Prognostic Value of LC3A Protein Expression Patterns in Rectal Cancer Tumors
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection
2.2. Sample Preparation and Tissue Microarrays
2.3. Immunohistochemistry
2.4. Statistical Analysis
3. Results
3.1. Study Population
3.2. Association Between LC3A Expression Patterns and Patient Clinicopathological Features
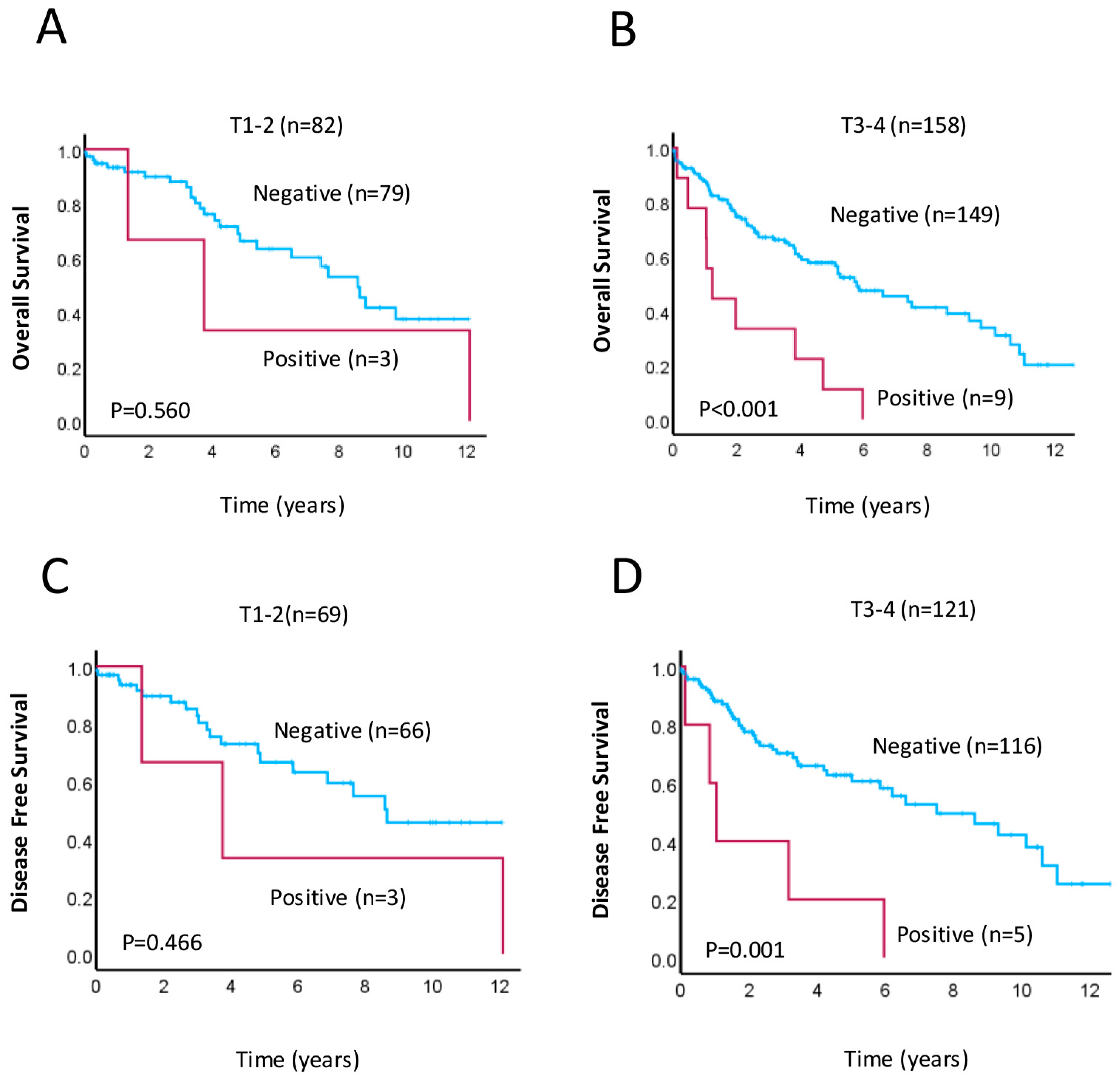
3.3. Prognostic Implications of LC3A Expression in Rectal Cancer
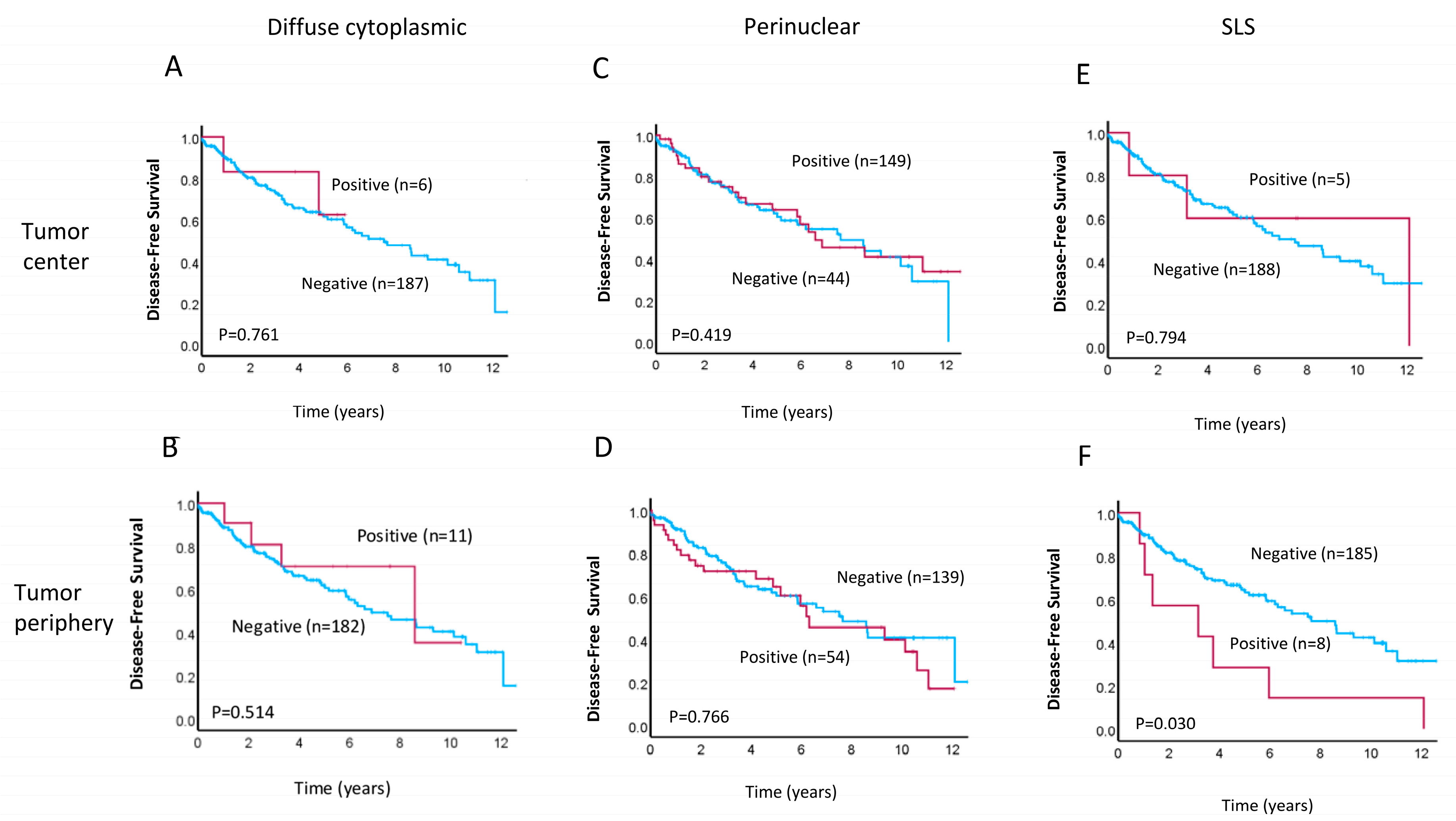
3.4. The SLS Pattern as Putative Prognostic Factor for Aggressive Tumors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CI | Confidence interval |
| CRC | Colorectal cancer |
| DFS | Disease-free survival |
| HR | Hazard ratio |
| LN | Lymph node |
| LVI | Lymphovascular invasion |
| MDPI | Multidisciplinary Digital Publishing Institute |
| OS | Overall survival |
| PNI | Perineural invasion |
| SLS | Stone-like structure |
| TC | Tumor center |
| TMA | Tissue microarray |
| TNM | Tumor-node-metastasis |
| TP | Tumor periphery |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal Cancer Statistics, 2023. CA A Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Lee, Y.-L.; Chuang, J.-P.; Lee, J.-C. Differences in Survival between Colon and Rectal Cancer from SEER Data. PLoS ONE 2013, 8, e78709. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, L.; Wan, J.; Zhang, H.; Wu, R.; Wang, J.; Wang, Y.; Xu, Y.; Cai, S.; Zhang, Z.; et al. Neoadjuvant Chemoradiotherapy Combined with Immunotherapy for Locally Advanced Rectal Cancer: A New Era for Anal Preservation. Front. Immunol. 2022, 13, 1067036. [Google Scholar] [CrossRef]
- Martins, S.; Veiga, P.; Tralhão, J.G.; Carreira, I.M.; Ribeiro, I.P. Rectal Cancer: Exploring Predictive Biomarkers Through Molecular Pathways Involved in Carcinogenesis. Biology 2024, 13, 1007. [Google Scholar] [CrossRef]
- Mondaca, S.; Yaeger, R. Genetics of Rectal Cancer and Novel Therapies; Primer for Radiologists. Abdom Radiol. 2019, 44, 3743–3750. [Google Scholar] [CrossRef]
- Petrişor, O.; Giuşcă, S.E.; Sajin, M.; Dobrescu, G.; Căruntu, I.-D. Ki-67, P53 and Bcl-2 Analysis in Colonic versus Rectal Adenocarcinoma. Rom. J. Morphol. Embryol. 2008, 49, 163–171. [Google Scholar]
- Ho, V.; Chung, L.; Revoltar, M.; Lim, S.H.; Tut, T.-G.; Abubakar, A.; Henderson, C.J.; Chua, W.; Ng, W.; Lee, M.; et al. MRE11 and ATM Expression Levels Predict Rectal Cancer Survival and Their Association with Radiotherapy Response. PLoS ONE 2016, 11, e0167675. [Google Scholar] [CrossRef]
- Dayde, D.; Tanaka, I.; Jain, R.; Tai, M.C.; Taguchi, A. Predictive and Prognostic Molecular Biomarkers for Response to Neoadjuvant Chemoradiation in Rectal Cancer. Int. J. Mol. Sci. 2017, 18, 573. [Google Scholar] [CrossRef]
- Ho, V.; Chung, L.; Singh, A.; Lea, V.; Revoltar, M.; Lim, S.H.; Tut, T.-G.; Ng, W.; Lee, M.; De Souza, P.; et al. Early Postoperative Low Expression of RAD50 in Rectal Cancer Patients Associates with Disease-Free Survival. Cancers 2017, 9, 163. [Google Scholar] [CrossRef]
- Bottarelli, L.; de’ Angelis, G.L.; Azzoni, C.; Mario, F.D.; de’ Angelis, N.; Leandro, G.; Fornaroli, F.; Gaiani, F.; Negri, F. Potential Predictive Biomarkers in Locally Advanced Rectal Cancer Treated with Preoperative Chemo-Radiotherapy. Acta Biomed. 2018, 89, 102–106. [Google Scholar] [CrossRef]
- Ho, V.; Chung, L.; Singh, A.; Lea, V.; Abubakar, A.; Lim, S.H.; Ng, W.; Lee, M.; de Souza, P.; Shin, J.-S.; et al. Overexpression of the MRE11-RAD50-NBS1 (MRN) Complex in Rectal Cancer Correlates with Poor Response to Neoadjuvant Radiotherapy and Prognosis. BMC Cancer 2018, 18, 869. [Google Scholar] [CrossRef]
- Ho, V.; Chung, L.; Singh, A.; Lea, V.; Abubakar, A.; Lim, S.H.; Chua, W.; Ng, W.; Lee, M.; Roberts, T.L.; et al. Aberrant Expression of RAD52, Its Prognostic Impact in Rectal Cancer and Association with Poor Survival of Patients. Int. J. Mol. Sci. 2020, 21, 1768. [Google Scholar] [CrossRef]
- Chou, C.-L.; Lin, C.-Y.; Li, W.-S.; Lee, S.-W.; Yang, C.-C.; Tian, Y.-F.; Shiue, Y.-L.; Tsai, H.-H.; Lai, H.-Y. Low CXCL11 Expression Is Indicative of Poor Prognosis in Rectal Cancer Patients Undergoing Preoperative Chemoradiotherapy: A Retrospective Cohort Study. Virchows Arch. 2025, 486, 803–815. [Google Scholar] [CrossRef]
- Kimmelman, A.C. The Dynamic Nature of Autophagy in Cancer. Genes Dev. 2011, 25, 1999–2010. [Google Scholar] [CrossRef]
- Mowers, E.E.; Sharifi, M.N.; Macleod, K.F. Autophagy in Cancer Metastasis. Oncogene 2017, 36, 1619–1630. [Google Scholar] [CrossRef]
- Li, X.; Zhou, Y.; Li, Y.; Yang, L.; Ma, Y.; Peng, X.; Yang, S.; Liu, J.; Li, H. Autophagy: A Novel Mechanism of Chemoresistance in Cancers. Biomed. Pharmacother. 2019, 119, 109415. [Google Scholar] [CrossRef]
- Chavez-Dominguez, R.; Perez-Medina, M.; Lopez-Gonzalez, J.S.; Galicia-Velasco, M.; Aguilar-Cazares, D. The Double-Edge Sword of Autophagy in Cancer: From Tumor Suppression to Pro-Tumor Activity. Front. Oncol. 2020, 10, 578418. [Google Scholar] [CrossRef]
- Ariosa, A.R.; Lahiri, V.; Lei, Y.; Yang, Y.; Yin, Z.; Zhang, Z.; Klionsky, D.J. A Perspective on the Role of Autophagy in Cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166262. [Google Scholar] [CrossRef]
- Rakesh, R.; PriyaDharshini, L.C.; Sakthivel, K.M.; Rasmi, R.R. Role and Regulation of Autophagy in Cancer. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2022, 1868, 166400. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Emr, S.D. Autophagy as a Regulated Pathway of Cellular Degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Autophagy in the Pathogenesis of Disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Marinković, M.; Šprung, M.; Buljubašić, M.; Novak, I. Autophagy Modulation in Cancer: Current Knowledge on Action and Therapy. Oxid. Med. Cell Longev. 2018, 2018, 8023821. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Yu, W.; Guo, M.; Zhu, N.; Chen, X.; Li, N.; Zhong, C.; Wang, G. Autophagy Regulates Apoptosis of Colorectal Cancer Cells Based on Signaling Pathways. Discov. Oncol. 2024, 15, 367. [Google Scholar] [CrossRef]
- Jin, C.; Wang, T.; Yang, Y.; Zhou, P.; Li, J.; Wu, W.; Lv, X.; Ma, G.; Wang, A. Rational Targeting of Autophagy in Colorectal Cancer Therapy: From Molecular Interactions to Pharmacological Compounds. Environ. Res. 2023, 227, 115721. [Google Scholar] [CrossRef]
- Bodmer, W.F.; Cottrell, S.; Frischauf, A.M.; Kerr, I.B.; Murday, V.A.; Rowan, A.J.; Smith, M.F.; Solomon, E.; Thomas, H.; Varesco, L. Genetic Analysis of Colorectal Cancer. Princess Takamatsu Symp. 1989, 20, 49–59. [Google Scholar]
- Liang, C.; Feng, P.; Ku, B.; Dotan, I.; Canaani, D.; Oh, B.-H.; Jung, J.U. Autophagic and Tumour Suppressor Activity of a Novel Beclin1-Binding Protein UVRAG. Nat. Cell Biol. 2006, 8, 688–699. [Google Scholar] [CrossRef]
- Kang, M.R.; Kim, M.S.; Oh, J.E.; Kim, Y.R.; Song, S.Y.; Kim, S.S.; Ahn, C.H.; Yoo, N.J.; Lee, S.H. Frameshift Mutations of Autophagy-Related Genes ATG2B, ATG5, ATG9B and ATG12 in Gastric and Colorectal Cancers with Microsatellite Instability. J. Pathol. 2009, 217, 702–706. [Google Scholar] [CrossRef]
- Manzoor, S.; Muhammad, J.S.; Maghazachi, A.A.; Hamid, Q. Autophagy: A Versatile Player in the Progression of Colorectal Cancer and Drug Resistance. Front. Oncol. 2022, 12, 924290. [Google Scholar] [CrossRef]
- Ma, T.-F.; Fan, Y.-R.; Zhao, Y.-H.; Liu, B. Emerging Role of Autophagy in Colorectal Cancer: Progress and Prospects for Clinical Intervention. World J. Gastrointest. Oncol. 2023, 15, 979–987. [Google Scholar] [CrossRef]
- Hu, D.; Huo, Y.; Xue, Y.; Feng, H.; Sun, W.; Wang, H.; Wu, J.; Wang, X. Clinical Application of Autophagy Proteins as Prognostic Biomarkers in Colorectal Cancer: A Meta-Analysis. Future Oncol. 2022, 18, 3537–3549. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Wang, L.; Wu, J.; Chen, X.; Hu, F.; Su, Y. Meta-analysis of the Autophagy-associated Protein LC3 as a Prognostic Marker in Colorectal Cancer. Exp. Ther. Med. 2023, 26, 492. [Google Scholar] [CrossRef]
- Li, J.; Yan, Q.; Liu, N.; Zheng, W.; Hu, M.; Yu, Z.; Zhou, Y.; Wang, X.; Liang, F.; Chen, R. The Prognostic Value of Autophagy-Related Markers Bclin-1 and LC-3 in Colorectal Cancers: A Systematic Review and Meta-Analysis. Evid. Based Complement. Alternat Med. 2020, 2020, 8475840. [Google Scholar] [CrossRef]
- Giatromanolaki, A.; Koukourakis, M.I.; Harris, A.L.; Polychronidis, A.; Gatter, K.C.; Sivridis, E. Prognostic Relevance of Light Chain 3 (LC3A) Autophagy Patterns in Colorectal Adenocarcinomas. J. Clin. Pathol. 2010, 63, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Ho, V.; Chung, L.; Wilkinson, K.; Lea, V.; Lim, S.H.; Abubakar, A.; Ng, W.; Lee, M.; Roberts, T.L.; Chua, W.; et al. Prognostic Significance of MRE11 Overexpression in Colorectal Cancer Patients. Cancers 2023, 15, 2438. [Google Scholar] [CrossRef]
- Klarskov, L.; Ladelund, S.; Holck, S.; Roenlund, K.; Lindebjerg, J.; Elebro, J.; Halvarsson, B.; Salomé, J.V.; Bernstein, I.; Nilbert, M. Interobserver variability in the evaluation of mismatch repair protein immunostaining. Hum. Pathol. 2010, 41, 1387–1396. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, L. Autophagy Is a Double-Edged Sword in the Therapy of Colorectal Cancer. Oncol. Lett. 2021, 21, 378. [Google Scholar] [CrossRef] [PubMed]
- de Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef]
- Sivridis, E.; Koukourakis, M.I.; Zois, C.E.; Ledaki, I.; Ferguson, D.J.; Harris, A.L.; Gatter, K.C.; Giatromanolaki, A. LC3A-positive light microscopy detected patterns of autophagy and prognosis in operable breast carcinomas. Am. J. Pathol. 2010, 176, 2477–2489. [Google Scholar] [CrossRef]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and Autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; White, E. Autophagy and Metabolism. Science 2010, 330, 1344–1348. [Google Scholar] [CrossRef] [PubMed]
- Shim, B.Y.; Sun, D.S.; Won, H.S.; Lee, M.A.; Hong, S.U.; Jung, J.-H.; Cho, H.-M.; Ko, Y.H. Role of Autophagy-Related Protein Expression in Patients with Rectal Cancer Treated with Neoadjuvant Chemoradiotherapy. BMC Cancer 2016, 16, 207. [Google Scholar] [CrossRef] [PubMed]
- Niklaus, M.; Adams, O.; Berezowska, S.; Zlobec, I.; Graber, F.; Slotta-Huspenina, J.; Nitsche, U.; Rosenberg, R.; Tschan, M.P.; Langer, R. Expression Analysis of LC3B and P62 Indicates Intact Activated Autophagy Is Associated with an Unfavorable Prognosis in Colon Cancer. Oncotarget 2017, 8, 54604–54615. [Google Scholar] [CrossRef]
- Paschke, S.; Jafarov, S.; Staib, L.; Kreuser, E.D.; Maulbecker-Armstrong, C.; Roitman, M.; Holm, T.; Harris, C.C.; Link, K.H.; Kornmann, M. Are Colon and Rectal Cancer Two Different Tumor Entities? A Proposal to Abandon the Term Colorectal Cancer. Int. J. Mol. Sci. 2018, 19, 2577. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, X.; Wang, X.; Sun, B. Autophagy Enhances the Aggressiveness of Human Colorectal Cancer Cells and Their Ability to Adapt to Apoptotic Stimulus. Cancer Biol. Med. 2012, 9, 105–110. [Google Scholar] [CrossRef]
- Zhang, C.; Zeng, C.; Xiong, S.; Zhao, Z.; Wu, G. A Mitophagy-Related Gene Signature Associated with Prognosis and Immune Microenvironment in Colorectal Cancer. Sci. Rep. 2022, 12, 18688. [Google Scholar] [CrossRef]
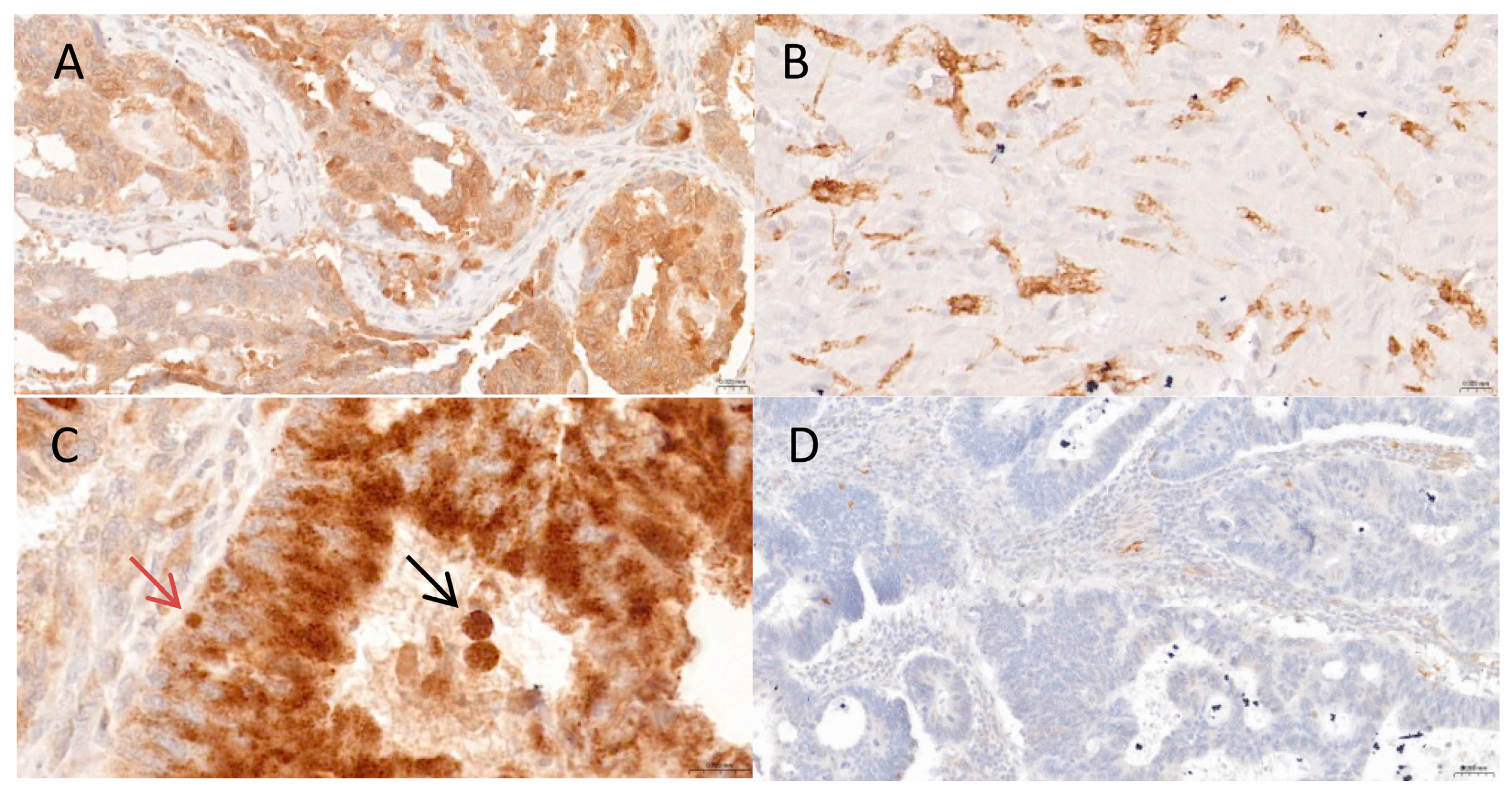

| Characteristics | Patients (n = 243) | % |
| Age median | 72 (35–100 years) | |
| <72 | 113 | 46.5 |
| ≥72 | 130 | 53.5 |
| Sex | ||
| Male | 161 | 66.6 |
| Female | 83 | 33.4 |
| Tumor stage | ||
| T1, T2 | 82 | 34.2 |
| T3, T4 | 158 | 65.8 |
| Node stage | ||
| N0 | 126 | 52.9 |
| N1, N2 | 112 | 47.1 |
| Metastasis stage | ||
| M0 | 215 | 93.5 |
| M1 | 15 | 6.5 |
| Grade | ||
| 1, 2 | 225 | 92.6 |
| 3 | 18 | 7.4 |
| Lymphovascular invasion | ||
| Absent | 185 | 76.8 |
| Present | 56 | 23.2 |
| Perineural invasion | ||
| Absent | 200 | 83 |
| Present | 41 | 17 |
| Treatment | ||
| Neoadjuvant therapy | ||
| No | 186 | 78.5 |
| Yes | 51 | 21.5 |
| Adjuvant therapy | ||
| No | 155 | 69.6 |
| Yes | 66 | 30.4 |
| Diffuse Cytoplasmic | Perinuclear | SLS | ||||||||
| Negative (%) | Positive (%) | p-Value | Negative (%) | Positive (%) | p-Value | Negative (%) | Positive (%) | p-Value | ||
| Sex | Male | 66.7 | 58.3 | 0.552 | 62.9 | 77.2 | 0.046 | 66.8 | 54.5 | 0.401 |
| Female | 33.3 | 41.7 | 37.1 | 22.8 | 33.2 | 45.5 | ||||
| Age | <72 | 45.5 | 66.7 | 0.151 | 48.9 | 38.6 | 0.171 | 45.7 | 63.6 | 0.244 |
| ≥72 | 54.5 | 33.3 | 51.1 | 61.4 | 54.3 | 36.4 | ||||
| Tumor stage | T1–T2 | 33.3 | 34.2 | 0.235 | 34.9 | 31.5 | 0.636 | 33.2 | 45.5 | 0.145 |
| T3–T4 | 66.7 | 65.8 | 65.1 | 68.5 | 66.8 | 54.5 | ||||
| Node stage | Negative | 51.8 | 75 | 0.116 | 56.8 | 39.6 | 0.028 | 52.0 | 72.7 | 0.178 |
| Positive | 48.2 | 25 | 43.2 | 60.4 | 48.0 | 27.3 | ||||
| Metastasis stage | M0 | 93.6 | 90 | 0.629 | 93.2 | 94.4 | 0.742 | 93.7 | 88.9 | 0.569 |
| M1 | 6.4 | 10 | 6.8 | 5.6 | 6.3 | 11.1 | ||||
| Grade | 1, 2 | 92.2 | 100 | 0.315 | 94.1 | 87.7 | 0.108 | 92.2 | 100 | 0.337 |
| 3 | 7.8 | 0 | 5.9 | 12.3 | 7.8 | 0 | ||||
| Lymphovascular invasion | Absent | 76 | 91.7 | 0.21 | 77.4 | 74.5 | 0.658 | 76.1 | 90.9 | 0.255 |
| Present | 24 | 8.3 | 22.6 | 25.5 | 23.9 | 9.1 | ||||
| Perineural invasion | Absent | 82.5 | 91.7 | 0.412 | 83.9 | 80 | 0.502 | 82.6 | 90.9 | 0.474 |
| Present | 17.5 | 8.3 | 16.1 | 20 | 17.4 | 9.1 | ||||
| Adjuvant therapy | No | 68.9 | 81.8 | 0.365 | 70.6 | 66 | 0.541 | 69.1 | 80 | 0.464 |
| Yes | 31.1 | 18.2 | 29.4 | 34 | 30.9 | 20 | ||||
| Neoadjuvant therapy | No | 79.1 | 66.7 | 0.307 | 78.9 | 77.2 | 0.786 | 79.2 | 63.6 | 0.22 |
| Yes | 20.9 | 33.3 | 21.1 | 22.8 | 20.8 | 36.4 | ||||
| Diffuse Cytoplasmic | Perinuclear | SLS | ||||||||
| Negative (%) | Positive (%) | p-Value | Negative (%) | Positive (%) | p-Value | Negative (%) | Positive (%) | p-Value | ||
| Sex | Male | 66.8 | 57.1 | 0.458 | 66.3 | 66.2 | 0.987 | 66.7 | 58.3 | 0.552 |
| Female | 33.2 | 42.9 | 33.7 | 33.8 | 33.3 | 41.7 | ||||
| Age | <72 | 46.7 | 42.9 | 0.771 | 45.1 | 50 | 0.496 | 45.7 | 41.7 | 0.731 |
| ≥72 | 53.3 | 57.1 | 54.9 | 50 | 54.3 | 58.3 | ||||
| Tumor stage | T1–T2 | 32.6 | 61.5 | 0.023 | 36.4 | 28.4 | 0.238 | 34.5 | 25 | 0.492 |
| T3–T4 | 67.4 | 38.5 | 63.6 | 71.6 | 65.5 | 75 | ||||
| Node stage | Negative | 51.6 | 76.9 | 0.075 | 55.2 | 47 | 0.253 | 54.0 | 33.6 | 0.163 |
| Positive | 48.2 | 23.1 | 44.8 | 53 | 46.0 | 66.4 | ||||
| Metastasis stage | M0 | 93.1 | 100 | 0.347 | 92.2 | 96.9 | 0.195 | 94.0 | 83.3 | 0.144 |
| M1 | 6.9 | 0 | 7.8 | 3.1 | 6.0 | 16.7 | ||||
| Grade | 1, 2 | 92.1 | 100 | 0.276 | 93.1 | 91.2 | 0.599 | 93.9 | 66.7 | <0.001 |
| 3 | 7.9 | 0 | 6.9 | 8.8 | 6.1 | 33.3 | ||||
| Lymphovascular invasion | Absent | 76.3 | 84.7 | 0.491 | 77.5 | 75 | 0.684 | 72.8 | 50 | 0.024 |
| Present | 23.7 | 15.3 | 22.5 | 25 | 7.2 | 50 | ||||
| Perineural invasion | Absent | 82.5 | 92.3 | 0.358 | 83.2 | 82.4 | 0.869 | 83.8 | 66.7 | 0.123 |
| Present | 17.5 | 7.7 | 16.8 | 17.6 | 16.2 | 33.3 | ||||
| Adjuvant therapy | No | 69.5 | 71.4 | 0.877 | 72.2 | 62.7 | 0.179 | 70.3 | 50 | 0.22 |
| Yes | 30.5 | 28.6 | 27.8 | 37.3 | 29.7 | 50 | ||||
| Neoadjuvant therapy | No | 77.6 | 92.9 | 0.177 | 82.8 | 67.6 | 0.161 | 77.9 | 90.1 | 0.304 |
| Yes | 22.4 | 7.1 | 17.2 | 32.4 | 22.1 | 9.1 | ||||
| Univariate | Multivariate | ||||||
| n (%) | HR | 95% CI | p-Value | HR | 95% | p-Value | |
| SLS TP * | |||||||
| Positive | 4.9 | 2.712 | 1.470–5.009 | 0.001 | 2.6313 | 1.090–6.349 | 0.031 |
| Negative | 95.1 | ||||||
| Sex | |||||||
| Male | 66.6 | 1.03 | 0.693–1.530 | 0.885 | 1.127 | 0.697–1.822 | 0.685 |
| Female | 33.4 | ||||||
| Age | |||||||
| <72 | 46.5 | 1.416 | 0.954–2.101 | 0.084 | 1.271 | 0.770–2.100 | 0.349 |
| ≥72 | 53.5 | ||||||
| Tumor stage | |||||||
| T1, T2 | 34.2 | 1.426 | 1.047–2.460 | 0.03 | 1.426 | 0.842–2.415 | 0.187 |
| T3, T4 | 65.8 | ||||||
| Node stage | |||||||
| Negative | 52.9 | 1.405 | 0.956–2.064 | 0.083 | 1.489 | 0.891–2.489 | 0.128 |
| Positive | 47.1 | ||||||
| Metastasis stage | |||||||
| M0 | 93.5 | 5.102 | 2.699–9.643 | <0.001 | 2.914 | 1.103–7.696 | 0.031 |
| M1 | 6.5 | ||||||
| Grade | |||||||
| 1, 2 | 92.6 | 1.719 | 0.942–3.135 | 0.077 | 1.343 | 0.252–1.763 | 0.414 |
| 3 | 7.4 | ||||||
| Lymphovascular invasion | |||||||
| Absent | 76.8 | 2.011 | 1.333–3.033 | 0.001 | 1.688 | 0.919–3.101 | 0.092 |
| Present | 23.2 | ||||||
| Perineural invasion | |||||||
| Absent | 83 | 2.478 | 1.602–3.833 | <0.001 | 1.997 | 1.035–3.893 | 0.042 |
| Present | 17 | ||||||
| Adjuvant therapy | |||||||
| No | 78.5 | 0.567 | 0.344–0.934 | 0.026 | 0.306 | 0.163–0.577 | 0.065 |
| Yes | 21.5 | ||||||
| Neoadjuvant therapy | |||||||
| No | 69.6 | 0.984 | 0.614–1.576 | 0.945 | 1.254 | 0.672–2.340 | 0.476 |
| Yes | 30.4 | ||||||
| T1–T2 SLS | 51.2 | 1.529 | 0.361–6.472 | 0.564 | |||
| T3–T4 SLS | 48.8 | 3.347 | 1.657–6.760 | 0.001 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ho, V.; Chung, L.; Rutland, T.; Lea, V.; Lim, S.H.; Abubakar, A.; Ng, W.; Lee, M.; Roberts, T.L.; Chua, W.; et al. Prognostic Value of LC3A Protein Expression Patterns in Rectal Cancer Tumors. Cancers 2025, 17, 1568. https://doi.org/10.3390/cancers17091568
Ho V, Chung L, Rutland T, Lea V, Lim SH, Abubakar A, Ng W, Lee M, Roberts TL, Chua W, et al. Prognostic Value of LC3A Protein Expression Patterns in Rectal Cancer Tumors. Cancers. 2025; 17(9):1568. https://doi.org/10.3390/cancers17091568
Chicago/Turabian StyleHo, Vincent, Liping Chung, Tristan Rutland, Vivienne Lea, Stephanie H. Lim, Askar Abubakar, Weng Ng, Mark Lee, Tara L. Roberts, Wei Chua, and et al. 2025. "Prognostic Value of LC3A Protein Expression Patterns in Rectal Cancer Tumors" Cancers 17, no. 9: 1568. https://doi.org/10.3390/cancers17091568
APA StyleHo, V., Chung, L., Rutland, T., Lea, V., Lim, S. H., Abubakar, A., Ng, W., Lee, M., Roberts, T. L., Chua, W., Mackenzie, S., & Lee, C. S. (2025). Prognostic Value of LC3A Protein Expression Patterns in Rectal Cancer Tumors. Cancers, 17(9), 1568. https://doi.org/10.3390/cancers17091568








