PIAS1 Shapes a Tumor-Suppressive Microenvironment by Suppressing Immune Evasion in Oral Squamous Cell Carcinoma
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Patient Cohort for TMA Analysis
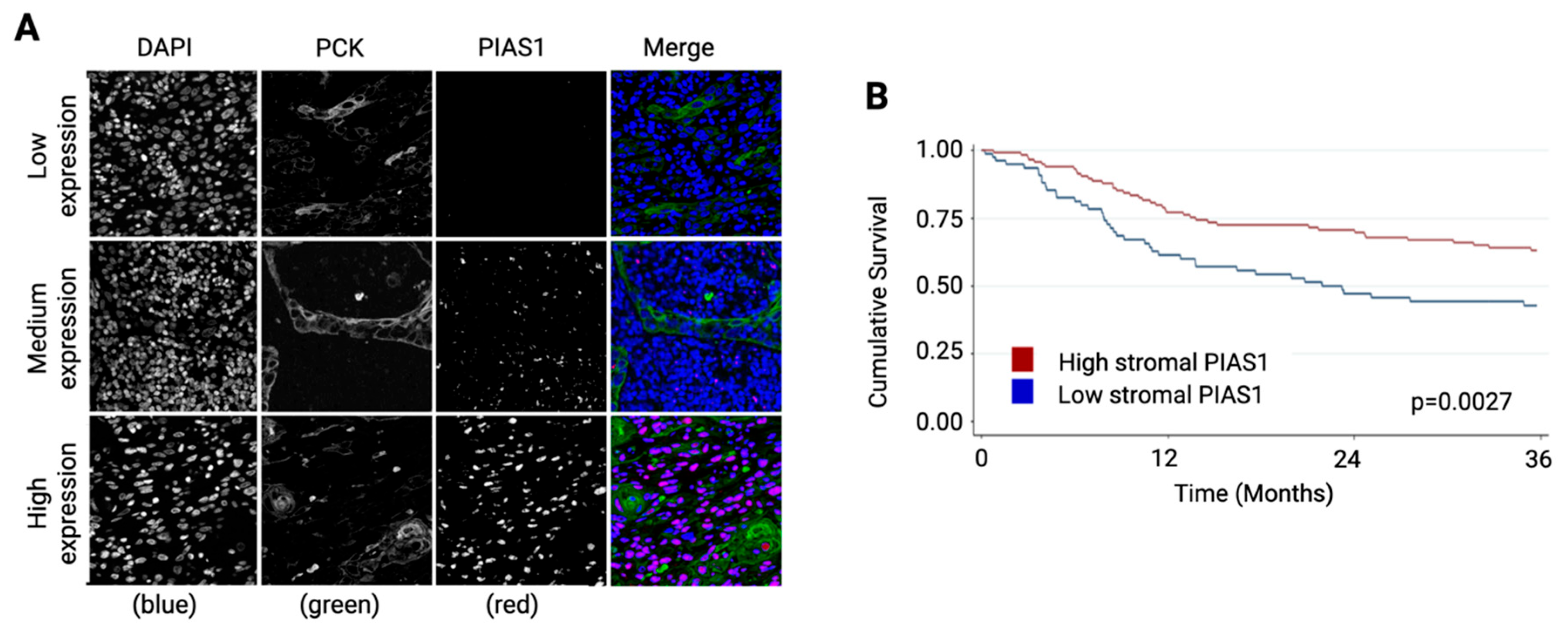
2.2. Fluorescence Immunohistochemistry for PIAS1
2.3. Quantitative Image Analysis
2.4. ScRNAseq Data Acquisition and Sample Selection
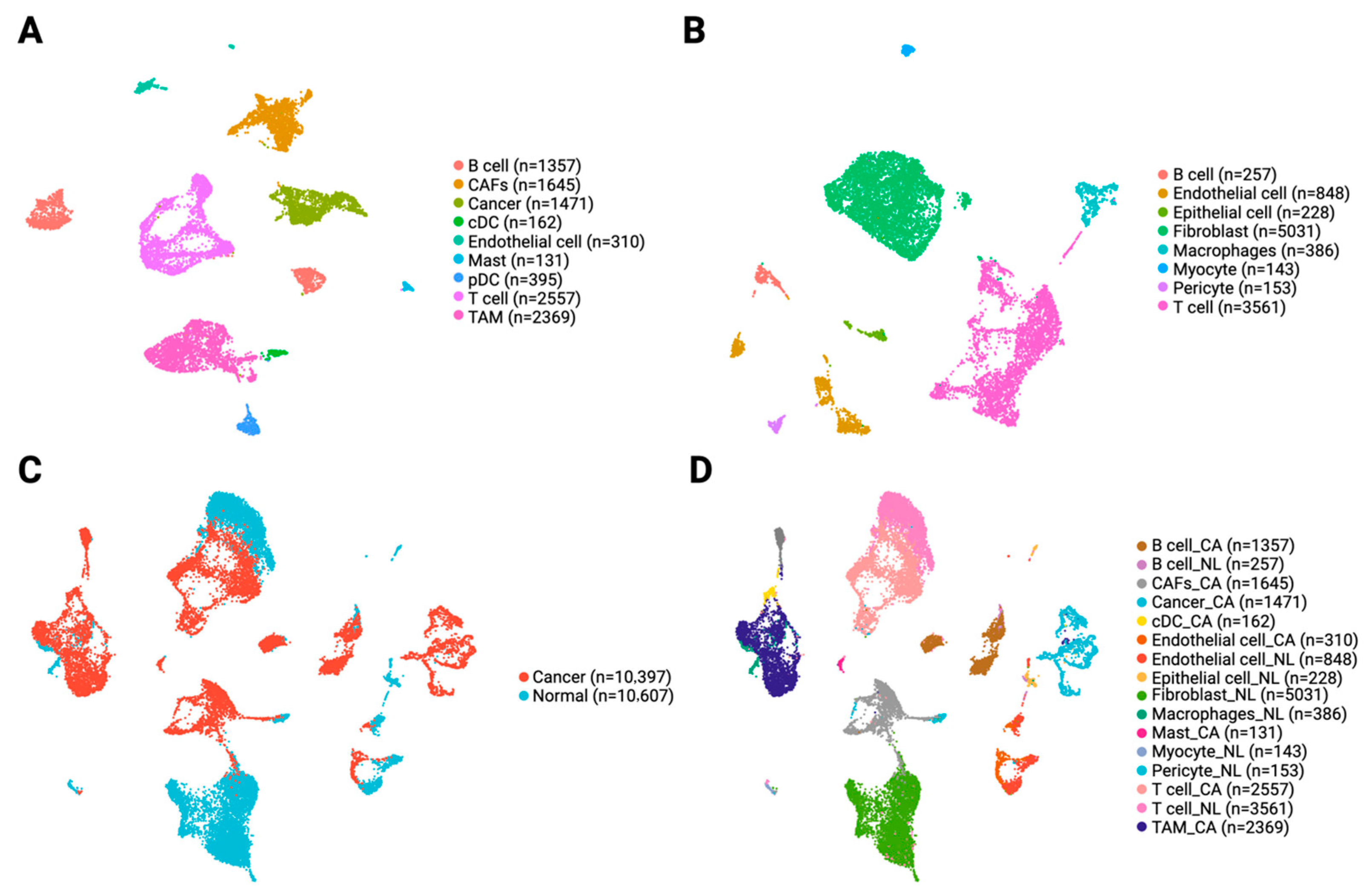
2.5. Quality Control, Data Normalization, Integration, and Clustering
2.6. Differential Expression and Cell Type Annotation
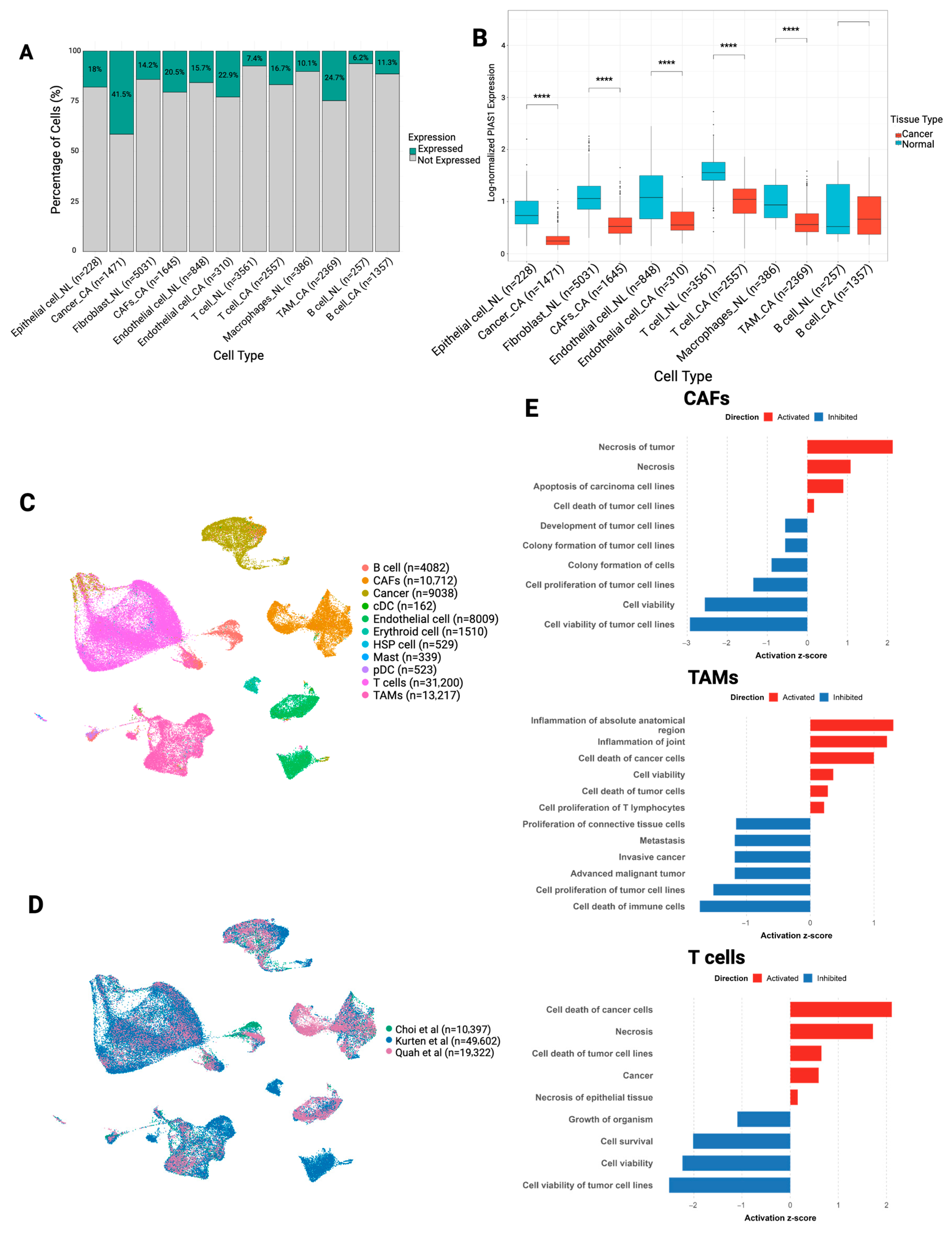
2.7. PIAS1 Expression Analysis Across Cell Types
2.8. Differential Expression Analysis Between PIAS1+ and PIAS1-Cells Across Cell Types
2.9. Ingenuity Pathway Analysis (IPA)
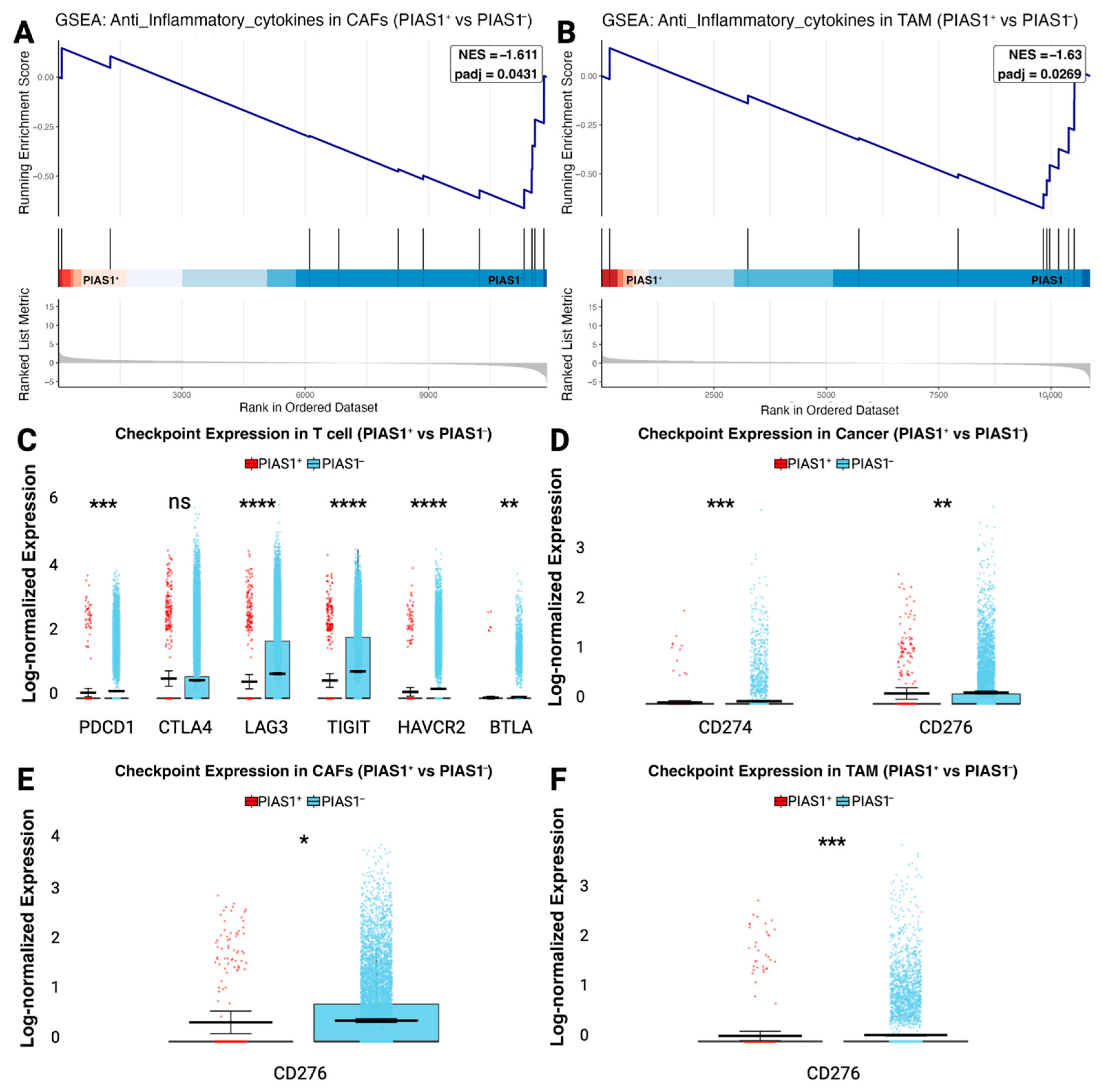
2.10. Cytokine Expression and Immune Checkpoint Analyses Between PIAS1+ and PIAS1-Cells
2.11. Cell–Cell Communication Analyses Between PIAS1+ and PIAS1− Cells
3. Results
3.1. Stromal PIAS1 Expression Correlates with Improved Survival in OSCC
3.2. Single-Cell Profiling Reveals Heterogeneity in Immune and Stromal Cells in the OSCC TME
3.3. PIAS1 Expression Is Broadly Reduced in the OSCC TME and Associated with Tumor-Suppressive Programs
3.4. PIAS1 Limits Immune Escape and Immune Checkpoint Expression
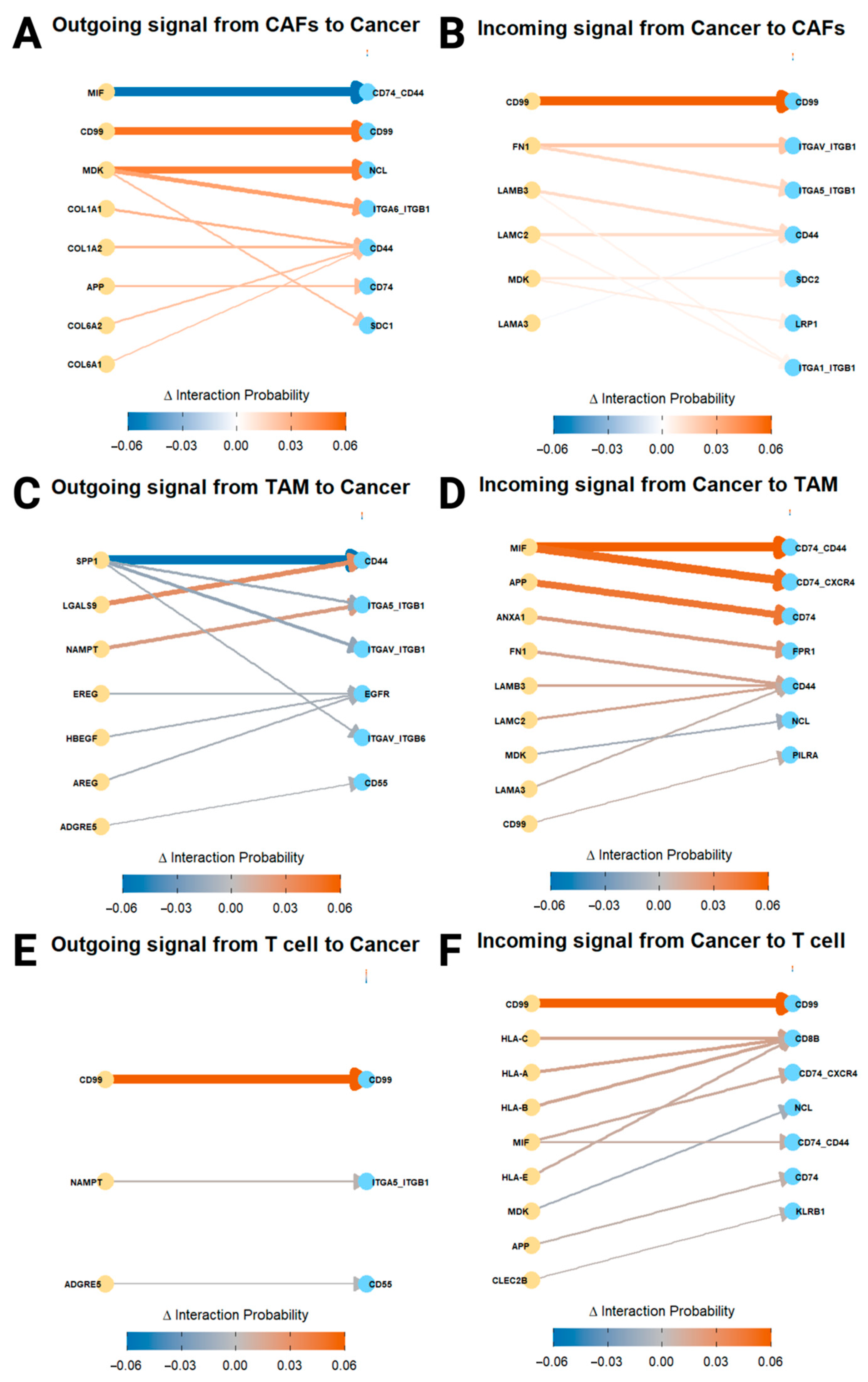
3.5. PIAS1 Modulates Cell–Cell Communication and Supports Anti-Tumor Immunity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CA | Cancer Tissue |
| CAFs | Cancer-Associated Fibroblasts |
| CD | Cluster of Differentiation (used in various CD markers) |
| DEGs | Differentially expressed gene(s) |
| ECM | Extracellular Matrix |
| EMT | Epithelial–Mesenchymal Transition |
| FN1 | Fibronectin 1 |
| GEO | Gene Expression Omnibus |
| GSEA | Gene set enrichment analysis |
| HNC | Head and Neck Cancer |
| HNSCC | Head and neck squamous cell carcinoma |
| HREBA | Health Research Ethics Board of Alberta |
| HRP | Horseradish Peroxidase |
| IHC | Immunohistochemistry |
| IPA | Ingenuity Pathway Analysis |
| ITGA5/ITGB1 | Integrin alpha-5/Integrin beta-1 |
| MIF | Macrophage Migration Inhibitory Factor |
| NL | Normal Tissue |
| OSCC | Oral Squamous Cell Carcinoma |
| PCA | Principal component analysis |
| PIAS1 | Protein Inhibitor of Activated STAT1 |
| REMARK | Reporting Recommendations for Tumor Marker Prognostic Studies |
| RNA-seq | RNA sequencing |
| SC3 | Single-cell Consensus Clustering |
| SNN | Shared Nearest Neighbor |
| SUMO | Small Ubiquitin-Related Modifier |
| TAMs | Tumor-Associated Macrophages |
| TMA | Tissue Microarray |
| TME | Tumor Microenvironment |
| UMAP | Uniform manifold approximation and projection |
| UMI | Unique Molecular Identifier |
| cDCs | Conventional Dendritic Cells |
| pDCs | Plasmacytoid Dendritic Cells |
| scRNA-seq | Single-cell RNA sequencing |
| Δ (Delta) | Change in Interaction Probability |
References
- Bellantoni, M.I.; Picciolo, G.; Pirrotta, I.; Irrera, N.; Vaccaro, M.; Vaccaro, F.; Squadrito, F.; Pallio, G. Oral Cavity Squamous Cell Carcinoma: An Update of the Pharmacological Treatment. Biomedicines 2023, 11, 1112. [Google Scholar] [CrossRef]
- Babu, S.; Manavalan, M.J.; Hifza Jasmine, S.; Krishnan, M. Tumor microenvironment in oral squamous cell carcinoma: Implications for novel therapies. Oral Oncol. Rep. 2024, 12, 100666. Available online: https://www.sciencedirect.com/science/article/pii/S2772906024005120?ref=pdf_download&fr=RR-2&rr=9417137aba7cc4fc (accessed on 27 May 2025). [CrossRef]
- Li, X.; Rasul, A.; Sharif, F.; Hassan, M. PIAS family in cancer: From basic mechanisms to clinical applications. Front. Oncol. 2024, 14, 1376633. [Google Scholar] [CrossRef]
- Puhr, M.; Hoefer, J.; Eigentler, A.; Dietrich, D.; van Leenders, G.; Uhl, B.; Hoogland, M.; Handle, F.; Schlick, B.; Neuwirt, H.; et al. PIAS1 is a determinant of poor survival and acts as a positive feedback regulator of AR signaling through enhanced AR stabilization in prostate cancer. Oncogene 2015, 35, 2322–2332. [Google Scholar] [CrossRef] [PubMed]
- Dadakhujaev, S.; Salazar-Arcila, C.; Netherton, S.J.; Chandhoke, A.S.; Singla, A.K.; Jirik, F.R.; Bonni, S. A novel role for the SUMO E3 ligase PIAS1 in cancer metastasis. Oncoscience 2014, 1, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Haynes, L.; Kumar, M.; McNeil, R.; Ashkani, J.; Nakoneshny, S.C.; Matthews, T.W.; Chandarana, S.; Hart, R.D.; Jones, S.J.M.; et al. NCBP2 and TFRC are novel prognostic biomarkers in oral squamous cell carcinoma. Cancer Gene Ther. 2023, 30, 752–765. [Google Scholar] [CrossRef] [PubMed]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M.; Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. Reporting recommendations for tumor marker prognostic studies (REMARK). J. Natl. Cancer Inst. 2005, 97, 1180–1184. Available online: https://pubmed.ncbi.nlm.nih.gov/16106022/ (accessed on 29 May 2025). [CrossRef] [PubMed]
- Chanda, A.; Chan, A.; Deng, L.; Kornaga, E.N.; Enwere, E.K.; Morris, D.G.; Bonni, S.; Agoulnik, I.U. Identification of the SUMO E3 ligase PIAS1 as a potential survival biomarker in breast cancer. PLoS ONE 2017, 12, e0177639. [Google Scholar] [CrossRef]
- GEO Accession Viewer. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181919 (accessed on 3 June 2025).
- Choi, J.-H.; Lee, B.-S.; Jang, J.Y.; Lee, Y.S.; Kim, H.J.; Roh, J.; Shin, Y.S.; Woo, H.G.; Kim, C.-H. Single-cell transcriptome profiling of the stepwise progression of head and neck cancer. Nat. Commun. 2023, 14, 1055. [Google Scholar] [CrossRef]
- Introduction to scRNA-Seq with R (Seurat)—Getting Started with scRNA-Seq Seminar Series. Available online: https://bioinformatics.ccr.cancer.gov/docs/getting-started-with-scrna-seq/IntroToR_Seurat/ (accessed on 7 August 2025).
- Split and Join Layers Together—JoinLayers SeuratObject. Available online: https://satijalab.github.io/seurat-object/reference/SplitLayers.html (accessed on 7 August 2025).
- Kürten, C.H.L.; Kulkarni, A.; Cillo, A.R.; Santos, P.M.; Roble, A.K.; Onkar, S.; Reeder, C.; Lang, S.; Chen, X.; Duvvuri, U.; et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat. Commun. 2021, 12, 7338. [Google Scholar] [CrossRef]
- Quah, H.S.; Cao, E.Y.; Suteja, L.; Li, C.H.; Leong, H.S.; Chong, F.T.; Gupta, S.; Arcinas, C.; Ouyang, J.F.; Ang, V.; et al. Single cell analysis in head and neck cancer reveals potential immune evasion mechanisms during early metastasis. Nat. Commun. 2023, 14, 1680. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- Analysis, Visualization, and Integration of Visium HD Spatial Datasets with Seurat. Available online: https://satijalab.org/seurat/articles/pbmc3k_tutorial.html (accessed on 3 June 2025).
- Márquez-Jurado, S.; Díaz-Colunga, J.; das Neves, R.P.; Martinez-Lorente, A.; Almazán, F.; Guantes, R.; Iborra, F.J. Mitochondrial levels determine variability in cell death by modulating apoptotic gene expression. Nat. Commun. 2018, 9, e85862. [Google Scholar] [CrossRef] [PubMed]
- Analysis, Visualization, and Integration of Visium HD Spatial Datasets with Seurat Seurat. Available online: https://satijalab.org/seurat/articles/integration_introduction.html (accessed on 3 June 2025).
- Haghverdi, L.; Lun, A.T.L.; Morgan, M.D.; Marioni, J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018, 36, 421–427. [Google Scholar] [CrossRef]
- Elbow Method for Optimal Value of k in KMeans|GeeksforGeeks. Available online: https://www.geeksforgeeks.org/elbow-method-for-optimal-value-of-k-in-kmeans/ (accessed on 3 June 2025).
- Plotting Clustering Trees. Available online: https://cloud.r-project.org/web/packages/clustree/vignettes/clustree.html#sc3-stability-index (accessed on 3 June 2025).
- Sperger, J.M.; Chen, X.; Draper, J.S.; Antosiewicz, J.E.; Chon, C.H.; Jones, S.B.; Brooks, J.D.; Andrews, P.W.; Brown, P.O.; Thomson, J.A. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 13350–13355. [Google Scholar] [CrossRef]
- CellTypist|Automated Cell Type Annotation for scRNA-Seq Datasets. Available online: https://www.celltypist.org/ (accessed on 3 June 2025).
- Normalize Data—NormalizeData Seurat. Available online: https://satijalab.org/seurat/reference/normalizedata (accessed on 3 June 2025).
- Stack Overlapping Objects on Top of Each Another—Position_stack ggplot2. Available online: https://ggplot2.tidyverse.org/reference/position_stack.html (accessed on 7 August 2025).
- A Box and Whiskers Plot (in the Style of Tukey)—Geom_boxplot ggplot2. Available online: https://ggplot2.tidyverse.org/reference/geom_boxplot.html (accessed on 7 August 2025).
- ggplot2 Based Publication Ready Plots ggpubr. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 3 June 2025).
- Gene Expression Markers of Identity Classes—FindMarkers Seurat. Available online: https://satijalab.org/seurat/reference/findmarkers (accessed on 3 June 2025).
- Arora, R.; Cao, C.; Kumar, M.; Sinha, S.; Chanda, A.; McNeil, R.; Samuel, D.; Arora, R.K.; Matthews, T.W.; Chandarana, S.; et al. Spatial transcriptomics reveals distinct and conserved tumor core and edge architectures that predict survival and targeted therapy response. Nat. Commun. 2023, 14, 5029. [Google Scholar] [CrossRef] [PubMed]
- Cartesian Coordinates with x and y Flipped—Coord_Flip Ggplot2. Available online: https://ggplot2.tidyverse.org/reference/coord_flip.html (accessed on 3 June 2025).
- QIAGEN. Undefined. Available online: https://geneglobe.qiagen.com/us/product-groups/qiaseq-targeted-rna-panels-and-indices/RHS-005Z (accessed on 3 June 2025).
- Chapter 15 Visualization of Functional Enrichment Result|Biomedical Knowledge Mining Using GOSemSim and ClusterProfiler. Available online: https://yulab-smu.top/biomedical-knowledge-mining-book/enrichplot.html (accessed on 7 August 2025).
- de Moura, R.G.; Covre, L.P.; Fantecelle, C.H.; Gajardo, V.A.T.; Cunha, C.B.; Stringari, L.L.; Belew, A.T.; Daniel, C.B.; Von Zeidler, S.V.; Tadokoro, C.E.; et al. PD-1 Blockade Modulates Functional Activities of Exhausted-like T Cell in Patients with Cutaneous Leishmaniasis. Front. Immunol. 2021, 12, 632667. [Google Scholar] [CrossRef]
- Hoskins, E.L.; Samorodnitsky, E.; Wing, M.R.; Reeser, J.W.; Hopkins, J.F.; Murugesan, K.; Kuang, Z.; Vella, R.; Stein, L.; Risch, Z.; et al. Pan-cancer Landscape of Programmed Death Ligand-1 and Programmed Death Ligand-2 Structural Variations. JCO Precis. Oncol. 2023, 7, e2200300. [Google Scholar] [CrossRef]
- Getu, A.A.; Tigabu, A.; Zhou, M.; Lu, J.; Fodstad, Ø.; Tan, M. New frontiers in immune checkpoint B7-H3 (CD276) research and drug development. Mol. Cancer 2023, 22, 43. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Zhao, X. B7-H3 in acute myeloid leukemia: From prognostic biomarker to immunotherapeutic target. Chin. Med. J. 2024, 137, 2540–2551. [Google Scholar] [CrossRef]
- Cheng, M.; Chen, S.; Li, K.; Wang, G.; Xiong, G.; Ling, R.; Zhang, C.; Zhang, Z.; Han, H.; Chen, Z.; et al. CD276-dependent efferocytosis by tumor-associated macrophages promotes immune evasion in bladder cancer. Nat. Commun. 2024, 15, 2818. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.-H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- ComputeCommunprob: Compute the Communication Probability/Strength Between Any… in Sqjin/CellChat: Inference and Analysis of Cell-Cell Communication from Single-Cell and Spatial Transcriptomics Data. Available online: https://rdrr.io/github/sqjin/CellChat/man/computeCommunProb.html (accessed on 7 August 2025).
- Nurmik, M.; Ullmann, P.; Rodriguez, F.; Haan, S.; Letellier, E. In search of definitions: Cancer-associated fibroblasts and their markers. Int. J. Cancer 2019, 146, 895–905. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, J.; Liu, H.; Li, A.; Wang, W.; Ni, Z.; Lin, M. MSR1 characterized by chromatin accessibility mediates M2 macrophage polarization to promote gastric cancer progression. Int. Immunopharmacol. 2022, 112, 109217. [Google Scholar] [CrossRef] [PubMed]
- Noe, J.T.; Mitchell, R.A. MIF-Dependent Control of Tumor Immunity. Front. Immunol. 2020, 11, 609949. [Google Scholar] [CrossRef] [PubMed]
- Presti, M.; Mazzon, E.; Basile, M.S.; Petralia, M.C.; Bramanti, A.; Colletti, G.; Bramanti, P.; Nicoletti, F.; Fagone, P. Overexpression of macrophage migration inhibitory factor and functionally-related genes, D-DT, CD74, CD44, CXCR2 and CXCR4, in glioblastoma. Oncol. Lett. 2018, 16, 2881–2886. [Google Scholar] [PubMed]
- Chen, C.; Hou, J.; Yu, S.; Li, W.; Wang, X.; Sun, H.; Qin, T.; Claret, F.X.; Guo, H.; Liu, Z. Role of cancer-associated fibroblasts in the resistance to antitumor therapy, and their potential therapeutic mechanisms in non-small cell lung cancer. Oncol. Lett. 2021, 21, 413. [Google Scholar] [CrossRef]
- Liu, X.-Y.; Liu, Y.-B.; Xu, J.-C.; Zhang, Y.-F.; Ruan, Y.-Y.; Zhao, Y.; Wu, L.-F.; Hu, J.-W.; Zhang, Z.; He, M.-J.; et al. Single-cell transcriptomic analysis deciphers key transitional signatures associated with oncogenic evolution in human intramucosal oesophageal squamous cell carcinoma. Clin. Transl. Med. 2023, 13, e1203. [Google Scholar] [CrossRef]
- Pang, L.; Sun, Q.; Wang, W.; Song, M.; Wu, Y.; Shi, X.; Shi, X. A novel gene signature for predicting outcome in colorectal cancer patients based on tumor cell-endothelial cell interaction via single-cell sequencing and machine learning. Heliyon 2025, 11, e42237. [Google Scholar] [CrossRef]
- Schwarzbauer, J. Basement membrane: Putting up the barriers. Curr. Biol. 1999, 9, R242–R244. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, R.; Deng, J.; Dai, X.; Zhu, X.; Fu, Q.; Zhang, H.; Tong, Z.; Zhao, P.; Fang, W.; et al. Construction of TME and Identification of crosstalk between malignant cells and macrophages by SPP1 in hepatocellular carcinoma. Cancer Immunol. Immunother. 2021, 71, 121–136. [Google Scholar] [CrossRef]
- Su, W.; Ye, Z.; Liu, J.; Deng, K.; Liu, J.; Zhu, H.; Duan, L.; Shi, C.; Wang, L.; Zhao, Y.; et al. Single-cell and spatial transcriptome analyses reveal tumor heterogeneity and immune remodeling involved in pituitary neuroendocrine tumor progression. Nat. Commun. 2025, 16, 5007. [Google Scholar] [CrossRef]
- Lv, Y.; Ma, X.; Ma, Y.; Du, Y.; Feng, J. A new emerging target in cancer immunotherapy: Galectin-9 (LGALS9). Genes Dis. 2023, 10, 2366–2382. [Google Scholar] [CrossRef]
- Nobumoto, A.; Nagahara, K.; Oomizu, S.; Katoh, S.; Nishi, N.; Takeshita, K.; Niki, T.; Tominaga, A.; Yamauchi, A.; Hirashima, M. Galectin-9 suppresses tumor metastasis by blocking adhesion to endothelium and extracellular matrices. Glycobiology 2008, 18, 735–744. [Google Scholar] [CrossRef]
- Xiang, B.; Wang, X.Y.; Liu, K.J. Dual Roles of Nicotinamide Phosphoribosyltransferase as a Promising Target for Cancer Radiotherapy. Radiat. Res. 2021, 196, 429–435. [Google Scholar] [CrossRef]
- Zeiner, P.S.; Preusse, C.; Blank, A.E.; Zachskorn, C.; Baumgarten, P.; Caspary, L.; Braczynski, A.K.; Weissenberger, J.; Bratzke, H.; Reiß, S.; et al. MIF Receptor CD74 is Restricted to Microglia/Macrophages, Associated with a M1-Polarized Immune Milieu and Prolonged Patient Survival in Gliomas. Brain Pathol. 2015, 25, 491–504. [Google Scholar] [CrossRef]
- Ma, C.; Chen, J.; Ji, J.; Zheng, Y.; Liu, Y.; Wang, J.; Chen, T.; Chen, H.; Chen, Z.; Zhou, Q.; et al. Therapeutic modulation of APP-CD74 axis can activate phagocytosis of TAMs in GBM. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2024, 1870, 167449. [Google Scholar] [CrossRef]
- Zhangsun, Z.; Dong, Y.; Tang, J.; Jin, Z.; Lei, W.; Wang, C.; Cheng, Y.; Wang, B.; Yang, Y.; Zhao, H. FPR1: A critical gatekeeper of the heart and brain. Pharmacol. Res. 2024, 202, 107125. [Google Scholar] [CrossRef]
- Manara, M.C.; Fiori, V.; Sparti, A.; Scotlandi, K. CD99: A Key Regulator in Immune Response and Tumor Microenvironment. Biomolecules 2025, 15, 632. [Google Scholar] [CrossRef]
- Cruz-Tapias, P.; Castiblanco, J.; Anaya, J.-M. Major Histocompatibility Complex: Antigen Processing and Presentation. Available online: https://www.ncbi.nlm.nih.gov/books/NBK459467/ (accessed on 3 June 2025).
- Netherton, S.J.; Bonni, S.; Vooijs, M. Suppression of TGFβ-Induced Epithelial-Mesenchymal Transition Like Phenotype by a PIAS1 Regulated Sumoylation Pathway in NMuMG Epithelial Cells. PLoS ONE 2010, 5, e13971. [Google Scholar] [CrossRef]
- Chanda, A.; Ikeuchi, Y.; Karve, K.; Sarkar, A.; Chandhoke, A.S.; Deng, L.; Bonni, A.; Bonni, S. PIAS1 and TIF1γ collaborate to promote SnoN SUMOylation and suppression of epithelial–mesenchymal transition. Cell Death Differ. 2020, 28, 267–282. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, J.; Lan, T.; He, J.; Lei, B.; Wang, H.; Mei, Z.; Lv, C. Integrative analysis revealed a correlation of PIAS family genes expression with prognosis, immunomodulation and chemotherapy. Eur. J. Med. Res. 2024, 29, 195. [Google Scholar] [CrossRef]
- Liu, B.; Tahk, S.; Yee, K.M.; Fan, G.; Shuai, K. The Ligase PIAS1 Restricts Natural Regulatory T Cell Differentiation by Epigenetic Repression. Science 2010, 330, 521–525. [Google Scholar] [CrossRef]
- Zhao, Y.; Shen, M.; Wu, L.; Yang, H.; Yao, Y.; Yang, Q.; Du, J.; Liu, L.; Li, Y.; Bai, Y. Stromal cells in the tumor microenvironment: Accomplices of tumor progression? Cell Death Dis. 2023, 14, 587. Available online: https://pmc.ncbi.nlm.nih.gov/articles/PMC10477351/ (accessed on 27 May 2025). [CrossRef]
- Huang, R.; Kang, T.; Chen, S. The role of tumor-associated macrophages in tumor immune evasion. J. Cancer Res. Clin. Oncol. 2024, 150, 238. [Google Scholar] [CrossRef]
- Ghahremanifard, P.; Chanda, A.; Bonni, S.; Bose, P. TGF-β Mediated Immune Evasion in Cancer—Spotlight on Cancer-Associated Fibroblasts. Cancers 2020, 12, 3650. [Google Scholar] [CrossRef]
- Tosi, A.; Parisatto, B.; Menegaldo, A.; Spinato, G.; Guido, M.; Del Mistro, A.; Bussani, R.; Zanconati, F.; Tofanelli, M.; Tirelli, G.; et al. The immune microenvironment of HPV-positive and HPV-negative oropharyngeal squamous cell carcinoma: A multiparametric quantitative and spatial analysis unveils a rationale to target treatment-naïve tumors with immune checkpoint inhibitors. J. Exp. Clin. Cancer Res. 2022, 41, 279. [Google Scholar] [CrossRef]
- Martínez-Barajas, M.G.; Jave-Suárez, L.F.; Ramírez-López, I.G.; García-Chagollán, M.; Zepeda-Nuño, J.S.; Ramírez-De-Arellano, A.; Ortiz-Lazareno, P.C.; Villegas-Pineda, J.C.; Pereira-Suárez, A.L. HPV-Negative and HPV-Positive Oral Cancer Cells Stimulate the Polarization of Neutrophils towards Different Functional Phenotypes In Vitro. Cancers 2023, 15, 5814. [Google Scholar] [CrossRef]
| Variable | n (%) Unless Otherwise Indicated |
|---|---|
| Number | 175 |
| Sex | |
| Female | 65 (37.1) |
| Male | 110 (62.9) |
| Median age at diagnosis, years (IQR) | 62.5 (53.9–74.8) |
| Pathologic T stage | |
| T1/T2 | 105 (60.0) |
| T3/T4 | 62 (35.4) |
| Missing | 8 (4.6) |
| Pathologic N stage | |
| Node-negative | 63 (36.0) |
| Node-positive | 79 (45.1) |
| Missing | 33 (18.9) |
| Overall Stage | |
| Stage I/II | 61 (34.9) |
| Stage III/IV | 107 (61.1) |
| Missing | 7 (4.0) |
| Dataset Accession | Study Title | Platform | No. of Samples/Cells | Cancer Type |
|---|---|---|---|---|
| GSE181919 | Single-cell transcriptome profiling of the stepwise progression of head and neck cancer | 10X Genomics Chromium | 23/54,239 | HNSCC |
| GSE164690 | Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing | 10X Genomics Chromium | 18/134,606 | HNSCC |
| GSE188737 | Single cell analysis in head and neck cancer reveals potential immune evasion mechanisms during early metastasis | 10X Genomics Chromium | 7/53,459 | HNSCC |
| Patient ID | HPV Status | Subsite | TN Stage |
|---|---|---|---|
| P4 | Negative | Oral cavity | T2N2a |
| P6 | Negative | Oral cavity | T2N1 |
| P7 | Negative | Oral cavity | T2N0 |
| P15 | Negative | Oral cavity | T1N0 |
| P21 | Negative | Oral cavity | T4aN1 |
| P26 | Negative | Oral cavity | T2N0 |
| P30 | Negative | Oral cavity | T2N0 |
| P31 | Negative | Oral cavity | T2N2 |
| P51 | Negative | Oral cavity | T2N1 |
| P60 | Negative | Oral cavity | T4aN0 |
| Patient ID | HPV Status | Subsite | TN Stage |
|---|---|---|---|
| 1 | Negative | Oral cavity | T4aN2b |
| 2 | Negative | Oral cavity | T3N2a |
| 3 | Negative | Oral cavity | T4aN0 |
| 4 | Negative | Oral cavity | T3N1 |
| 5 | Negative | Oral cavity | T3N3b |
| 6 | Negative | Oral cavity | T3N0 |
| 8 | Negative | Oral cavity | T1N0 |
| 9 | Negative | Oral cavity | T3N2b |
| 10 | Negative | Oral cavity | T3N0 |
| 11 | Negative | Oral cavity | T2N0 |
| 15 | Negative | Oral cavity | T2N0 |
| 22 | Negative | Oral cavity | T4aN2c |
| Patient ID | HPV Status | Subsite | TN Stage |
|---|---|---|---|
| HN237 | Negative | Oral cavity | T4aN1 |
| HN242 | Negative | Oral cavity | T3N3b |
| HN251 | Negative | Oral cavity | T3N3b |
| HN257 | Negative | Oral cavity | T4aN3b |
| HN263 | Negative | Oral cavity | T3N2bMx |
| HN272 | Negative | Oral cavity | T4aN2b |
| Cell Type | n Cells | Median | Mean | IQR (25th–75th Percentile) |
|---|---|---|---|---|
| Epithelial cell (NL) | 41 | 0.734 | 0.830 | 0.569–1.010 |
| Cancer (CA) | 610 | 0.244 | 0.279 | 0.171–0.335 |
| Fibroblast (NL) | 712 | 1.060 | 1.100 | 0.853–1.300 |
| CAFs (CA) | 337 | 0.525 | 0.563 | 0.390–0.689 |
| Endothelial cell (NL) | 133 | 1.080 | 1.110 | 0.669–1.500 |
| Endothelial cell (CA) | 71 | 0.552 | 0.630 | 0.451–0.804 |
| T cell (NL) | 263 | 1.560 | 1.570 | 1.410–1.760 |
| T cell (CA) | 428 | 1.040 | 0.993 | 0.774–1.240 |
| Macrophage (NL) | 39 | 0.937 | 1.010 | 0.689–1.320 |
| TAM (CA) | 586 | 0.560 | 0.616 | 0.420–0.770 |
| B cell (NL) | 16 | 0.521 | 0.807 | 0.381–1.330 |
| B cell (CA) | 154 | 0.664 | 0.736 | 0.372–1.100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghahremanifard, P.; An, J.; Chanda, A.; Chan, A.M.Y.; Nakoneshny, S.C.; Matthews, T.W.; Chandarana, S.P.; Hart, R.D.; Hyrcza, M.D.; Dort, J.C.; et al. PIAS1 Shapes a Tumor-Suppressive Microenvironment by Suppressing Immune Evasion in Oral Squamous Cell Carcinoma. Cancers 2025, 17, 2905. https://doi.org/10.3390/cancers17172905
Ghahremanifard P, An J, Chanda A, Chan AMY, Nakoneshny SC, Matthews TW, Chandarana SP, Hart RD, Hyrcza MD, Dort JC, et al. PIAS1 Shapes a Tumor-Suppressive Microenvironment by Suppressing Immune Evasion in Oral Squamous Cell Carcinoma. Cancers. 2025; 17(17):2905. https://doi.org/10.3390/cancers17172905
Chicago/Turabian StyleGhahremanifard, Parisa, Jinsu An, Ayan Chanda, Angela M. Y. Chan, Steven C. Nakoneshny, T. Wayne Matthews, Shamir P. Chandarana, Robert D. Hart, Martin D. Hyrcza, Joseph C. Dort, and et al. 2025. "PIAS1 Shapes a Tumor-Suppressive Microenvironment by Suppressing Immune Evasion in Oral Squamous Cell Carcinoma" Cancers 17, no. 17: 2905. https://doi.org/10.3390/cancers17172905
APA StyleGhahremanifard, P., An, J., Chanda, A., Chan, A. M. Y., Nakoneshny, S. C., Matthews, T. W., Chandarana, S. P., Hart, R. D., Hyrcza, M. D., Dort, J. C., Bonni, S., & Bose, P. (2025). PIAS1 Shapes a Tumor-Suppressive Microenvironment by Suppressing Immune Evasion in Oral Squamous Cell Carcinoma. Cancers, 17(17), 2905. https://doi.org/10.3390/cancers17172905










