Artificial Intelligence in Thyroid Cytopathology: Diagnostic and Technical Insights
Simple Summary
Abstract
1. Introduction
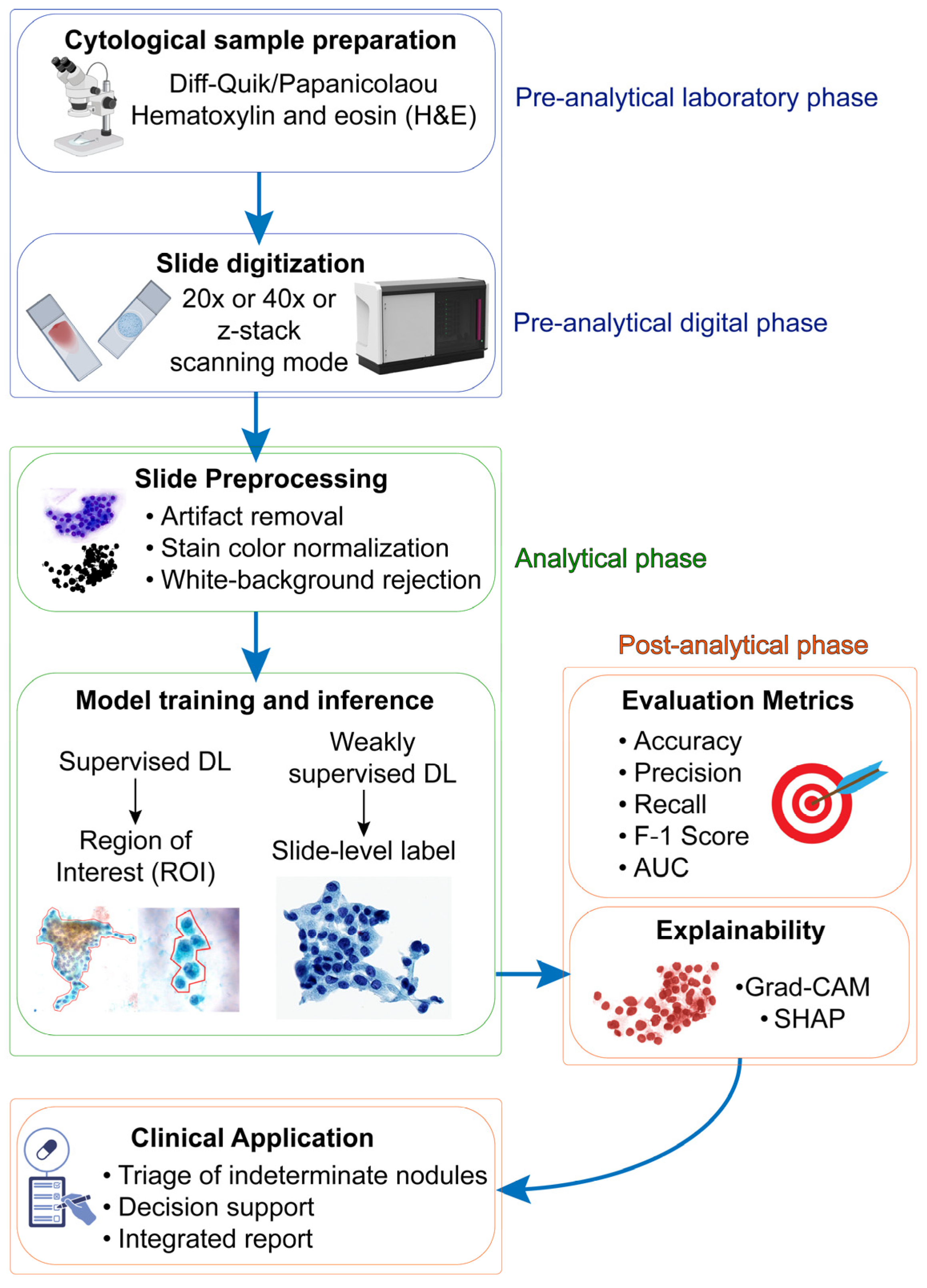
2. Preanalytical Considerations
2.1. Staining Quality
2.2. Specimen Preparation and Slide Digitalization
2.3. Human Variability in Region of Interest (ROI) Annotation
2.4. Data Quality, Inclusion Criteria, and Domain Shift
3. Architectural Variables
3.1. Convolutional Neural Networks (CNNs)
3.2. Multiple Instance Learning (MIL)
3.3. Hybrid DL Platforms
4. Toward a Reliable Digital Cytodiagnostic Pipeline
4.1. Co-Pilot
4.2. Bethesda Classifiers
4.3. Molecular Classifiers
5. Conclusions and Future Directions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zamora, E.A.; Khare, S.; Cassaro, S. Thyroid nodule. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Haugen, B.R. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: What is new and what has changed? Cancer 2016, 123, 372–381. [Google Scholar] [CrossRef]
- Bayrak, Y.B.; Eruyar, A.T. Malignancy rates for Bethesda III and IV thyroid nodules: A retrospective study of the correlation between fine-needle aspiration cytology and histopathology. BMC Endocr. Disord. 2020, 20, 48. [Google Scholar] [CrossRef] [PubMed]
- Piticchio, T.; Russ, G.; Radzina, M.; Frasca, F.; Durante, C.; Trimboli, P. Head-to-head comparison of American, European, and Asian TIRADSs in thyroid nodule assessment: Systematic review and meta-analysis. Eur. Thyroid. J. 2024, 13, e230242. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zeng, F.; Wang, Y.; Bai, Y.; Shan, X.; Kong, L. Prevalence and associated metabolic factors for thyroid nodules: A cross-sectional study in Southwest of China with more than 120 thousand populations. BMC Endocr. Disord. 2021, 21, 175. [Google Scholar] [CrossRef] [PubMed]
- Angerilli, V.; Galuppini, F.; Pagni, F.; Fusco, N.; Malapelle, U.; Fassan, M. The Role of the Pathologist in the Next-Generation Era of Tumor Molecular Characterization. Diagnostics 2021, 11, 339. [Google Scholar] [CrossRef]
- Cappello, F.; Angerilli, V.; Munari, G.; Ceccon, C.; Sabbadin, M.; Pagni, F.; Fusco, N.; Malapelle, U.; Fassan, M. FFPE-Based NGS Approaches into Clinical Practice: The Limits of Glory from a Pathologist Viewpoint. J. Pers. Med. 2022, 12, 750. [Google Scholar] [CrossRef]
- Fusco, N.; Pruneri, G.; Pagni, F.; Malapelle, U. Molecular Testing in Solid Tumors: Best Practices from the Molecular Pathology and Precision Medicine Study Group of the Italian Society of Pathology (PMMP/SIAPeC): Shaping Excellence in Molecular Diagnostics. Pathol. J. Ital. Soc. Anat. Pathol. Diagn. Cytopathol. 2025, 117, S1–S4. [Google Scholar] [CrossRef]
- Hirokawa, M.; Niioka, H.; Suzuki, A.; Abe, M.; Arai, Y.; Nagahara, H.; Miyauchi, A.; Akamizu, T. Application of deep learning as an ancillary diagnostic tool for thyroid FNA cytology. Cancer Cytopathol. 2022, 131, 217–225. [Google Scholar] [CrossRef]
- Wei, X.; Zhu, J.; Zhang, H.; Gao, H.; Yu, R.; Liu, Z.; Zheng, X.; Gao, M.; Zhang, S. Visual Interpretability in Computer-Assisted Diagnosis of Thyroid Nodules Using Ultrasound Images. Med Sci. Monit. 2020, 26, e927007-1–e927007-11. [Google Scholar] [CrossRef]
- Hou, L.; Samaras, D.; Kurc, T.M.; Gao, Y.; Davis, J.E.; Saltz, J.H. Patch-Based Convolutional Neural Network for Whole Slide Tissue Image Classification. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2424–2433. [Google Scholar] [CrossRef]
- Dimitriou, N.; Arandjelović, O.; Caie, P.D. Deep Learning for Whole Slide Image Analysis: An Overview. Front. Med. 2019, 6, 264. [Google Scholar] [CrossRef]
- IHC World. Diff-Quick (Diff-Quik) Staining Protocol. 2024. Available online: https://ihcworld.com/2024/01/26/diff-quick-diff-quik-staining-protocol/ (accessed on 15 June 2025).
- Sathawane, P.; Kamal, M.M.; Deotale, P.R.; Mankar, H. Nuances of the Papanicolaou stain. Cytojournal 2022, 19, 43. [Google Scholar] [CrossRef]
- IHC World. Hematoxylin and Eosin (H&E) Staining Protocol. 2024. Available online: https://ihcworld.com/2024/01/25/hematoxylin-and-eosin-he-staining-protocol/ (accessed on 15 June 2025).
- Ding, F.; Cai, C.; Li, J.; Liu, M.; Jiao, Y.; Wu, Z.; Xu, J. Classification of Whole-Slide Pathology Images Based on State Space Models and Graph Neural Networks. Electronics 2025, 14, 2056. [Google Scholar] [CrossRef]
- Reinhard, E.; Adhikhmin, M.; Gooch, B.; Shirley, P. Color transfer between images. IEEE Comput. Graph. Appl. 2001, 21, 34–41. [Google Scholar] [CrossRef]
- Kang, G. StainNet: A fast and robust stain normalization network. Front. Med. 2021, 8, 746307. [Google Scholar] [CrossRef] [PubMed]
- Zanjani, F. StainGAN: Stain Style Transfer for Digital Histological Images. Sci. Rep. 2023, 13, 13781. [Google Scholar]
- Kim, D.; Burkhardt, R.; Alperstein, S.A.; Gokozan, H.N.; Goyal, A.; Heymann, J.J.; Patel, A.; Siddiqui, M.T. Evaluating the role of Z-stack to improve the morphologic evaluation of urine cytology whole slide images for high-grade urothelial carcinoma: Results and review of a pilot study. Cancer Cytopathol. 2022, 130, 630–639. [Google Scholar] [CrossRef]
- Gu, H.; Onstott, E.; Yan, W.; Xu, T.; Wang, R.; Wu, Z.; Chen, X.A.; Haeri, M. Z-stack scanning can improve AI detection of mitosis: A case study of meningiomas. In Proceedings of the 2025 IEEE 22nd International Symposium on Biomedical Imaging (ISBI), Houston, TX, USA, 14–17 April 2025. [Google Scholar]
- Osamura, R.Y.; Matsui, N.; Kawashima, M.; Saiga, H.; Ogura, M.; Kiyuna, T. Digital/Computational Technology for Molecular Cytology Testing: A Short Technical Note with Literature Review. Acta Cytol. 2021, 65, 342–347. [Google Scholar] [CrossRef]
- Yao, K.; Shen, R.; Parwani, A.; Li, Z. Comprehensive Study of Telecytology Using Robotic Digital Microscope and Single Z-Stack Digital Scan for Fine-Needle Aspiration-Rapid On-Site Evaluation. J. Pathol. Inform. 2018, 9, 49. [Google Scholar] [CrossRef]
- Ji, X.; Salmon, R.; Mulliqi, N.; Khan, U.; Wang, Y.; Blilie, A.; Olsson, H.; Pedersen, B.G.; Sørensen, K.D.; Ulhøi, B.P.; et al. Physical Color Calibration of Digital Pathology Scanners for Robust Artificial Intelligence—Assisted Cancer Diagnosis. Modern Pathol. 2023, 38, 100715. [Google Scholar] [CrossRef]
- Brixtel, R.; Bougleux, S.; Lezoray, O.; Caillot, Y.; Lemoine, B.; Fontaine, M.; Nebati, D.; Renouf, A. Whole Slide Image Quality in Digital Pathology: Review and Perspectives. IEEE Access 2022, 10, 131005–131035. [Google Scholar] [CrossRef]
- Holub, P.; Müller, H.; Bíl, T.; Pireddu, L.; Plass, M.; Prasser, F.; Schlünder, I.; Zatloukal, K.; Nenutil, R.; Brázdil, T. Privacy risks of whole-slide image sharing in digital pathology. Nat. Commun. 2023, 14, 2577. [Google Scholar] [CrossRef]
- Hossain, M.S.; Shahriar, G.M.; Syeed, M.M.M.; Uddin, M.F.; Hasan, M.; Shivam, S.; Advani, S. Region of interest (ROI) selection using vision transformer for automatic analysis using whole slide images. Sci. Rep. 2023, 13, 11314. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, W.; Zhang, M.-L. Attention is not what you need: Revisiting multi-instance learning for whole slide image classification. arXiv 2024, arXiv:2408.09449. [Google Scholar] [CrossRef]
- Cazzaniga, G.; Del Carro, F.; Eccher, A.; Becker, J.U.; Gambaro, G.; Rossi, M.; Pieruzzi, F.; Fraggetta, F.; Pagni, F.; L’iMperio, V. Improving the Annotation Process in Computational Pathology: A Pilot Study with Manual and Semi-automated Approaches on Consumer and Medical Grade Devices. J. Imaging. Inform. Med. 2024, 38, 1112–1119. [Google Scholar] [CrossRef] [PubMed]
- Corradi, A.; Bonizzi, G.; Sajjadi, E.; Pavan, F.; Fumagalli, M.; Molendini, L.O.; Monturano, M.; Cassi, C.; Musico, C.R.; Leoni, L.; et al. The Regulatory Landscape of Biobanks in Europe: From Accreditation to Intellectual Property. Curr. Genom. 2025, 26, 15–23. [Google Scholar] [CrossRef]
- Lee, B.; Smola, B.; Roh, M.H.; Hughes, D.T.; Miller, B.S.; Jing, X. The impact of using the Bethesda System for reporting thyroid cytology diagnostic criteria on the follicular lesion of undetermined significance category. J. Am. Soc. Cytopathol. 2014, 3, 131–136. [Google Scholar] [CrossRef]
- Barthe, P.; Brixtel, R.; Caillot, Y.; Lemoine, B.; Renouf, A.; Thurotte, V.; Beniken, O.; Bougleux, S.; Lézoray, O. Assessing the quality of whole slide images in cytology from nuclei features. J. Pathol. Informatics 2025, 17, 100420. [Google Scholar] [CrossRef]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.-M.; Ciompi, F.; van der Laak, J. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 2019, 58, 101544. [Google Scholar] [CrossRef]
- Stacke, K.; Eilertsen, G.; Unger, J.; Lundstrom, C. Measuring Domain Shift for Deep Learning in Histopathology. IEEE J. Biomed. Health Inform. 2020, 25, 325–336. [Google Scholar] [CrossRef]
- Lin, Y.-J.; Chao, T.-K.; Khalil, M.-A.; Lee, Y.-C.; Hong, D.-Z.; Wu, J.-J.; Wang, C.-W. Deep Learning Fast Screening Approach on Cytological Whole Slides for Thyroid Cancer Diagnosis. Cancers 2021, 13, 3891. [Google Scholar] [CrossRef]
- Gadermayr, M.; Tschuchnig, M. Multiple instance learning for digital pathology: A review of the state-of-the-art, limitations & future potential. Comput. Med. Imaging Graph. 2024, 112, 102337. [Google Scholar]
- Pittaro, A.; Del Gobbo, A.; Iofrida, E.; Fusco, N. Chondroid Differentiation in Thyroid Nodular Hyperplasia: An Innocent Bystander? Int. J. Surg. Pathol. 2018, 27, 274. [Google Scholar] [CrossRef]
- Smith, A.; Galli, M.; Piga, I.; Denti, V.; Stella, M.; Chinello, C.; Fusco, N.; Leni, D.; Manzoni, M.; Roversi, G.; et al. Molecular signatures of medullary thyroid carcinoma by matrix-assisted laser desorption/ionisation mass spectrometry imaging. J. Proteom. 2019, 191, 114–123. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Boehm, K.M.; El Nahhas, O.S.M.; Marra, A.; Waters, M.; Jee, J.; Braunstein, L.; Schultz, N.; Selenica, P.; Wen, H.Y.; Weigelt, B.; et al. Multimodal histopathologic models stratify hormone receptor-positive early breast cancer. Nat. Commun. 2025, 16, 2106. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2012; Volume 25. [Google Scholar]
- Rawat, R.R.; Ortega, I.; Roy, P.; Sha, F.; Shibata, D.; Ruderman, D.; Agus, D.B. Deep learned tissue “fingerprints” classify breast cancers by ER/PR/Her2 status from H&E images. Sci. Rep. 2020, 10, 7275. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Talo, M. Automated classification of histopathology images using transfer learning. Artif. Intell. Med. 2019, 101, 101743. [Google Scholar] [CrossRef]
- Gupta, P.; Huang, Y.; Sahoo, P.K.; You, J.-F.; Chiang, S.-F.; Onthoni, D.D.; Chern, Y.-J.; Chao, K.-Y.; Chiang, J.-M.; Yeh, C.-Y.; et al. Colon Tissues Classification and Localization in Whole Slide Images Using Deep Learning. Diagnostics 2021, 11, 1398. [Google Scholar] [CrossRef]
- Matias-Guiu, X.; Temprana-Salvador, J.; Lopez, P.G.; Kammerer-Jacquet, S.-F.; Rioux-Leclercq, N.; Clark, D.; Schürch, C.M.; Fend, F.; Mattern, S.; Snead, D.; et al. Implementing digital pathology: Qualitative and financial insights from eight leading European laboratories. Virchows Arch. 2025, 487, 815–826. [Google Scholar] [CrossRef]
- Campanella, G.; Chen, S.; Singh, M.; Verma, R.; Muehlstedt, S.; Zeng, J.; Stock, A.; Croken, M.; Veremis, B.; Elmas, A.; et al. A clinical benchmark of public self-supervised pathology foundation models. Nat. Commun. 2025, 16, 3640. [Google Scholar] [CrossRef]
- Ilse, M.; Tomczak, J.; Welling, M. Attention-based deep multiple instance learning. In Proceedings of the 35th International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; PMLR: Norfolk, MA, USA, 2018. [Google Scholar]
- Myronenko, A.; Xu, Z.; Yang, D.; Roth, H.R.; Xu, D. Accounting for dependencies in deep learning based multiple instance learning for whole slide imaging. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; Springer International Publishing: Cham, Switzerland, 2021. [Google Scholar]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Oh, S.Y.; Lee, Y.M.; Kang, D.J.; Kwon, H.J.; Chakraborty, S.; Park, J.H. Breaking Barriers in Thyroid Cytopathology: Harnessing Deep Learning for Accurate Diagnosis. Bioengineering 2025, 12, 293. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. arXiv 2016, arXiv:1610.02391. [Google Scholar]
- Kurita, Y.; Meguro, S.; Tsuyama, N.; Kosugi, I.; Enomoto, Y.; Kawasaki, H.; Uemura, T.; Kimura, M.; Iwashita, T. Accurate deep learning model using semi-supervised learning and Noisy Student for cervical cancer screening in low magnification images. PLoS ONE 2023, 18, e0285996. [Google Scholar] [CrossRef]
- Duan, W.; Gao, L.; Liu, J.; Li, C.; Jiang, P.; Wang, L.; Chen, H.; Sun, X.; Cao, D.; Pang, B.; et al. Computer-Assisted Fine-Needle Aspiration Cytology of Thyroid Using Two-Stage Refined Convolutional Neural Network. Electronics 2022, 11, 4089. [Google Scholar] [CrossRef]
- Pham-Ngoc, H.; Nguyen-Van, D.; Vu-Tien, D.; Le-Hong, P. ThyroidEffi 1.0: A cost-effective system for high-performance multi-class thyroid carcinoma classifi-cation. arXiv 2025, arXiv:2504.14139. [Google Scholar]
- Sadeghi, Z.; Alizadehsani, R.; Cifci, M.A.; Kausar, S.; Rehman, R.; Mahanta, P.; Bora, P.K.; Almasri, A.; Alkhawaldeh, R.S.; Hussain, S.; et al. A review of Explainable Artificial Intelligence in healthcare. Comput. Electr. Eng. 2024, 118, 109370. [Google Scholar] [CrossRef]
- Sorrenti, S.; Dolcetti, V.; Radzina, M.; Bellini, M.I.; Frezza, F.; Munir, K.; Grani, G.; Durante, C.; D’andrea, V.; David, E.; et al. Artificial Intelligence for Thyroid Nodule Characterization: Where Are We Standing? Cancers 2022, 14, 3357. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Shi, L.; Lai, J.; Hu, Y.; Gu, L. Combined model integrating clinical, radiomics, BRAFV600E and ultrasound for differentiating between benign and malignant indeterminate cytology (Bethesda III) thyroid nodules: A bi-center retrospective study. Gland. Surg. 2024, 13, 1954–1964. [Google Scholar] [CrossRef]
- Issa, P.P.; McCarthy, C.; Hussein, M.; Albuck, A.L.; Emad, E.; Shama, M.; Moroz, K.; Toraih, E.; Kandil, E. Assessing Adequacy: A Meta-Analysis of Rapid Onsite Evaluation of Thyroid Nodules. J. Surg. Res. 2024, 296, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Frascarelli, C.; Venetis, K.; Marra, A.; Mane, E.; Ivanova, M.; Cursano, G.; Porta, F.M.; Concardi, A.; Ceol, A.G.M.; Farina, A.; et al. Deep learning algorithm on H&E whole slide images to characterize TP53 alterations frequency and spatial distribution in breast cancer. Comput. Struct. Biotechnol. J. 2024, 23, 4252–4259. [Google Scholar] [CrossRef]
- Ha, E.J.; Baek, J.H. Applications of machine learning and deep learning to thyroid imaging: Where do we stand? Ultrasonography 2021, 40, 23–29. [Google Scholar] [CrossRef]
- Lu, Q.; Wu, Y.; Chang, J.; Zhang, L.; Lv, Q.; Sun, H. Application progress of artificial intelligence in managing thyroid disease. Front. Endocrinol. 2025, 16, 1578455. [Google Scholar] [CrossRef]
- Yang, Y.; Guan, S.; Ou, Z.; Li, W.; Yan, L.; Situ, B. Advances in AI-based cancer cytopathology. Interdiscip. Med. 2023, 1, e20230013. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Zhu, Y.; Sang, Q.; Jia, S.; Wang, Y.; Deyer, T. Deep neural networks could differentiate Bethesda class III versus class IV/V/VI. Ann. Transl. Med. 2019, 7, 231. [Google Scholar] [CrossRef]
- Poursina, O.; Khayyat, A.; Maleki, S.; Amin, A. Artificial Intelligence and Whole Slide Imaging Assist in Thyroid Indeterminate Cytology: A Systematic Review. Acta Cytol. 2025, 69, 161–170. [Google Scholar] [CrossRef]
- Jassal, K.; Edwards, M.; Koohestani, A.; Brown, W.; Serpell, J.W.; Lee, J.C. Beyond genomics: Artificial intelligence-powered diagnostics for indeterminate thyroid nodules—A systematic review and meta-analysis. Front. Endocrinol. 2025, 16, 1506729. [Google Scholar] [CrossRef]
- Gharib, H.; Papini, E.; Garber, J.R.; Duick, D.S.; Harrell, R.M.; Hegedüs, L.; Paschke, R.; Valcavi, R.; Vitti, P.; AACE/ACE/AME Task Force on Thyroid Nodules. American Association of Clinical Endocrinologists, American College of Endocrinology, and Associazione Medici Endocrinologi Medical Guidelines for Clinical Practice for the Diagnosis and Management of Thyroid Nodules—2016 Update. Endocr. Pract. 2016, 22, 622–639. [Google Scholar] [CrossRef]
- Capitoli, G.; Alviano, A.M.; Monza, N.; Pagani, L.; Piga, I.; Bernasconi, D.P.; Greco, A.; Leni, D.; Maggioni, A.; Gatti, A.-V.; et al. Biomarker identification through spatial proteomics for the characterization of indeterminate thyroid nodules. Endocrine 2025, 90, 800–809. [Google Scholar] [CrossRef]
- Denti, V.; Greco, A.; Alviano, A.M.; Capitoli, G.; Monza, N.; Smith, A.; Pilla, D.; Maggioni, A.; Ivanova, M.; Venetis, K.; et al. Spatially Resolved Molecular Characterization of Noninvasive Follicular Thyroid Neoplasms with Papillary-like Nuclear Features (NIFTPs) Identifies a Distinct Proteomic Signature Associated with RAS-Mutant Lesions. Int. J. Mol. Sci. 2024, 25, 13115. [Google Scholar] [CrossRef]
- L’iMperio, V.; Coelho, V.; Cazzaniga, G.; Papetti, D.M.; Del Carro, F.; Capitoli, G.; Marino, M.; Ceku, J.; Fusco, N.; Ivanova, M.; et al. Machine Learning Streamlines the Morphometric Characterization and Multiclass Segmentation of Nuclei in Different Follicular Thyroid Lesions: Everything in a Nutshell. Mod. Pathol. 2024, 37, 100608. [Google Scholar] [CrossRef]
- Piga, I.; L’imperio, V.; Principi, L.; Bellevicine, C.; Fusco, N.; Maffini, F.; Venetis, K.; Ivanova, M.; Seminati, D.; Casati, G.; et al. Spatially Resolved Molecular Approaches for the Characterisation of Non-Invasive Follicular Tumours with Papillary-like Features (NIFTPs). Int. J. Mol. Sci. 2023, 24, 2567. [Google Scholar] [CrossRef]
- Seminati, D.; Mane, E.; Ceola, S.; Casati, G.; Putignano, P.; Garancini, M.; Gatti, A.; Leni, D.; Pincelli, A.I.; Fusco, N.; et al. An Indeterminate for Malignancy FNA Report Does Not Increase the Surgical Risk of Incidental Thyroid Carcinoma. Cancers 2022, 14, 5427. [Google Scholar] [CrossRef] [PubMed]
- Cazzaniga, G.; Seminati, D.; Smith, A.; Piga, I.; Capitoli, G.; Garancini, M.; L’Imperio, V.; Fusco, N.; Pagni, F. Lights on HBME-1: The elusive biomarker in thyroid cancer pathology. J. Clin. Pathol. 2022, 75, 588–592. [Google Scholar] [CrossRef]
- Baloch, Z.W.; Asa, S.L.; Barletta, J.A.; Ghossein, R.A.; Juhlin, C.C.; Jung, C.K.; LiVolsi, V.A.; Papotti, M.G.; Sobrinho-Simões, M.; Tallini, G.; et al. Overview of the 2022 WHO Classification of Thyroid Neoplasms. Endocr. Pathol. 2022, 33, 27–63. [Google Scholar] [CrossRef]
- Vora, A.; Holt, S.; Haque, W.; Lingvay, I. Long-Term Outcomes of Thyroid Nodule AFIRMA GEC Testing and Literature Review: An Institutional Experience. Otolaryngol. Head Neck Surg. 2020, 162, 634–640. [Google Scholar] [CrossRef]
- Chen, T.; Gilfix, B.M.; Rivera, J.A.; Sadeghi, N.; Richardson, K.; Hier, M.P.; Forest, V.-I.; Fishman, D.; Caglar, D.; Pusztaszeri, M.; et al. The Role of the ThyroSeq v3 Molecular Test in the Surgical Management of Thyroid Nodules in the Canadian Public Health Care Setting. Thyroid 2020, 30, 1280–1287. [Google Scholar] [CrossRef]
- Wang, C.-W.; Muzakky, H.; Lee, Y.-C.; Lin, Y.-J.; Chao, T.-K. Annotation-Free Deep Learning-Based Prediction of Thyroid Molecular Cancer Biomarker BRAF (V600E) from Cytological Slides. Int. J. Mol. Sci. 2023, 24, 2521. [Google Scholar] [CrossRef]
- Anand, B.; Ramdas, A.; Ambroise, M.M.; Kumar, N.P. The Bethesda System for Reporting Thyroid Cytopathology: A Cytohistological Study. J. Thyroid. Res. 2020, 2020, 8095378. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Negrelli, M.; Frascarelli, C.; Maffini, F.; Mangione, E.; Di Tonno, C.; Lombardi, M.; Porta, F.M.; Urso, M.; L’Imperio, V.; Pagni, F.; et al. Artificial Intelligence in Thyroid Cytopathology: Diagnostic and Technical Insights. Cancers 2025, 17, 3525. https://doi.org/10.3390/cancers17213525
Negrelli M, Frascarelli C, Maffini F, Mangione E, Di Tonno C, Lombardi M, Porta FM, Urso M, L’Imperio V, Pagni F, et al. Artificial Intelligence in Thyroid Cytopathology: Diagnostic and Technical Insights. Cancers. 2025; 17(21):3525. https://doi.org/10.3390/cancers17213525
Chicago/Turabian StyleNegrelli, Mariachiara, Chiara Frascarelli, Fausto Maffini, Elisa Mangione, Clementina Di Tonno, Mariano Lombardi, Francesca Maria Porta, Mario Urso, Vincenzo L’Imperio, Fabio Pagni, and et al. 2025. "Artificial Intelligence in Thyroid Cytopathology: Diagnostic and Technical Insights" Cancers 17, no. 21: 3525. https://doi.org/10.3390/cancers17213525
APA StyleNegrelli, M., Frascarelli, C., Maffini, F., Mangione, E., Di Tonno, C., Lombardi, M., Porta, F. M., Urso, M., L’Imperio, V., Pagni, F., Bellevicine, C., Nacchio, M., Malapelle, U., Troncone, G., Marra, A., Curigliano, G., Venetis, K., Guerini-Rocco, E., & Fusco, N. (2025). Artificial Intelligence in Thyroid Cytopathology: Diagnostic and Technical Insights. Cancers, 17(21), 3525. https://doi.org/10.3390/cancers17213525










