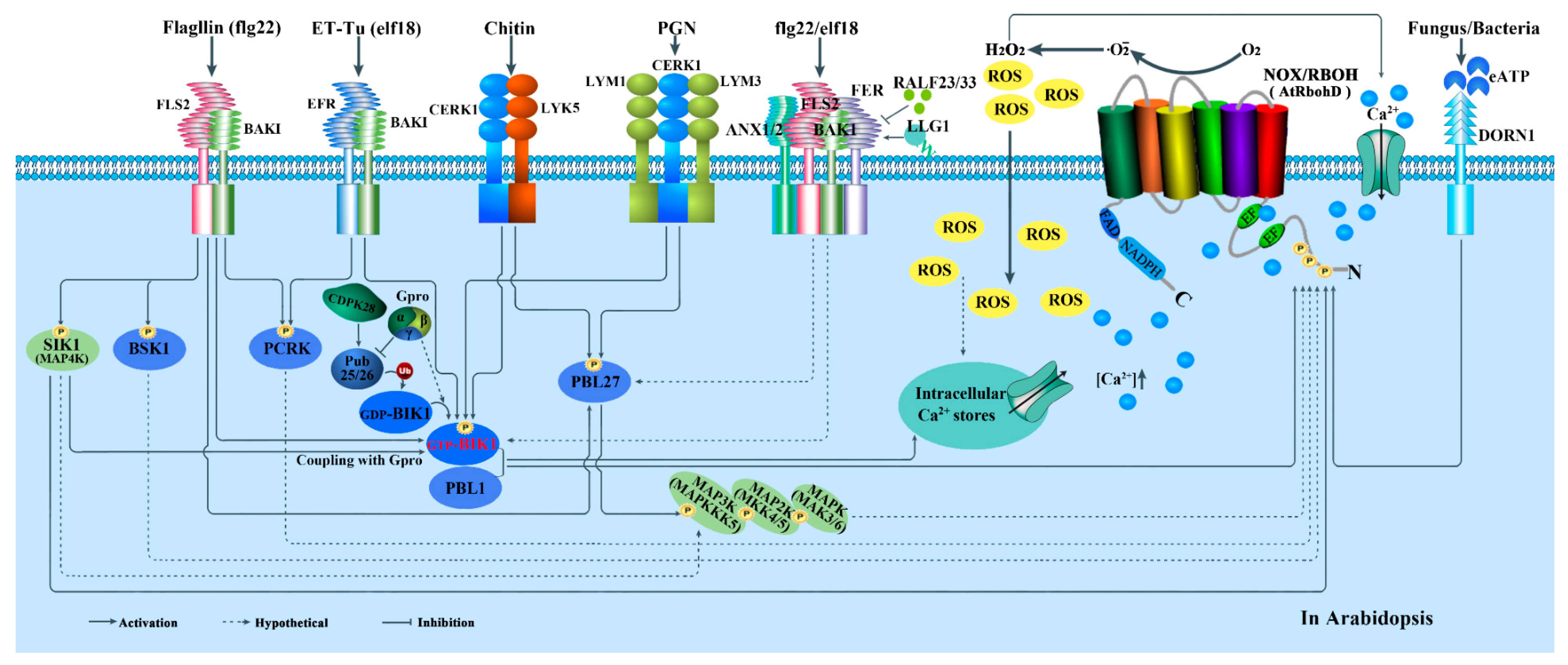
4.1. Roles in Plant Immunity
4.2. Roles in Abiotic Stress Tolerance
4.2.1. Salinity Stress
4.2.2. Hypoxic Stress
4.2.3. Heavy Metal Stress
4.2.4. Wound Response
4.2.5. Temperature Response
4.2.6. Drought Stress
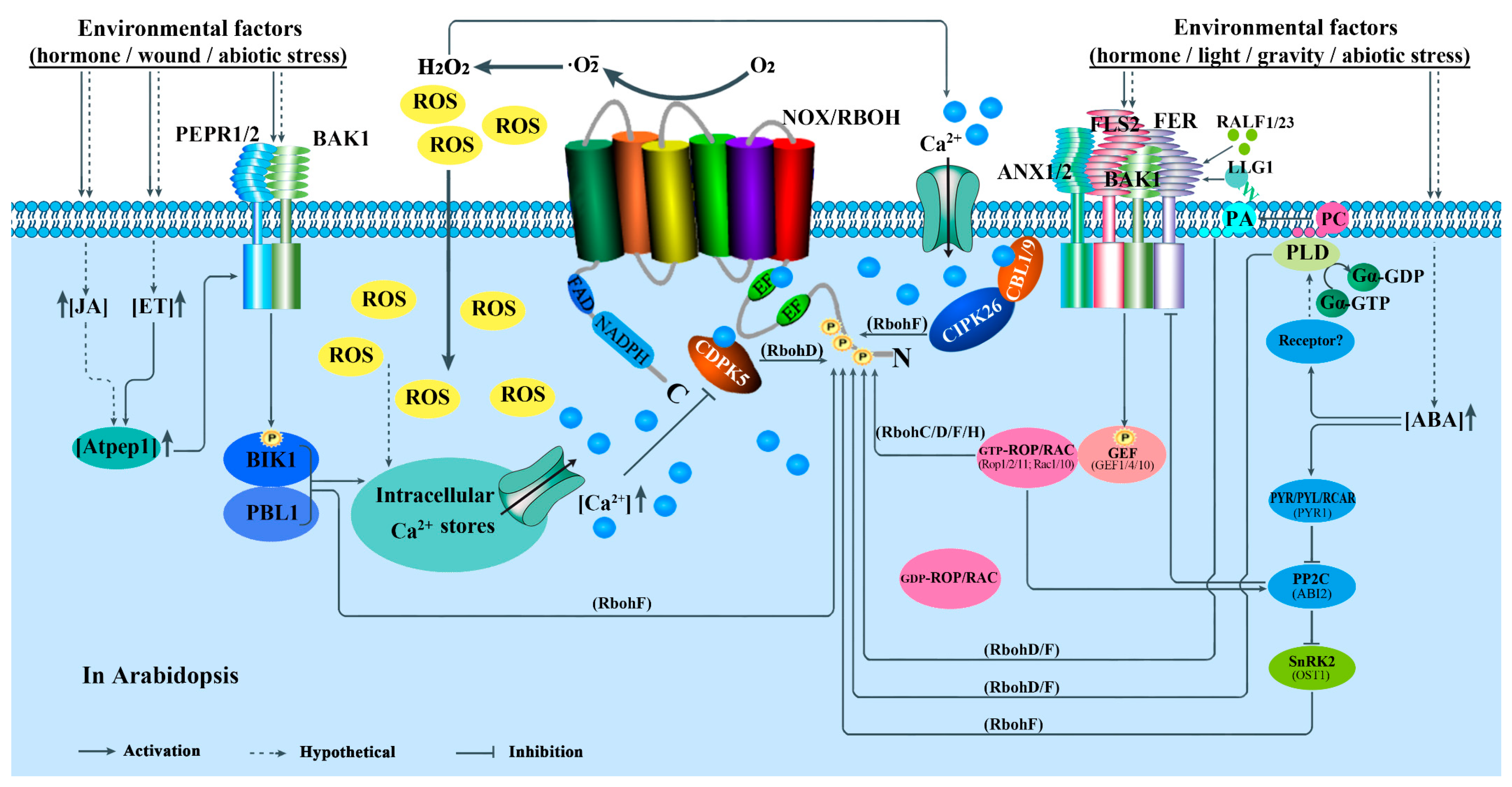
4.2.7. Hormonal Response
5. Regulations of NOXs/RBOHs in Plants
5.1. Transcriptional Regulation
5.2. Activity Regulation
5.2.1. Ca2+-Regulated Protein Kinases-Mediated Regulation
5.2.2. RLKs and RLCKs-Mediated Regulation
5.2.3. Rho-Type GTPases-Mediated Regulation
5.2.4. MAPK Cascades-Mediated Regulation
5.2.5. Hormone-Mediated Regulation
5.2.6. Other Kinds of Particles-Mediated Regulation
6. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Full-Name | Abbreviation |
| abscisic acid | ABA |
| ABA insensitive | ABI |
| ABA-responsive element | ABRE |
| arbuscular mycorrhizal fungal | AMF |
| ANXUR | ANX |
| α-naphthaleneacetic acid | α-NAA |
| BRI1-associated receptor kinase 1 | BAK1 |
| botrytis-induced kinase1 | BIK1 |
| brassinosteroid | BR |
| brassinolide-signaling kinase | BSK |
| cis-acting regulatory element | CARE |
| calcineurin B-like | CBL |
| calcium-dependent protein kinase | CDPK |
| chitin elicitor-binding protein | CEBiP |
| chitin-elicitor receptor kinase | CERK |
| calcineurin B-like-interacting protein kinase | CIPK |
| coronatine-insensitive | COI |
| damage associated molecular pattern | DAMP |
| dominant negative form | DN |
| diphenylene iodonium | DPI |
| dual oxidase | DUOX |
| elongation factor-tu receptor | EFR |
| ethylene | ET |
| effector triggered immunity | ETI |
| ethylene receptor | ETR |
| flavin adenine dinucleotide | FAD |
| FERONIA | FER |
| flagellin sensing 2 | FLS2 |
| ferric reduction oxidase | FRO |
| guanine nucleotide exchange factors | GEF |
| glycosylphosphatidylinositol-anchored protein | GPI-AP |
| the α subunit of G protein | GPA1 |
| guanosine triphosphate | GTP |
| hypersensitive type of resistance | HR |
| a H+ pump by coupling with enerty of ATP hydrolysis on plasma membranes | H+-ATPases |
| indole-3-acetic acid | IAA |
| jasmonate | JA |
| L-type lectin receptor-like kinase | LecRK |
| lorelei-like GPI-anchored protein 1 | LLG1 |
| leucine-rich repeat | LRR |
| LysM-containing receptor-like kinase | LYK |
| mitogen-activated protein kinase | MAPK |
| methyl jasmonate | MeJA |
| leucine-rich repeat containing receptor | NLR |
| nitric oxide | NO |
| NADPH oxidase | NOX |
| superoxide anion | ·O2- |
| open stomata | OST |
| phosphatidic acid | PA |
| pathogen-associated molecular pattern | PAMP |
| PBS1-like kinase | PBL1 |
| phosphatidylcholine | PC |
| programmed cell death | PCD |
| pattern-triggered immunity compromised receptor-like cytoplasmic kinase | PCRK |
| phosphoenolpyruvic acid | PEP |
| PEP 1 receptor | PEPR1 |
| plasma membrane | PM |
| phospholipase D alpha | PLDA |
| peptidoglycan | PGN |
| protein phosphatase 2C | PP2C |
| pattern recognition receptor | PRR |
| PAMP-triggered immunity | PTI |
| PYR-LIKE | PYL |
| pyrabactin resistance protein | PYR |
| receptor for activated C-kinase | RACK |
| rapid alkalinization factors | RALF |
| respiratory burst oxidase homolog | RBOH |
| regularity components of ABA receptor | RCAR |
| receptor-like cytoplasmic kinases | RLCK |
| receptor-like protein kinase | RLK |
| receptor-like kinase-like proteins | RLKL |
| reactive oxygen species | ROS |
| Rho-type GTPases belong to the Rat sarcoma (Ras) superfamily of small GTP-binding proteins, and plants have a sole subfamily of Rho-type GTPases, called ROPs (Rho of plants) or RACs (for the sequence similarity they share with animal Racs, a Rho subfamily). | ROP/RAC |
| salicylic acid | SA |
| systemic acquired acclimation | SAA |
| salt inducible kinase | SIK |
| somatic embryogenesis receptor kinase | SERK |
| salicylic acid-induced protein kinase | SIPK |
| sucrose non-fermenting | SNF |
| SNF1-related protein kinase family | SnRK |
| transcription factor | TF |
| wound-induced protein kinase | WIPK |
| a protein that it has an atypical zinc-finger structure at the C-terminus and bind preferentially to the core sequence of W box (TTGACC/T) on the DNA sequence of some promoters | WRKY |
| 2,4-dichlorophenoxyacetic acid | 2,4-D |
References
- Gapper, C.; Dolan, L. Control of Plant Development by Reactive Oxygen Species. Plant Physiol. 2006, 141, 341–345. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.A. Ros in Biotic Interactions. Physiol. Plant. 2010, 138, 414–429. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.V.; Suzuki, S.; Miller, N.; Tognetti, G.; Vandepoele, V.B.; Gollery, K.; Shulaev, M.; Van Breusegem, V.F. Ros Signaling: The New Wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A Burst of Plant NADPH Oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Sharma, A.; Guruprasad, K.; Pati, P.K. Versatile Roles of Plant NADPH Oxidases and Emerging Concepts. Biotechnol. Adv. 2014, 32, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Sagi, M.; Fluhr, R. Production of Reactive Oxygen Species by Plant NADPH Oxidases. Plant Physiol. 2006, 141, 336–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.L.; Li, W.Y.; Miao, H.; Yang, S.Q.; Li, R.; Wang, X.; Li, W.Q.; Chen, K.M. Comprehensive Genomic Analysis and Expression Profiling of the Nox Gene Families under Abiotic Stresses and Hormones in Plants. Genome Biol. Evol. 2016, 8, 791–810. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.H.; Wei, X.Y.; Yuan, B.; Yao, L.B.; Ma, T.T.; Zhang, P.P.; Wang, X.; Wang, P.Q.; Liu, W.; Tai, L.; et al. Genome-Wide Identification and Functional Analysis of NADPH Oxidase Family Genes in Wheat during Development and Environmental Stress Responses. Front. Plant Sci. 2018, 9, 906. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.A.; Dangl, J.L. Functions of the Respiratory Burst Oxidase in Biotic Interactions, Abiotic Stress and Development. Curr. Opin. Plant Biol. 2005, 8, 397–403. [Google Scholar] [CrossRef]
- Bedard, K.; Krause, K.H. The Nox Family of Ros-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Bedard, K.; Lardy, B.; Krause, K.H. Nox Family NADPH Oxidases: Not Just in Mammals. Biochimie 2007, 89, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, J.; Rios-Momberg, M.; Hewitt, D.; Hansberg, W. Reactive Oxygen Species and Development in Microbial Eukaryotes. Trends Microbiol. 2005, 13, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Babior, B.M.; Lambeth, J.D.; Nauseef, W. The Neutrophil NADPH Oxidase. Arch. Biochem. Biophys. 2002, 397, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, J.D. Nox Enzymes and the Biology of Reactive Oxygen. Nat. Rev. Immunol. 2004, 4, 181–189. [Google Scholar] [CrossRef]
- Cross, A.R.; Segal, A.W. The NADPH Oxidase of Professional Phagocytes—Prototype of the Nox Electron Transport Chain Systems. BBA Bioenerg. 2004, 1657, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leto, T.L.; Geiszt, M. Role of Nox Family NADPH Oxidases in Host Defense. Antioxid. Redox Sign. 2006, 8, 1549–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glyan’ko, A.K.; Ischenko, A.A. Structural and Functional Characteristics of Plant NADPH Oxidase: A Review. Appl. Biochem. Microbiol. 2010, 46, 463–471. [Google Scholar] [CrossRef]
- Rada, B.; Leto, T. Oxidative Innate Immune Defenses by Nox/Duox Family NADPH Oxidases. In Contributions to Microbiology; Egesten, A., Schmidt, A., Herwald, H., Eds.; Karger: Basel, Switzerland, 2008; pp. 164–187. [Google Scholar]
- Wang, G.F.; Li, W.Q.; Li, W.Y.; Wu, L.C.; Zhou, Y.; Chen, K.M. Characterization of Rice NADPH Oxidase Genes and Their Expression under Various Environmental Conditions. Int. J. Mol. Sci. 2013, 14, 9440–9458. [Google Scholar] [CrossRef] [Green Version]
- Xing, T.; Higgins, V.J.; Blumwald, E. Race-Specific Elicitors of Cladosporium Fulvum Promote Translocation of Cytosolic Components of NADPH Oxidase to the Plasma Membrane of Tomato Cells. Plant Cell 1997, 9, 249–259. [Google Scholar]
- Wang, X.; Zhang, M.M.; Wang, Y.J.; Gao, Y.T.; Li, R.; Wang, G.F.; Li, W.Q.; Liu, W.T.; Chen, K.M. The Plasma Membrane NADPH Oxidase Osrboha Plays a Crucial Role in Developmental Regulation and Drought-Stress Response in Rice. Physiol. Plant. 2016, 156, 421–443. [Google Scholar] [CrossRef]
- Morgante, M.; Brunner, S.; Pea, G.; Fengler, K.; Zuccolo, A.; Rafalski, A. Gene Duplication and Exon Shuffling by Helitron-Like Transposons Generate Intraspecies Diversity in Maize. Nat. Genet. 2005, 37, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Fawcett, J.A.; Proost, S.; Sterck, L.; Vandepoele, K. The Flowering World: A Tale of Duplications. Trends Plant Sci. 2009, 14, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Kaessmann, H. Origins, Evolution, and Phenotypic Impact of New Genes. Genome Res. 2010, 20, 1313–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene Duplication as a Major Force in Evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Potocky, M.; Jones, M.A.; Bezvoda, R.; Smirnoff, N.; Zarsky, V. Reactive Oxygen Species Produced by NADPH Oxidase Are Involved in Pollen Tube Growth. New Phytol. 2007, 174, 742–751. [Google Scholar] [CrossRef]
- Wang, S.S.; Zhu, X.N.; Lin, J.X.; Zheng, W.J.; Zhang, B.T.; Zhou, J.Q.; Ni, J.; Pan, Z.C.; Zhu, S.H.; Ding, W.N. OsNOX3, encoding a NADPH oxidase, regulates root hair initiation and elongation in rice. Biol. Plant. 2018, 62, 732–740. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Lituiev, D.S.; Nestorova, A.; Franck, C.M.; Thirugnanarajah, S.; Grossniklaus, U. Anxur Receptor-Like Kinases Coordinate Cell Wall Integrity with Growth at the Pollen Tube Tip via NADPH Oxidases. PLoS Biol. 2013, 11, e1001719. [Google Scholar] [CrossRef] [Green Version]
- Potocky, M.; Pejchar, P.; Gutkowska, M.; Jimenez-Quesada, M.J.; Potocka, A.; Alche Jde, D.; Kost, B.; Zarsky, V. NADPH Oxidase Activity in Pollen Tubes Is Affected by Calcium Ions, Signaling Phospholipids and Rac/Rop Gtpases. J. Plant Physiol. 2012, 169, 1654–1663. [Google Scholar] [CrossRef]
- Wang, X.L.; Takai, T.; Kamijo, S.; Gunawan, H.; Ogawa, H.; Okumura, K. NADPH Oxidase Activity in Allergenic Pollen Grains of Different Plant Species. Biochem. Biophys. Res. Commun. 2009, 387, 430–434. [Google Scholar] [CrossRef]
- Pasqualini, S.; Tedeschini, E.; Frenguelli, G.; Wopfner, N.; Ferreira, F.; D’Amato, G.; Ederli, L. Ozone Affects Pollen viability and Nad(P)H Oxidase Release from Ambrosia Artemisiifolia Pollen. Environ. Pollut. 2011, 159, 2823–2830. [Google Scholar] [CrossRef] [Green Version]
- Kaya, H.; Nakajima, R.; Iwano, M.; Kanaoka, M.M.; Kimura, S.; Takeda, S.; Kawarazaki, T.; Senzaki, E.; Hamamura, Y.; Higashiyama, T.; et al. Ca2+-Activated Reactive Oxygen Species Production by Arabidopsis Rbohh and Rbohj Is Essential for Proper Pollen Tube Tip Growth. Plant Cell 2014, 26, 1069–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaya, H.; Iwano, M.; Takeda, S.; Kanaoka, M.M.; Kimura, S.; Abe, M.; Kuchitsu, K. Apoplastic Ros Production Upon Pollination by Rbohh and Rbohj in Arabidopsis. Plant Signal. Behav. 2015, 10, e989050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaya, H.; Takeda, S.; Kobayashi, M.J.; Kimura, S.; Iizuka, A.; Imai, A.; Hishinuma, H.; Kawarazaki, T.; Mori, K.; Yamamoto, Y.; et al. Comparative Analysis of the Reactive Oxygen Species-Producing Enzymatic Activity of Arabidopsis NADPH Oxidases. Plant J. 2019, 98, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.T.; Wan, Z.Y.; Li, S.; Zhang, Y. Spatiotemporal Production of Reactive Oxygen Species by NADPH Oxidase Is Critical for Tapetal Programmed Cell Death and Pollen Development in Arabidopsis. Plant Cell 2014, 26, 2007–2023. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Quesada, M.J.; Traverso, J.A.; Potocky, M.; Zarsky, V.; Alche, J.D. Generation of Superoxide by Oerbohh, a NADPH Oxidase Activity During Olive (Olea Europaea L.) Pollen Development and Germination. Front. Plant Sci. 2019, 10, 1149. [Google Scholar] [CrossRef] [Green Version]
- Cramer, G.R.; Jones, R.L. Osmotic Stress and Abscisic Acid Reduce Cytosolic Calcium Activities in Roots of Arabidopsis Thaliana. Plant Cell Environ. 1996, 19, 1291–1298. [Google Scholar] [CrossRef]
- Majumdar, A.; Kar, R.K. Congruence between Pm H+-Atpase and NADPH Oxidase During Root Growth: A Necessary Probability. Protoplasma 2019, 255, 1129–1137. [Google Scholar] [CrossRef]
- Bacon, E.; Morre, D.J. Plasma Membrane Nadh Oxidase of Maize Roots Responds to Gravity and Imposed Centrifugal Forces. Plant Physiol. Biochem. 2001, 39, 487–494. [Google Scholar] [CrossRef]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D. Reactive Oxygen Species Produced by NADPH Oxidase Regulate Plant Cell Growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef]
- Jones, M.A.; Raymond, M.J.; Yang, Z.; Smirnoff, N. NADPH Oxidase-Dependent Reactive Oxygen Species Formation Required for Root Hair Growth Depends on Rop Gtpase. J. Exp. Bot. 2007, 58, 1261–1270. [Google Scholar] [CrossRef] [Green Version]
- Nestler, J.; Liu, S.Z.; Wen, T.J.; Paschold, A.; Marcon, C.; Tang, H.M.; Li, D.L.; Li, L.; Meeley, R.B.; Sakai, H.; et al. Roothairless5, Which Functions in Maize (Zea Mays L.) Root Hair Initiation and Elongation Encodes a Monocot-Specific NADPH Oxidase. Plant J. 2014, 79, 729–740. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Sun, L.; Song, Y.; Wang, L.; Liu, L.; Zhang, L.; Liu, B.; Li, N.; Miao, C.; Hao, F. Atrbohd and Atrbohf Positively Regulate Abscisic Acid-Inhibited Primary Root Growth by Affecting Ca2+ Signalling and Auxin Response of Roots in Arabidopsis. J. Exp. Bot. 2013, 64, 4183–4192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Sun, L.; Zhang, L.; Song, Y.; Hu, P.; Li, C.; Hao, F.S. Atrbohd and Atrbohf Negatively Regulate Lateral Root Development by Changing the Localized Accumulation of Superoxide in Primary Roots of Arabidopsis. Planta 2015, 241, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Montiel, J.; Arthikala, M.K.; Quinto, C. Phaseolus Vulgaris Rbohb Functions in Lateral Root Development. Plant Signal. Behav. 2013, 8, e22694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montiel, J.; Fonseca-García, C.; Quinto, C. Phylogeny and Expression of NADPH Oxidases during Symbiotic Nodule Formation. Agriculture 2018, 8, 179. [Google Scholar] [CrossRef] [Green Version]
- Tewari, R.K.; Kim, S.; Hahn, E.J.; Paek, K.Y. Involvement of Nitric Oxide-Induced NADPH Oxidase in Adventitious Root Growth and Antioxidant Defense in Panax Ginseng. Plant Biotechnol. Rep. 2008, 2, 113–122. [Google Scholar] [CrossRef]
- Muller, K.; Carstens, A.C.; Linkies, A.; Torres, M.A.; Leubner-Metzger, G. The Nadph-Oxidase Atrbohb Plays a Role in Arabidopsis Seed after-Ripening. New Phytol. 2009, 184, 885–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.Y.; Chen, B.X.; Chen, Z.J.; Gao, Y.T.; Chen, Z.; Liu, J. Reactive Oxygen Species Generated by NADPH Oxidases Promote Radicle Protrusion and Root Elongation During Rice Seed Germination. Int. J. Mol. Sci. 2017, 18, 110. [Google Scholar] [CrossRef]
- Tanaka, A.; Christensen, M.J.; Takemoto, D.; Park, P.; Scott, B. Reactive Oxygen Species Play a Role in Regulating a Fungus-Perennial Ryegrass Mutualistic Interaction. Plant Cell 2006, 18, 1052–1066. [Google Scholar] [CrossRef] [Green Version]
- Yushi, I.; Tawaratsumida, T.; Zheng, S.J.; Yuasa, T.; Iwaya-Inoue, M. NADPH Oxidases Act as Key Enzyme on Germination and Seedling Growth in Barley (Hordeum Vulgare L.). Plant Prod. Sci. 2010, 13, 45–52. [Google Scholar]
- Ishibashi, Y.; Kasa, S.; Sakamoto, M.; Aoki, N.; Kai, K.; Yuasa, T.; Hanada, A.; Yamaguchi, S.; Iwaya-Inoue, M. A Role for Reactive Oxygen Species Produced by NADPH Oxidases in the Embryo and Aleurone Cells in Barley Seed Germination. PLoS ONE 2015, 10, e0143173. [Google Scholar] [CrossRef] [PubMed]
- Kai, K.; Kasa, S.; Sakamoto, M.; Aoki, N.; Watabe, G.; Yuasa, T.; Iwaya-Inoue, M.; Ishibashi, Y. Role of Reactive Oxygen Species Produced by NADPH Oxidase in Gibberellin Biosynthesis During Barley Seed Germination. Plant Signal. Behav. 2016, 11, e1180492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montiel, J.; Nava, N.; Cardenas, L.; Sanchez-Lopez, R.; Arthikala, M.K.; Santana, O.; Sanchez, F.; Quinto, C. A Phaseolus Vulgaris NADPH Oxidase Gene Is Required for Root Infection by Rhizobia. Plant Cell Physiol. 2012, 53, 1751–1767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arthikala, M.K.; Sanchez-Lopez, R.; Nava, N.; Santana, O.; Cardenas, L.; Quinto, C. Rbohb, a Phaseolus Vulgaris NADPH Oxidase Gene, Enhances Symbiosome Number, Bacteroid Size, and Nitrogen Fixation in Nodules and Impairs Mycorrhizal Colonization. New Phytol. 2014, 202, 886–900. [Google Scholar] [CrossRef]
- Marino, D.; Andrio, E.; Danchin, E.G.; Oger, E.; Gucciardo, S.; Lambert, A.; Puppo, A.; Pauly, N. A Medicago Truncatula NADPH Oxidase Is Involved in Symbiotic Nodule Functioning. New Phytol. 2011, 189, 580–592. [Google Scholar] [CrossRef] [Green Version]
- Kayano, Y.; Tanaka, A.; Akano, F.; Scott, B.; Takemoto, D. Differential Roles of NADPH Oxidases and Associated Regulators in Polarized Growth, Conidiation and Hyphal Fusion in the Symbiotic Fungus Epichloe Festucae. Fungal Genet. Biol. 2013, 56, 87–97. [Google Scholar] [CrossRef]
- Nordzieke, D.E.; Fernandes, T.R.; El Ghalid, M.; Turra, D.; Di Pietro, A. NADPH Oxidase Regulates Chemotropic Growth of the Fungal Pathogen Fusarium Oxysporum Towards the Host Plant. New Phytol. 2019, 224, 1600–1612. [Google Scholar] [CrossRef]
- Zhang, Z.B.; Zhao, Y.L.; Feng, X.B.; Luo, Z.Y.; Kong, S.W.; Zhang, C.; Gong, A.D.; Yuan, H.; Cheng, L.; Wang, X.N. Genomic, Molecular Evolution, and Expression Analysis of Nox Genes in Soybean (Glycine Max). Genomics 2019, 111, 619–628. [Google Scholar] [CrossRef]
- Lara-Ortíz, T.; Riveros-Rosas, H.; Aguirre, J. Reactive Oxygen Species Generated by Microbial NADPH Oxidase Noxa Regulate Sexual Development in Aspergillus Nidulans. Mol. Microbiol. 2003, 50, 1241–1255. [Google Scholar] [CrossRef] [Green Version]
- Dirschnabel, D.E.; Nowrousian, M.; Cano-Dominguez, N.; Aguirre, J.; Teichert, I.; Kuck, U. New Insights into the Roles of NADPH Oxidases in Sexual Development and Ascospore Germination in Sordaria Macrospora. Genetics 2014, 196, 729–744. [Google Scholar] [CrossRef] [Green Version]
- Cano-Dominguez, N.; Alvarez-Delfin, L.; Hansberg, W.; Aguirre, J. NADPH Oxidases Nox-1 and Nox-2 Require the Regulatory Subunit nor-1 to Control Cell Differentiation and Growth in Neurospora Crassa. Eukaryot. Cell 2018, 7, 1352–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roca, M.G.; Weichert, M.; Siegmund, U.; Tudzynski, P.; Fleissner, A. Germling Fusion via Conidial Anastomosis Tubes in the Grey Mould Botrytis Cinerea Requires NADPH Oxidase Activity. Fungal Biol. 2012, 116, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, C.; Zhang, D.; Li, H.; Li, P.; Ma, F. Reactive Oxygen Species Produced via Plasma Membrane NADPH Oxidase Regulate Anthocyanin Synthesis in Apple Peel. Planta 2014, 240, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Tewari, R.K.; Watanabe, D.; Watanabe, M. Chloroplastic NADPH Oxidase-Like Activity-Mediated Perpetual Hydrogen Peroxide Generation in the Chloroplast Induces Apoptotic-Like Death of Brassica Napus Leaf Protoplasts. Planta 2012, 235, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhao, S.; Tan, F.; Zhao, H.; Wang, D.; Si, H.; Chen, Q. Changes in Ros Production and Antioxidant Capacity during Tuber Sprouting in Potato. Food Chem. 2017, 237, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH Oxidase Rbohd During Plant Immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.J.; Wei, X.Y.; Jing, X.Q.; Chang, Y.L.; Hu, C.H.; Wang, X.; Chen, K.M. The Fundamental Role of Nox Family Proteins in Plant Immunity and Their Regulation. Int. J. Mol. Sci. 2016, 17, 805. [Google Scholar] [CrossRef] [Green Version]
- Morales, J.; Kadota, Y.; Zipfel, C.; Molina, A.; Torres, M.A. The Arabidopsis NADPH Oxidases Rbohd and Rbohf Display Differential Expression Patterns and Contributions During Plant Immunity. J. Exp. Bot. 2016, 67, 1663–1676. [Google Scholar] [CrossRef] [Green Version]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jones, J.D.; Shirasu, K.; Menke, F.; Jones, A.; et al. Direct Regulation of the NADPH Oxidase Rbohd by the Prr-Associated Kinase Bik1 During Plant Immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef] [Green Version]
- Pogany, M.; von Rad, U.; Grun, S.; Dongo, A.; Pintye, A.; Simoneau, P.; Bahnweg, G.; Kiss, L.; Barna, B.; Durner, J. Dual Roles of Reactive Oxygen Species and NADPH Oxidase Rbohd in an Arabidopsis-Alternaria Pathosystem. Plant Physiol. 2009, 151, 1459–1475. [Google Scholar] [CrossRef] [Green Version]
- Chaouch, S.; Queval, G.; Noctor, G. Atrbohf Is a Crucial Modulator of Defence-Associated Metabolism and a Key Actor in the Interplay between Intracellular Oxidative Stress and Pathogenesis Responses in Arabidopsis. Plant J. 2012, 69, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.B.; Wang, X.D.; Zhang, Y.Y.; Dong, J.J.; Ma, C.; Chen, W.L. NADPH Oxidase Rbohd Contributes to Autophagy and Hypersensitive Cell Death During the Plant Defense Response in Arabidopsis Thaliana. Biol. Plant. 2015, 59, 570–580. [Google Scholar] [CrossRef]
- Foley, R.C.; Gleason, C.A.; Anderson, J.P.; Hamann, T.; Singh, K.B. Genetic and Genomic Analysis of Rhizoctonia Solani Interactions with Arabidopsis; Evidence of Resistance Mediated through NADPH Oxidases. PLoS ONE 2013, 8, e56814. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Islam, M.R.; Badawi, M.A.; El-Bebany, A.F.; Daayf, F. Overexpression of Strboha in Arabidopsis Thaliana Enhances Defence Responses against Verticillium Dahliae. Physiol. Mol. Plant Pathol. 2015, 90, 105–114. [Google Scholar]
- Zhang, Z.; Yang, F.; Na, R.; Zhang, X.; Yang, S.; Gao, J.; Fan, M.; Zhao, Y.; Zhao, J. Atrop1 Negatively Regulates Potato Resistance to Phytophthora Infestans via NADPH Oxidase-Mediated Accumulation of H2O2. BMC Plant Biol. 2014, 14, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshioka, H.; Adachi, H.; Nakano, T.; Miyagawa, N.; Asai, S.; Ishihama, N.; Yoshioka, M. Hierarchical Regulation of NADPH Oxidase by Protein Kinases in Plant Immunity. Physiol. Mol. Plant Pathol. 2016, 95, 20–26. [Google Scholar] [CrossRef]
- Lherminier, J.; Elmayan, T.; Fromentin, J.; Elaraqui, K.T.; Vesa, S.; Morel, J.; Verrier, J.L.; Cailleteau, B.; Blein, J.P.; Simon-Plas, F. NADPH Oxidase-Mediated Reactive Oxygen Species Production: Subcellular Localization and Reassessment of Its Role in Plant Defense. Mol. Plant-Microbe Interact. 2009, 22, 868–881. [Google Scholar] [CrossRef] [Green Version]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.P.; Schulze, W.X.; Romeis, T. Calcium-Dependent Protein Kinase/NADPH Oxidase Activation Circuit Is Required for Rapid Defense Signal Propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef] [Green Version]
- Yoshiaki, Y.; Goto, K.; Takai, R.; Iwano, M.; Takayama, S.; Isogai, A.; Che, F.S. Function of the Rice gp91phox Homologs OsrbohA and OsrbohE Genes in Ros-Dependent Plant Immune Responses. Plant Biotechnol. 2015, 22, 127–135. [Google Scholar]
- Egan, M.J.; Wang, Z.Y.; Jones, M.A.; Smirnoff, M.; Talbot, N.J. Generation of Reactive Oxygen Species by Fungal NADPH Oxidases Is Required for Rice Blast Disease. Proc. Natl. Acad. Sci. USA 2007, 104, 11772–11777. [Google Scholar] [CrossRef] [Green Version]
- Proels, R.K.; Oberhollenzer, K.; Pathuri, I.P.; Hensel, G.; Kumlehn, J.; Huckelhoven, R. Rbohf2 of Barley Is Required for Normal Development of Penetration Resistance to the Parasitic Fungus Blumeria Graminis F. sp. Hordei. Mol. Plant-Microbe Interact. 2010, 23, 1143–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trujillo, M.; Altschmied, L.; Schweizer, P.; Kogel, K.H.; Huckelhoven, R. Respiratory Burst Oxidase Homologue a of Barley Contributes to Penetration by the Powdery Mildew Fungus Blumeria Graminis F. sp. Hordei. J. Exp. Bot. 2006, 57, 3781–3891. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Fang, Q.; Zhang, Z.; Wang, Y.; Zheng, X. The Role of Respiratory Burst Oxidase Homologues in Elicitor-Induced Stomatal Closure and Hypersensitive Response in Nicotiana Benthamiana. J. Exp. Bot. 2009, 60, 3109–3122. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Cao, Y.; Li, H.; Kim, D.; Ahsan, N.; Thelen, J.; Stacey, G. Extracellular Atp Elicits Dorn1-Mediated Rbohd Phosphorylation to Regulate Stomatal Aperture. Nat. Commun. 2017, 8, 2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toum, L.; Torres, P.S.; Gallego, S.M.; Benavides, M.P.; Vojnov, A.A.; Gudesblat, G.E. Coronatine Inhibits Stomatal Closure through Guard Cell-Specific Inhibition of NADPH Oxidase-Dependent Ros Production. Front. Plant Sci. 2016, 7, 1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.J.; Steinebrunner, I.; Wang, X.; Stout, S.C.; Roux, S.J. Extracellular Atp Induces the Accumulation of Superoxide via NADPH Oxidases in Arabidopsis. Plant Physiol. 2006, 140, 1222–1232. [Google Scholar] [CrossRef] [Green Version]
- Clark, G.; Wu, M.; Wat, N.; Onyirimba, J.; Pham, T.; Herz, N.; Ogoti, J.; Gomez, D.; Canales, A.A.; Aranda, G.; et al. Both the Stimulation and Inhibition of Root Hair Growth Induced by Extracellular Nucleotides in Arabidopsis Are Mediated by Nitric Oxide and Reactive Oxygen Species. Plant Mol. Biol. 2010, 74, 423–435. [Google Scholar] [CrossRef]
- Clark, G.; Fraley, D.; Steinebrunner, I.; Cervantes, A.; Onyirimba, J.; Liu, A.; Torres, J.; Tang, W.; Kim, J.; Roux, S.J. Extracellular Nucleotides and Apyrases Regulate Stomatal Aperture in Arabidopsis. Plant Physiol. 2011, 156, 1740–1753. [Google Scholar] [CrossRef] [Green Version]
- Clark, G.; Darwin, C.; Mehta, V.; Jackobs, F.; Perry, T.; Hougaard, K.; Roux, S. Effects of Chemical Inhibitors and Apyrase Enzyme Further Document a Role for Apyrases and Extracellular Atp in the Opening and Closing of Stomates in Arabidopsis. Plant Signal. Behav. 2013, 8, e26093. [Google Scholar] [CrossRef] [Green Version]
- Giesbert, S.; Schurg, T.; Scheele, S.; Tudzynski, P. The NADPH Oxidase Cpnox1 Is Required for Full Pathogenicity of the Ergot Fungus Claviceps Purpurea. Mol. Plant Pathol. 2008, 9, 317–327. [Google Scholar] [CrossRef]
- Chung, K.R. Reactive Oxygen Species in the Citrus Fungal Pathogen Alternaria Alternata: The Roles of Nadph-Dependent Oxidase. Physiol. Mol. Plant Pathol. 2014, 88, 10–17. [Google Scholar] [CrossRef]
- Lima, M.R.M.; Alberto, C.P.D. Phaeomoniella Chlamydospora-Induced Oxidative Burst in Vitis Vinifera Cell Suspensions: Role of NADPH Oxidase and Ca2+. J. Phytopathol. 2012, 160, 129–134. [Google Scholar] [CrossRef] [Green Version]
- Arthikala, M.K.; Montiel, J.; Sanchez-Lopez, R.; Nava, N.; Cardenas, L.; Quinto, C. Respiratory Burst Oxidase Homolog Gene a Is Crucial for Rhizobium Infection and Nodule Maturation and Function in Common Bean. Front. Plant Sci. 2017, 8, 2003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montiel, J.; Arthikala, M.K.; Cardenas, L.; Quinto, C. Legume NADPH Oxidases Have Crucial Roles at Different Stages of Nodulation. Int. J. Mol. Sci. 2016, 17, 680. [Google Scholar] [CrossRef] [Green Version]
- Kurusu, T.; Kuchitsu, K.; Tada, Y. Plant Signaling Networks Involving Ca2+ and Rboh/Nox-Mediated Ros Production under Salinity Stress. Front. Plant Sci. 2015, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, D.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.H.; Schroeder, J.I. Plant Salt-Tolerance Mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar]
- Ma, L.; Zhang, H.; Sun, L.; Jiao, Y.; Zhang, G.; Miao, C.; Hao, F. NADPH Oxidase Atrbohd and Atrbohf Function in Ros-Dependent Regulation of Na+/K+ Homeostasis in Arabidopsis under Salt Stress. J. Exp. Bot. 2012, 63, 305–317. [Google Scholar] [CrossRef]
- Ben Rejeb, K.; Benzarti, M.; Debez, A.; Bailly, C.; Savoure, A.; Abdelly, C. NADPH Oxidase-Dependent H2O2 Production Is Required for Salt-Induced Antioxidant Defense in Arabidopsis Thaliana. J. Plant Physiol. 2015, 174, 5–15. [Google Scholar] [CrossRef]
- Ben Rejeb, K.; Lefebvre-De Vos, D.; Le Disquet, I.; Leprince, A.S.; Bordenave, M.; Maldiney, R.; Jdey, A.; Abdelly, C.; Savoure, A. Hydrogen Peroxide Produced by NADPH Oxidases Increases Proline Accumulation During Salt or Mannitol Stress in Arabidopsis Thaliana. New Phytol. 2015, 208, 1138–1148. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, H.; Matsuda, O.; Iba, K. Itn1, a Novel Gene Encoding an Ankyrin-Repeat Protein That Affects the Aba-Mediated Production of Reactive Oxygen Species and Is Involved in Salt-Stress Tolerance in Arabidopsis Thaliana. Plant J. 2008, 56, 411–422. [Google Scholar] [CrossRef]
- Liu, Y.; He, C. Regulation of Plant Reactive Oxygen Species (ROS) in Stress Responses: Learning from Atrbohd. Plant Cell Rep. 2016, 35, 995–1007. [Google Scholar] [CrossRef]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The Plant NADPH Oxidase Rbohd Mediates Rapid Systemic Signaling in Response to Diverse Stimuli. Sci. Signal. 2009, 2, ra45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittler, R.; Blumwald, E. The Roles of Ros and Aba in Systemic Acquired Acclimation. Plant Cell 2015, 27, 64–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Xing, D.; Zhang, L. Involvement of NADPH Oxidase in Sulfur Dioxide-Induced Oxidative Stress in Plant Cells. Photochem. Photobiol. Sci. 2007, 6, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Huang, Y.; Sun, S.; Sun, J.; Cao, H.; Shabala, S.; Bie, Z. Root Respiratory Burst Oxidase Homologue-Dependent H2O2 Production Confers Salt Tolerance on a Grafted Cucumber by Controlling Na+ Exclusion and Stomatal Closure. J. Exp. Bot. 2018, 69, 3465–3476. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhou, J.; Qin, Y.; Liu, L.; Chen, J. Water-Forming Nadh Oxidase Protects Torulopsis Glabrata against Hyperosmotic Stress. Yeast 2010, 27, 207–216. [Google Scholar] [PubMed] [Green Version]
- Rodriguez, A.A.; Lascano, H.R.; Bustos, D.; Taleisnik, E. Salinity-Induced Decrease in NADPH Oxidase Activity in the Maize Leaf Blade Elongation Zone. J. Plant Physiol. 2007, 164, 223–230. [Google Scholar] [CrossRef]
- Shiuan, L.I.; Wu, Y.S.; Chen, C.T.; Chen, G.H.; Hwang, S.G.; Jauh, G.Y.; Tzen, J.; Yang, C.Y. Atrboh I Confers Submergence Tolerance and Is Involved in Auxin-Mediated Signaling Pathways under Hypoxic Stress. Plant Growth Regul. 2017, 83, 277–285. [Google Scholar]
- Yang, C.Y.; Hong, C.P. The NADPH Oxidase Rboh D Is Involved in Primary Hypoxia Signalling and Modulates Expression of Hypoxia-Inducible Genes under Hypoxic Stress. Environ. Exp. Bot. 2015, 115, 63–72. [Google Scholar] [CrossRef]
- Liu, B.; Sun, L.; Ma, L.; Hao, F.S. Both Atrbohd and Atrbohf Are Essential for Mediating Responses to Oxygen Deficiency in Arabidopsis. Plant Cell Rep. 2017, 36, 947–957. [Google Scholar] [CrossRef]
- Wu, Y.S.; Yang, C.Y. Physiological Responses and Expression Profile of NADPH Oxidase in Rice (Oryza Sativa) Seedlings under Different Levels of Submergence. Rice 2016, 9, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, T.; Yoshioka, M.; Fukazawa, A.; Mori, H.; Nishizawa, N.K.; Tsutsumi, N.; Yoshioka, H.; Nakazono, M. An NADPH Oxidase Rboh Functions in Rice Roots During Lysigenous Aerenchyma Formation under Oxygen-Deficient Conditions. Plant Cell 2017, 29, 775–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, D.K.; Pena, L.B.; Romero-Puertas, M.C.; Hernandez, A.; Inouhe, M.; Sandalio, L.M. NADPH Oxidases Differentially Regulate Ros Metabolism and Nutrient Uptake under Cadmium Toxicity. Plant Cell Environ. 2017, 40, 509–526. [Google Scholar] [CrossRef]
- Chaoui, A.; Jarrar, B.; Ferjani, E.E. Effects of Cadmium and Copper on Peroxidase, Nadh Oxidase and Iaa Oxidase Activities in Cell Wall, Soluble and Microsomal Membrane Fractions of Pea Roots. J. Plant Physiol. 2004, 161, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Groppa, M.D.; Ianuzzo, M.P.; Rosales, E.P.; Vazquez, S.C.; Benavides, M.P. Cadmium Modulates NADPH Oxidase Activity and Expression in Sunflower Leaves. Biol. Plant. 2012, 56, 167–171. [Google Scholar] [CrossRef]
- Jakubowska, D.; Janicka-Russak, M.; Kabala, K.; Migocka, M.; Reda, M. Modification of Plasma Membrane NADPH Oxidase Activity in Cucumber Seedling Roots in Response to Cadmium Stress. Plant Sci. 2015, 234, 50–59. [Google Scholar] [CrossRef]
- Jakubowska, D.; Janicka, M. The Role of Brassinosteroids in the Regulation of the Plasma Membrane H+-Atpase and NADPH Oxidase under Cadmium Stress. Plant Sci. 2017, 264, 37–47. [Google Scholar] [CrossRef]
- Pourrut, B.; Perchet, G.; Silvestre, J.; Cecchi, M.; Guiresse, M.; Pinelli, E. Potential Role of Nadph-Oxidase in Early Steps of Lead-Induced Oxidative Burst in Vicia Faba Roots. J. Plant Physiol. 2008, 165, 571–579. [Google Scholar] [CrossRef] [Green Version]
- Hao, F.S.; Wang, X.C.; Chen, J. Involvement of Plasma-Membrane NADPH Oxidase in Nickel-Induced Oxidative Stress in Roots of Wheat Seedlings. Plant Sci. 2006, 170, 151–158. [Google Scholar] [CrossRef]
- Gupta, D.K.; Inouhe, M.; Rodriguez-Serrano, M.; Romero-Puertas, M.C.; Sandalio, L.M. Oxidative Stress and Arsenic Toxicity: Role of NADPH Oxidases. Chemosphere 2013, 90, 1987–1996. [Google Scholar] [CrossRef]
- Sagi, M.; Davydov, O.; Orazova, S.; Yesbergenova, Z.; Ophir, R.; Stratmann, J.W.; Fluhr, R. Plant Respiratory Burst Oxidase Homologs Impinge on Wound Responsiveness and Development in Lycopersicon Esculentum. Plant Cell 2004, 16, 616–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, G.N.; Iyer, S.; Knowles, N.R. Strboh a Homologue of NADPH Oxidase Regulates Wound-Induced Oxidative Burst and Facilitates Wound-Healing in Potato Tubers. Planta 2007, 227, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, L.; Wunsche, H.; Baldwin, I.T. Narboh D, a Respiratory Burst Oxidase Homolog in Nicotiana Attenuata, Is Required for Late Defense Responses after Herbivore Attack. J. Integr. Plant Biol. 2013, 55, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Piotrovskii, M.S.; Shevyreva, T.A.; Zhestkova, I.M.; Trofimova, M.S. Activation of Plasmalemmal NADPH Oxidase in Etiolated Maize Seedlings Exposed to Chilling Temperatures. Russ. J. Plant Physiol. 2011, 58, 290–298. [Google Scholar] [CrossRef]
- Kawarazaki, T.; Kimura, S.; Iizuka, A.; Hanamata, S.; Nibori, H.; Michikawa, M.; Imai, A.; Abe, M.; Kaya, H.; Kuchitsu, K. A Low Temperature-Inducible Protein Atsrc2 Enhances the Ros-Producing Activity of NADPH Oxidase Atrbohf. Biochim. Biophys. Acta 2013, 1833, 2775–2780. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, Y.; He, Y.; Hu, W.; Zhang, Y.; Wang, X.; Tang, H. Identification of NADPH Oxidase Family Members Associated with Cold Stress in Strawberry. FEBS Open Bio 2018, 8, 593–605. [Google Scholar] [CrossRef]
- Yoshioka, H.; Numata, N.; Nakajima, K.; Katou, S.; Kawakita, K.; Rowland, O.; Jones, J.D.G.; Doke, N. Nicotiana Benthamiana gp91(Phox) Homologs Nbrboha and Nbrbohb Participate in H2O2 Accumulation and Resistance to Phytophthora Infestans. Plant Cell 2003, 15, 706–718. [Google Scholar] [CrossRef] [Green Version]
- Kiraly, L.; Hafez, Y.M.; Fodor, J.; Kiraly, Z. Suppression of Tobacco Mosaic Virus-Induced Hypersensitive-Type Necrotization in Tobacco at High Temperature Is Associated with Downregulation of NADPH Oxidase and Superoxide and Stimulation of Dehydroascorbate Reductase. J. Gen. Virol. 2008, 89, 799–808. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J. Involvement of Plasma-Membrane NADPH Oxidase in Abscisic Acid- and Water Stress-Induced Antioxidant Defense in Leaves of Maize Seedlings. Planta 2002, 215, 1022–1030. [Google Scholar] [CrossRef]
- Duan, Z.Q.; Bai, L.; Zhao, Z.G.; Zhang, G.P.; Cheng, F.M.; Jiang, L.X.; Chen, K.M. Drought-Stimulated Activity of Plasma Membrane Nicotinamide Adenine Dinucleotide Phosphate Oxidase and Its Catalytic Properties in Rice. J. Integr. Plant Biol. 2009, 51, 1104–1115. [Google Scholar] [CrossRef]
- Rohollahi, I.; Khoshkholghsima, N.; Nagano, H.; Hoshino, Y.; Yamada, T. Respiratory Burst Oxidase-D Expression and Biochemical Responses in Festuca Arundinacea under Drought Stress. Crop. Sci. 2018, 58, 435–442. [Google Scholar] [CrossRef] [Green Version]
- Brightman, A.O.; Barr, R.; Crane, F.L.; Morre, D.J. Auxin-Stimulated Nadh Oxidase Purified from Plasma Membrane of Soybean. Plant Physiol. 1988, 86, 1264–1269. [Google Scholar] [CrossRef] [PubMed]
- Morre, D.J.; Gomez-Rey, M.L.; Schramke, C.; Em, O.; Lawler, J.; Hobeck, J.; Morre, D.M. Use of Dipyridyl-Dithio Substrates to Measure Directly the Protein Disulfide-Thiol Interchange Activity of the Auxin Stimulated Nadh: Protein Disulfide Reductase (Nadh Oxidase) of Soybean Plasma Membranes. Mol. Cell. Biochem. 1999, 200, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, G.; Manzoor, M.A.; Wang, H.; Abdullah, M.; Su, X.; Zhang, J.; Jiang, T.; Jin, Q.; Cai, Y.; et al. In Silico Genome-Wide Analysis of Respiratory Burst Oxidase Homolog (Rboh) Family Genes in Five Fruit-Producing Trees, and Potential Functional Analysis on Lignification of Stone Cells in Chinese White Pear. Cells 2019, 8, 520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Wang, F.; Zhao, Q.; Liu, J.; Cheng, F. Involvement of NADPH Oxidase Isoforms in the Production of O2- Manipulated by Aba in the Senescing Leaves of Early-Senescence-Leaf (Esl) Mutant Rice (Oryza Sativa). PLoS ONE 2018, 13, e0190161. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.S.; Zhang, J.G.; Yu, Z.L.; Chen, J. Involvement of NADPH Oxidase Ntrbohd in the Rapid Production of H2O2 Induced by Aba in Cultured Tobacco Cell Line by-2. Prog. Nat. Sci. 2008, 18, 267–271. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J. Crosstalk between Calcium and Reactive Oxygen Species Originated from NADPH Oxidase in Abscisic Acid-Induced Antioxidant Defence in Leaves of Maize Seedlings. Plant Cell Environ. 2003, 26, 929–939. [Google Scholar] [CrossRef]
- Chen, H.J.; Huang, C.S.; Huang, G.J.; Chow, T.J.; Lin, Y.H. NADPH Oxidase Inhibitor Diphenyleneiodonium and Reduced Glutathione Mitigate Ethephon-Mediated Leaf Senescence, H2O2 Elevation and Senescence-Associated Gene Expression in Sweet Potato (Ipomoea Batatas). J. Plant Physiol. 2013, 170, 1471–1483. [Google Scholar] [CrossRef]
- Kaur, G.; Pati, P.K. Analysis of Cis-Acting Regulatory Elements of Respiratory Burst Oxidase Homolog (Rboh) Gene Families in Arabidopsis and Rice Provides Clues for Their Diverse Functions. Comput. Biol. Chem. 2016, 62, 104–118. [Google Scholar] [CrossRef]
- Gadjev, I.; Vanderauwera, S.; Gechev, T.S.; Laloi, C.; Minkov, I.N.; Shulaev, V.; Apel, K.; Inze, D.; Mittler, R.; Van Breusegem, F. Transcriptomic Footprints Disclose Specificity of Reactive Oxygen Species Signaling in Arabidopsis. Plant Physiol. 2006, 141, 436–445. [Google Scholar] [CrossRef] [Green Version]
- Petrov, V.; Vermeirssen, V.; De Clercq, I.; Van Breusegem, F.; Minkov, I.; Vandepoele, K.; Gechev, T.S. Identification of Cis-Regulatory Elements Specific for Different Types of Reactive Oxygen Species in Arabidopsis Thaliana. Gene 2012, 499, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.S.; Ley, S.C. Mitogen-Activated Protein Kinases in Innate Immunity. Nat. Rev. Immunol. 2013, 13, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, S. Mapk Cascades in Plant Disease Resistance Signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Somssich, I.E. Transcriptional Networks in Plant Immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Nakano, T.; Miyagawa, N.; Ishihama, N.; Yoshioka, M.; Katou, Y.; Yaeno, T.; Shirasu, K.; Yoshioka, H. Wrky Transcription Factors Phosphorylated by Mapk Regulate a Plant Immune NADPH Oxidase in Nicotiana Benthamiana. Plant Cell 2015, 27, 2645–2663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishihama, N.; Yamada, R.; Yoshioka, M.; Katou, S.; Yoshioka, H. Phosphorylation of the Nicotiana Benthamiana Wrky8 Transcription Factor by Mapk Functions in the Defense Response. Plant Cell 2011, 23, 1153–1170. [Google Scholar] [CrossRef] [Green Version]
- Mersmann, S.; Bourdais, G.; Rietz, S.; Robatzek, S. Ethylene Signaling Regulates Accumulation of the Fls2 Receptor and Is Required for the Oxidative Burst Contributing to Plant Immunity. Plant Physiol. 2010, 154, 391–400. [Google Scholar] [CrossRef] [Green Version]
- Boudsocq, M.; Willmann, M.R.; McCormack, M.; Lee, H.; Shan, L.; He, P.; Bush, J.; Cheng, S.H.; Sheen, J. Differential Innate Immune Signalling via Ca2+ Sensor Protein Kinases. Nature 2010, 464, 418–422. [Google Scholar] [CrossRef] [Green Version]
- Asai, S.; Ohta, K.; Yoshioka, H. Mapk Signaling Regulates Nitric Oxide and NADPH Oxidase-Dependent Oxidative Bursts in Nicotiana Benthamiana. Plant Cell 2008, 20, 1390–1406. [Google Scholar] [CrossRef] [Green Version]
- Kovtun, Y.; Chiu, W.L.; Tena, G.; Sheen, J. Functional Analysis of Oxidative Stress-Activated Mitogen-Activated Protein Kinase Cascade in Plants. Proc. Natl. Acad. Sci. USA 2000, 97, 2940–2945. [Google Scholar] [CrossRef] [Green Version]
- Lin, F.; Ding, H.; Wang, J.; Zhang, H.; Zhang, A.; Zhang, Y.; Tan, M.; Dong, W.; Jiang, M. Positive Feedback Regulation of Maize NADPH Oxidase by Mitogen-Activated Protein Kinase Cascade in Abscisic Acid Signalling. J. Exp. Bot. 2009, 60, 3221–3238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirichandra, C.; Gu, D.; Hu, H.C.; Davanture, M.; Lee, S.; Djaoui, M.; Valot, B.; Zivy, M.; Leung, J.; Merlot, S.; et al. Phosphorylation of the Arabidopsis Atrbohf NADPH Oxidase by Ost1 Protein Kinase. FEBS Lett. 2009, 583, 2982–2986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nibau, C.; Wu, H.M.; Cheung, A.Y. Rac/Rop Gtpases: ‘Hubs’ for Signal Integration and Diversification in Plants. Trends Plant Sci. 2006, 11, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Shin, L.J.; Huang, H.E.; Chang, H.; Lin, Y.H.; Feng, T.Y.; Ger, M.J. Ectopic Ferredoxin I Protein Promotes Root Hair Growth through Induction of Reactive Oxygen Species in Arabidopsis Thaliana. J. Plant Physiol. 2011, 168, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Oda, T.; Hashimoto, H.; Kuwabara, N.; Akashi, S.; Hayashi, K.; Kojima, C.; Wong, H.L.; Kawasaki, T.; Shimamoto, K.; Sato, M.; et al. Structure of the N-Terminal Regulatory Domain of a Plant NADPH Oxidase and Its Functional Implications. J. Biol. Chem. 2010, 285, 1435–1445. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, M.; Ohura, I.; Kawakita, K.; Yokota, N.; Fujiwara, M.; Shimamoto, K.; Doke, N.; Yoshioka, H. Calcium-Dependent Protein Kinases Regulate the Production of Reactive Oxygen Species by Potato NADPH Oxidase. Plant Cell 2007, 19, 1065–1080. [Google Scholar] [CrossRef] [Green Version]
- Ogasawara, Y.; Kaya, H.; Hiraoka, G.; Yumoto, F.; Kimura, S.; Kadota, Y.; Hishinuma, H.; Senzaki, E.; Yamagoe, S.; Nagata, K.; et al. Synergistic Activation of the Arabidopsis NADPH Oxidase Atrbohd by Ca2+ and Phosphorylation. J. Biol. Chem. 2008, 283, 8885–8892. [Google Scholar] [CrossRef] [Green Version]
- Waszczak, C.; Carmody, M.; Kangasjarvi, J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [Green Version]
- McAinsh, M.R.; Clayton, H.; Mansfield, T.A.; Hetherington, A.M. Changes in Stomatal Behavior and Guard Cell Cytosolic Free Calcium in Response to Oxidative Stress. Plant Physiol. 1996, 111, 1031–1042. [Google Scholar] [CrossRef] [Green Version]
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; KluÈsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium Channels Activated by Hydrogen Peroxide Mediate Abscisic Acid Signalling in Guard Cells. Nature 2000, 406, 732–734. [Google Scholar] [CrossRef]
- Robert, F.; Moshe, S. Superoxide Production by Plant Homologues of the gp91(Phox) NADPH Oxidase: Mmodulation of Activity by Calcium and by Tobacco Mosaic Virus Infection. Plant Physiol. Biochem. 2001, 126, 1281–1290. [Google Scholar]
- Xing, T.; Wang, X.J.; Malik, K.; Miki, B.L. Ectopic Expression of an Arabidopsis Calmodulin-Like Domain Protein Kinase-Enhanced NADPH Oxidase Activity and Oxidative Burst in Tomato Protoplasts. Mol. Plant-Microbe Interact. 2001, 14, 1261–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, A.A.; Saitoh, H.; Felix, G.; Freymark, G.; Miersch, O.; Wasternack, C.; Boller, T.; Jones, J.D.G.; Romeis, T. Ethylene-Mediated Crosstalk between Calcium-Dependent Protein Kinase and Mapk Signaling Controls Stress Responses in Plants. Proc. Natl. Acad. Sci. USA 2005, 102, 10736–10741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S.; et al. A Rice Calcium-Dependent Protein Kinase Oscpk12 Oppositely Modulates Salt-Stress Tolerance and Blast Disease Resistance. Plant J. 2012, 69, 26–36. [Google Scholar] [CrossRef]
- Boudsocq, M.; Sheen, J. Cdpks in Immune and Stress Signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Kudla, J.; Batistic, O.; Hashimoto, K. Calcium Signals: The Lead Currency of Plant Information Processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef]
- Luan, S. The Cbl-Cipk Network in Plant Calcium Signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef]
- De la Torre, F.; Gutierrez-Beltran, E.; Pareja-Jaime, Y.; Chakravarthy, S.; Martin, G.B.; del Pozo, O. The Tomato Calcium Sensor Cbl10 and Its Interacting Protein Kinase Cipk6 Define a Signaling Pathway in Plant Immunity. Plant Cell 2013, 25, 2748–2764. [Google Scholar] [CrossRef] [Green Version]
- Drerup, M.M.; Schlucking, K.; Hashimoto, K.; Manishankar, P.; Steinhorst, L.; Kuchitsu, K.; Kudla, J. The Calcineurin B-Like Calcium Sensors Cbl1 and Cbl9 together with Their Interacting Protein Kinase Cipk26 Regulate the Arabidopsis NADPH Oxidase Rbohf. Mol. Plant 2013, 6, 559–569. [Google Scholar] [CrossRef] [Green Version]
- Batistič, O.; Waadt, R.; Steinhorst, L.; Held, K.; Kudla, J. Cbl-Mediated Targeting of Cipks Facilitates the Decoding of Calcium Signals Emanating from Distinct Cellular Stores. Plant J. 2009, 61, 211–222. [Google Scholar] [CrossRef]
- Weinl, S.; Kudla, J. The Cbl-Cipk Ca2+-Decoding Signaling Network: Function and Perspectives. New Phytol. 2009, 184, 517–528. [Google Scholar] [CrossRef]
- Hashimoto, K.; Kudla, J. Calcium Decoding Mechanisms in Plants. Biochimie 2011, 93, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Kaya, H.; Kawarazaki, T.; Hiraoka, G.; Senzaki, E.; Michikawa, M.; Kuchitsu, K. Protein Phosphorylation Is a Prerequisite for the Ca2+-Dependent Activation of Arabidopsis NADPH Oxidases and May Function as a Trigger for the Positive Feedback Regulation of Ca2+ and Reactive Oxygen Species. BBA Mol. Cell Res. 2012, 1823, 398–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antolin-Llovera, M.; Ried, M.K.; Binder, A.; Parniske, M. Receptor Kinase Signaling Pathways in Plant-Microbe Interactions. Annu. Rev. Phytopathol. 2012, 50, 451–473. [Google Scholar] [CrossRef] [PubMed]
- Vij, S.; Giri, J.; Dansana, P.K.; Kapoor, S.; Tyagi, A.K. The Receptor-Like Cytoplasmic Kinase (Osrlck) Gene Family in Rice: Organization, Phylogenetic Relationship, and Expression During Development and Stress. Mol. Plant 2008, 1, 732–750. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Ma, X.; Shan, L.; He, P. Big Roles of Small Kinases: The Complex Functions of Receptor-Like Cytoplasmic Kinases in Plant Immunity and Development. J. Integr. Plant Biol. 2013, 55, 1188–1197. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Li, M.; Yu, L.; Zhou, Z.; Liang, X.; Liu, Z.; Cai, G.; Gao, L.; Zhang, X.; Wang, Y.; et al. The Fls2-Associated Kinase Bik1 Directly Phosphorylates the NADPH Oxidase Rbohd to Control Plant Immunity. Cell Host Microbe 2014, 15, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Akamatsu, A.; Wong, H.L.; Fujiwara, M.; Okuda, J.; Nishide, K.; Uno, K.; Imai, K.; Umemura, K.; Kawasaki, T.; Kawano, Y.; et al. An Oscebip/Oscerk1-Osracgef1-Osrac1 Module Is an Essential Early Component of Chitin-Induced Rice Immunity. Cell Host Microbe 2013, 13, 465–476. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A Receptor-Like Cytoplasmic Kinase, Bik1, Associates with a Flagellin Receptor Complex to Initiate Plant Innate Immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-Like Cytoplasmic Kinases Integrate Signaling from Multiple Plant Immune Receptors and Are Targeted by a Pseudomonas Syringae Effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wu, Y.; Yang, F.; Zhang, Y.; Chen, S.; Xie, Q.; Tian, X.; Zhou, J.M. Bik1 Interacts with Peprs to Mediate Ethylene-Induced Immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 6205–6210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the Bacterial Pamp Ef-Tu by the Receptor Efr Restricts Agrobacterium-Mediated Transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef]
- Tang, J.; Han, Z.; Sun, Y.; Zhang, H.; Gong, X.; Chai, J. Structural Basis for Recognition of an Endogenous Peptide by the Plant Receptor Kinase Pepr1. Cell Res. 2015, 25, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Hann, D.R.; Ntoukakis, V.; Petutschnig, E.; Lipka, V.; Rathjen, J.P. Avrptob Targets the Lysm Receptor Kinase Cerk1 to Promote Bacterial Virulence on Plants. Curr. Biol. 2009, 19, 423–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-Induced Dimerization Activates a Plant Immune Receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, K.; Yamada, K.; Ishikawa, K.; Yoshimura, S.; Hayashi, N.; Uchihashi, K.; Ishihama, N.; Kishi-Kaboshi, M.; Takahashi, A.; Tsuge, S.; et al. A Receptor-Like Cytoplasmic Kinase Targeted by a Plant Pathogen Effector Is Directly Phosphorylated by the Chitin Receptor and Mediates Rice Immunity. Cell Host Microbe 2013, 13, 347–357. [Google Scholar] [CrossRef] [Green Version]
- Nissen, K.S.; Willats, W.G.T.; Malinovsky, F.G. Understanding Crrlk1l Function: Cell Walls and Growth Control. Trends Plant Sci. 2016, 21, 516–527. [Google Scholar] [CrossRef]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The Receptor Kinase Fer Is a Ralf-Regulated Scaffold Controlling Plant Immune Signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef] [Green Version]
- Franck, C.M.; Westermann, J.; Boisson-Dernier, A. Plant Malectin-Like Receptor Kinases: From Cell Wall Integrity to Immunity and Beyond. Annu. Rev. Plant Biol. 2018, 69, 301–328. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of Pattern Recognition Receptor Signalling in Plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Shi, H.; Shen, Q.; Qi, Y.; Yan, H.; Nie, H.; Chen, Y.; Zhao, T.; Katagiri, F.; Tang, D. Br-Signaling Kinase1 Physically Associates with Flagellin Sensing2 and Regulates Plant Innate Immunity in Arabidopsis. Plant Cell 2013, 25, 1143–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nurnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A Flagellin-Induced Complex of the Receptor Fls2 and Bak1 Initiates Plant Defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Schulze, B.; Mentzel, T.; Jehle, A.K.; Mueller, K.; Beeler, S.; Boller, T.; Felix, G.; Chinchilla, D. Rapid Heteromerization and Phosphorylation of Ligand-Activated Plant Transmembrane Receptors and Their Associated Kinase Bak1. J. Biol. Chem. 2010, 285, 9444–9451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Grubb, L.E.; Wang, J.; Liang, X.; Li, L.; Gao, C.; Ma, M.; Feng, F.; Li, M.; Li, L.; et al. A Regulatory Module Controlling Homeostasis of a Plant Immune Kinase. Mol. Cell 2018, 69, 493–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, F.; Yang, F.; Rong, W.; Wu, X.; Zhang, J.; Chen, S.; He, C.; Zhou, J.M. A Xanthomonas Uridine 5′-Monophosphate Transferase Inhibits Plant Immune Kinases. Nature 2012, 485, 114–118. [Google Scholar] [CrossRef]
- Ranf, S.; Eschen-Lippold, L.; Frohlich, K.; Westphal, L.; Scheel, D.; Lee, J. Microbe-Associated Molecular Pattern-Induced Calcium Signaling Requires the Receptor-Like Cytoplasmic Kinases, Pbl1 and Bik1. BMC Plant Biol. 2014, 14, 374. [Google Scholar] [CrossRef] [Green Version]
- Monaghan, J.; Matschi, S.; Romeis, T.; Zipfel, C. The Calcium-Dependent Protein Kinase Cpk28 Negatively Regulates the Bik1-Mediated Pamp-Induced Calcium Burst. Plant Signal. Behav. 2015, 10, e1018497. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Ding, P.; Lian, K.; Wang, J.; Ma, M.; Li, L.; Li, L.; Li, M.; Zhang, X.; Chen, S.; et al. Arabidopsis Heterotrimeric G Proteins Regulate Immunity by Directly Coupling to the Fls2 Receptor. eLife 2016, 5, e13568. [Google Scholar] [CrossRef]
- Zhang, M.X.; Chiang, Y.; Toruño, T.Y.; Lee, D.; Ma, M.; Liang, X.; Lal, N.K.; Lemos, M.; Lu, Y.; Ma, S.; et al. The Map4 Kinase Sik1 Ensures Robust Extracellular Ros Burst and Antibacterial Immunity in Plants. Cell Host Microbe 2018, 24, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Shinya, T.; Motoyama, N.; Ikeda, A.; Wada, M.; Kamiya, K.; Hayafune, M.; Kaku, H.; Shibuya, N. Functional Characterization of Cebip and Cerk1 Homologs in Arabidopsis and Rice Reveals the Presence of Different Chitin Receptor Systems in Plants. Plant Cell Physiol. 2012, 53, 1696–1706. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The Kinase Lyk5 Is a Major Chitin Receptor in Arabidopsis and Forms a Chitin-Induced Complex with Related Kinase Cerk1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef] [PubMed]
- Shinya, T.; Yamaguchi, K.; Desaki, Y.; Yamada, K.; Narisawa, T.; Kobayashi, Y.; Maeda, K.; Suzuki, M.; Tanimoto, T.; Takeda, J.; et al. Selective Regulation of the Chitin-Induced Defense Response by the Arabidopsis Receptor-Like Cytoplasmic Kinase Pbl27. Plant J. 2014, 79, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Yamada, K.; Yoshimura, S.; Yamaguchi, K. Chitin Receptor-Mediated Activation of Map Kinases and Ros Production in Rice and Arabidopsis. Plant Signal. Behav. 2017, 12, e1361076. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.; Brechmann, M.; Röhling, S.; Tapernoux, M.; Mock, T.; Winter, D.; Lehmann, W.D.; Kiefer, F.; Thome, M.; Krammer, P.H.; et al. Phosphorylation of Carma1 by Hpk1 Is Critical for Nf-Kb Activation in T Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 14508–14513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leberer, E.; Thomas, D.Y.; Whiteway, M. Pheromone Signalling and Polarized Morphogenesis in Yeast. Curr. Opin. Genet. Dev. 1997, 7, 59–66. [Google Scholar] [CrossRef]
- Yu, X.; Feng, B.; He, P.; Shan, L. From Chaos to Harmony: Responses and Signaling Upon Microbial Pattern Recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef]
- Willmanna, R.; Lajunena, H.M.; Erbsb, G.; Newmanb, M.; Kolba, D.; Tsudac, K.; Katagiric, F.; Fliegmannd, J.; Bonof, J.; Cullimoref, J.V.; et al. Arabidopsis Lysin-Motif Proteins Lym1 Lym3 Cerk1 Mediate Bacterial Peptidoglycan Sensing and Immunity to Bacterial Infection. Proc. Natl. Acad. Sci. USA 2011, 108, 19824–19829. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Tanaka, K.; Zhang, X.C.; Son, G.H.; Brechenmacher, L.; Nguyen, T.H.; Stacey, G. Lyk4, a Lysin Motif Receptor-Like Kinase, Is Important for Chitin Signaling and Plant Innate Immunity in Arabidopsis. Plant Physiol. 2012, 160, 396–406. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, K.; Yamada, K.; Kawasaki, T. Receptor-Like Cytoplasmic Kinases Are Pivotal Components in Pattern Recognition Receptor-Mediated Signaling in Plant Immunity. Plant Signal. Behav. 2013, 8, e25662. [Google Scholar] [CrossRef] [Green Version]
- Ao, Y.; Li, Z.; Feng, D.; Xiong, F.; Liu, J.; Li, J.F.; Wang, M.; Wang, J.; Liu, B.; Wang, H.B. Oscerk1 and Osrlck176 Play Important Roles in Peptidoglycan and Chitin Signaling in Rice Innate Immunity. Plant J. 2014, 80, 1072–1084. [Google Scholar] [CrossRef]
- Liu, B.; Li, J.F.; Ao, Y.; Qu, J.; Li, Z.; Su, J.; Zhang, Y.; Liu, J.; Feng, D.; Qi, K.; et al. Lysin Motif-Containing Proteins Lyp4 and Lyp6 Play Dual Roles in Peptidoglycan and Chitin Perception in Rice Innate Immunity. Plant Cell 2012, 24, 3406–3419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two Lysm Receptor Molecules, Cebip and Oscerk1, Cooperatively Regulate Chitin Elicitor Signaling in Rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [Green Version]
- Hayafunea, M.; Berisiob, R.; Marchettic, R.; Silipoc, A.; Kayamaa, M.; Desakia, Y.; Arimaa, S.; Squegliab, F.; Ruggierob, A.; Tokuyasud, K.; et al. Chitin-Induced Activation of Immune Signaling by the Rice Receptor Cebip Relies on a Unique Sandwich-Type Dimerization. Proc. Natl. Acad. Sci. USA 2013, 111, e404–e413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishi-Kaboshi, M.; Okada, K.; Kurimoto, L.; Murakami, S.; Umezawa, T.; Shibuya, N.; Yamane, H.; Miyao, A.; Takatsuji, H.; Takahashi, A.; et al. A Rice Fungal Mamp-Responsive Mapk Cascade Regulates Metabolic Flow to Antimicrobial Metabolite Synthesis. Plant J. 2010, 63, 599–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.; Yamaguchi, K.; Yoshimura, S.; Terauchi, A.; Kawasaki, T. Conservation of Chitin-Induced Mapk Signaling Pathways in Rice and Arabidopsis. Plant Cell Physiol. 2017, 58, 993–1002. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Zhang, C.; Zhu, P.; Dai, H.; Yu, N.; He, Z.; Xu, L.; Wang, E. Oscerk1-Mediated Chitin Perception and Immune Signaling Requires Receptor-Like Cytoplasmic Kinase 185 to Activate an Mapk Cascade in Rice. Mol. Plant 2017, 10, 619–633. [Google Scholar] [CrossRef] [Green Version]
- Van Breusegem, F.; Bailey-Serres, J.; Mittler, R. Unraveling the Tapestry of Networks Involving Reactive Oxygen Species in Plants. Plant Physiol. 2008, 147, 978–984. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, A.; Chen, L.; Thao, N.P.; Fujiwara, M.; Wong, H.L.; Kuwano, M.; Umemura, K.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. Rack1 Functions in Rice Innate Immunity by Interacting with the Rac1 Immune Complex. Plant Cell 2008, 20, 2265–2279. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Li, J.F.; Niu, Y.; Zhang, X.C.; Woody, O.Z.; Xiong, Y.; Djonovic, S.; Millet, Y.; Bush, J.; McConkey, B.J.; et al. Pathogen-Secreted Proteases Activate a Novel Plant Immune Pathway. Nature 2015, 521, 213–216. [Google Scholar] [CrossRef]
- Wong, H.L.; Kawasaki, T.; Shimamoto, K. Rac Gtpase and the Regulation of NADPH Oxidase in Rice Innate Immunity Response. In Advances in Genetics, Genomics and Control of Rice Blast Disease; Wang, G.L., Valent, B., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 173–178. [Google Scholar]
- Wong, H.L.; Pinontoan, R.; Hayashi, K.; Tabata, R.; Yaeno, T.; Hasegawa, K.; Kojima, C.; Yoshioka, H.; Iba, K.; Kawasaki, T.; et al. Regulation of Rice NADPH Oxidase by Binding of Rac Gtpase to Its N-Terminal Extension. Plant Cell 2007, 19, 4022–4034. [Google Scholar] [CrossRef] [Green Version]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.M. Feronia Receptor-Like Kinase Regulates Rho Gtpase Signaling of Root Hair Development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef] [Green Version]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras Superfamily at a Glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaothien, P.; Ok, S.H.; Shuai, B.; Wengier, D.; Cotter, R.; Kelley, D.; Kiriakopolos, S.; Muschietti, J.; McCormick, S. Kinase Partner Protein Interacts with the Leprk1 and Leprk2 Receptor Kinases and Plays a Role in Polarized Pollen Tube Growth. Plant J. 2005, 42, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yeh, F.L.; Cheung, A.Y.; Duan, Q.; Kita, D.; Liu, M.C.; Maman, J.; Luu, E.J.; Wu, B.W.; Gates, L.; et al. Glycosylphosphatidylinositol-Anchored Proteins as Chaperones and Co-Receptors for Feronia Receptor Kinase Signaling in Arabidopsis. eLife 2015, 4, e06587. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A Peptide Hormone and Its Receptor Protein Kinase Regulate Plant Cell Expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zayed, O.; Yu, Z.; Jiang, W.; Zhu, P.; Hsu, C.C.; Zhang, L.; Tao, W.A.; Lozano-Duran, R.; Zhu, J.K. Leucine-Rich Repeat Extensin Proteins Regulate Plant Salt Tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef] [Green Version]
- Suharsono, U.; Fujisawa, Y.; Kawasaki, T.; Iwasaki, Y.; Satoh, H.; Shimamoto, K. The Heterotrimeric G Protein A Subunit Acts Upstream of the Small Gtpase Rac in Disease Resistance of Rice. Proc. Natl. Acad. Sci. USA 2002, 99, 13307–13312. [Google Scholar] [CrossRef] [Green Version]
- Daugaard, M.; Nitsch, R.; Razaghi, B.; McDonald, L.; Jarrar, A.; Torrino, S.; Castillo-Lluva, S.; Rotblat, B.; Li, L.; Malliri, A.; et al. Hace1 Controls Ros Generation of Vertebrate Rac1-Dependent NADPH Oxidase Complexes. Nat. Commun. 2013, 4, 2180. [Google Scholar] [CrossRef] [Green Version]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.G.; Schroeder, J.I. NADPH Oxidase Atrbohd and Atrbohf Genes Function in Ros-Dependent Aba Signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant Stomata Function in Innate Immunity against Bacterial Invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [Green Version]
- Mustilli, A.C. Arabidopsis Ost1 Protein Kinase Mediates the Regulation of Stomatal Aperture by Abscisic Acid and Acts Upstream of Reactive Oxygen Species Production. Plant Cell 2002, 14, 3089–3099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Qian, L.; Nibau, C.; Duan, Q.; Kita, D.; Levasseur, K.; Li, X.; Lu, C.; Li, H.; Hou, C.; et al. Feronia Receptor Kinase Pathway Suppresses Abscisic Acid Signaling in Arabidopsis by Activating Abi2 Phosphatase. Proc. Natl. Acad. Sci. USA 2012, 109, 14693–14698. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic Acid Inhibits Type 2c Protein Phosphatases via the Pyr/Pyl Family of Start Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Yu, F.; Liu, Y.; Du, C.; Li, X.; Zhu, S.; Wang, X.; Lan, W.; Rodriguez, P.L.; Liu, X.; et al. Feronia Interacts with Abi2-Type Phosphatases to Facilitate Signaling Crosstalk between Abscisic Acid and Ralf Peptide in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, e5519–e5527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase Dalpha1 and Phosphatidic Acid Regulate NADPH Oxidase Activity and Production of Reactive Oxygen Species in Aba-Mediated Stomatal Closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girish, M.; Zhang, W.H.; Deng, F.; Zhao, J.; Wang, X.M. A Bifurcating Pathway Directs Abscisic Acid Effects on Stomatal Closure and Opening in Arabidopsis. Science 2006, 312, 264–266. [Google Scholar]
- Desikan, R.; Last, K.; Harrett-Williams, R.; Tagliavia, C.; Harter, K.; Hooley, R.; Hancock, J.T.; Neill, S.J. Ethylene-Induced Stomatal Closure in Arabidopsis Occurs via Atrbohf-Mediated Hydrogen Peroxide Synthesis. Plant J. 2006, 47, 907–916. [Google Scholar] [CrossRef]
- Alisa, H.; Gregory, P.; Ryan Clarence, A. An Endogenous Peptide Signal in Arabidopsis Activates Components of the Innate Immune Response. Proc. Natl. Acad. Sci. USA 2006, 103, 10098–10103. [Google Scholar]
- Bartels, S.; Lori, M.; Mbengue, M.; van Verk, M.; Klauser, D.; Hander, T.; Boni, R.; Robatzek, S.; Boller, T. The Family of Peps and Their Precursors in Arabidopsis: Differential Expression and Localization but Similar Induction of Pattern-Triggered Immune Responses. J. Exp. Bot. 2013, 64, 5309–5321. [Google Scholar] [CrossRef] [Green Version]
- Maruta, T.; Inoue, T.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. Arabidopsis NADPH Oxidases, Atrbohd and Atrbohf, Are Essential for Jasmonic Acid-Induced Expression of Genes Regulated by Myc2 Transcription Factor. Plant Sci. 2011, 180, 655–660. [Google Scholar] [CrossRef]
- Hettenhausen, C.; Schuman, M.C.; Wu, J.Q. Mapk Signaling: A Key Element in Plant Defense Response to Insects. Insect Sci. 2015, 22, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, M.C.; Petersen, M.; Mundy, J. Mitogen-Activated Protein Kinase Signaling in Plants. Annu. Rev. Plant Biol. 2010, 61, 621–649. [Google Scholar]
- Liu, Y.K. Roles of Mitogen-Activated Protein Kinase Cascades in Aba Signaling. Plant Cell Rep. 2012, 31, 1–12. [Google Scholar] [CrossRef]
- Liu, Y.; Kong, X.; Pan, J.; Li, D. Vip1: Linking Agrobacterium-Mediated Transformation to Plant Immunity? Plant Cell Rep. 2010, 29, 805–812. [Google Scholar] [CrossRef]
- Teige, M.; Scheikl, E.; Eulgem, T.; Doczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The Mkk2 Pathway Mediates Cold and Salt Stress Signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef]
- Pitzschke, A.; Djamei, A.; Bitton, F.; Hirt, H. A Major Role of the Mekk1-Mkk1/2-Mpk4 Pathway in Ros Signalling. Mol. Plant 2009, 2, 120–137. [Google Scholar] [CrossRef] [Green Version]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. Map Kinase Signalling Cascade in Arabidopsis Innate Immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Zhang, J.; Shao, F.; Li, Y.; Cui, H.; Chen, L.; Li, H.; Zou, Y.; Long, C.; Lan, L.; Chai, J.; et al. A Pseudomonas Syringae Effector Inactivates Mapks to Suppress Pamp-Induced Immunity in Plants. Cell Host Microbe 2007, 1, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Macho, A.P.; Boutrot, F.; Rathjen, J.P.; Zipfel, C. Aspartate Oxidase Plays an Important Role in Arabidopsis Stomatal Immunity. Plant Physiol. 2012, 159, 1845–1856. [Google Scholar] [CrossRef] [Green Version]
- Durrant, W.E.; Dong, X. Systemic Acquired Resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional Machineries in Jasmonate-Elicited Plant Secondary Metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef]
- Overmyer, K.; Tuominen, H.; Kettunen, R.; Betz, C.; Langebartels, C.; Sandermann, H.; Kangasjarvi, J. Ozone-Sensitive Arabidopsis Rcd1 Mutant Reveals Opposite Roles for Ethylene and Jasmonate Signaling Pathways in Regulating Superoxide-Dependent Cell Death. Plant Cell 2000, 12, 1849–1862. [Google Scholar] [CrossRef] [Green Version]
- Tintor, N.; Ross, A.; Kanehara, K.; Yamada, K.; Fan, L.; Kemmerling, B.; Nurnberger, T.; Tsuda, K.; Saijo, Y. Layered Pattern Receptor Signaling via Ethylene and Endogenous Elicitor Peptides During Arabidopsis Immunity to Bacterial Infection. Proc. Natl. Acad. Sci. USA 2013, 110, 6211–6216. [Google Scholar] [CrossRef] [Green Version]
- Flury, P.; Klauser, D.; Schulze, B.; Boller, T.; Bartels, S. The Anticipation of Danger: Microbe-Associated Molecular Pattern Perception Enhances Atpep-Triggered Oxidative Burst. Plant Physiol. 2013, 161, 2023–2035. [Google Scholar] [CrossRef] [Green Version]
- Browse, J. Jasmonate Passes Muster: A Receptor and Targets for the Defense Hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef]
- Xia, X.J.; Wang, Y.J.; Zhou, Y.H.; Tao, Y.; Mao, W.H.; Shi, K.; Asami, T.; Chen, Z.; Yu, J.Q. Reactive Oxygen Species Are Involved in Brassinosteroid-Induced Stress Tolerance in Cucumber. Plant Physiol. 2009, 150, 801–814. [Google Scholar] [CrossRef] [Green Version]
- Elmayan, T.; Fromentin, J.; Riondet, C.; Alcaraz, G.; Blein, J.P.; Simon-Plas, F. Regulation of Reactive Oxygen Species Production by a 14-3-3 Protein in Elicited Tobacco Cells. Plant Cell Environ. 2007, 30, 722–732. [Google Scholar] [CrossRef]
- Tsuruhara, A.; Suzuki, H.; Tezuka, T. Inhibition of the Activity of Nad(P)H-Dependent Oxidase in Pistils of Lilium Longiflorum by Camp. Plant Cell Physiol. 1999, 40, 1093–1098. [Google Scholar] [CrossRef] [Green Version]
- Souabni, H.; Thoma, V.; Bizouarn, T.; Chatgilialoglu, C.; Siafaka-Kapadai, A.; Baciou, L.; Ferreri, C.; Houee-Levin, C.; Ostuni, M.A. trans Arachidonic Acid Isomers Inhibit Nadph-Oxidase Activity by Direct Interaction with Enzyme Components. Biochim. Biophys. Acta 2012, 1818, 2314–2324. [Google Scholar] [CrossRef] [Green Version]
- Arnaud, D.; Hwang, I. A Sophisticated Network of Signaling Pathways Regulates Stomatal Defenses to Bacterial Pathogens. Mol. Plant 2015, 8, 566–581. [Google Scholar] [CrossRef] [Green Version]
- Yun, B.W.; Feechan, A.; Yin, M.; Saidi, N.B.; Bihan, T.L.; Yu, M.; Moore, J.W.; Kang, J.G.; Kwon, E.; Spoel, S.H.; et al. S-Nitrosylation of NADPH Oxidase Regulates Cell Death in Plant Immunity. Nature 2011, 478, 264–268. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, C.-H.; Wang, P.-Q.; Zhang, P.-P.; Nie, X.-M.; Li, B.-B.; Tai, L.; Liu, W.-T.; Li, W.-Q.; Chen, K.-M. NADPH Oxidases: The Vital Performers and Center Hubs during Plant Growth and Signaling. Cells 2020, 9, 437. https://doi.org/10.3390/cells9020437
Hu C-H, Wang P-Q, Zhang P-P, Nie X-M, Li B-B, Tai L, Liu W-T, Li W-Q, Chen K-M. NADPH Oxidases: The Vital Performers and Center Hubs during Plant Growth and Signaling. Cells. 2020; 9(2):437. https://doi.org/10.3390/cells9020437
Chicago/Turabian StyleHu, Chun-Hong, Peng-Qi Wang, Peng-Peng Zhang, Xiu-Min Nie, Bin-Bin Li, Li Tai, Wen-Ting Liu, Wen-Qiang Li, and Kun-Ming Chen. 2020. "NADPH Oxidases: The Vital Performers and Center Hubs during Plant Growth and Signaling" Cells 9, no. 2: 437. https://doi.org/10.3390/cells9020437
APA StyleHu, C.-H., Wang, P.-Q., Zhang, P.-P., Nie, X.-M., Li, B.-B., Tai, L., Liu, W.-T., Li, W.-Q., & Chen, K.-M. (2020). NADPH Oxidases: The Vital Performers and Center Hubs during Plant Growth and Signaling. Cells, 9(2), 437. https://doi.org/10.3390/cells9020437




