Elevated HDAC4 Expression Is Associated with Reduced T-Cell Inflamed Tumor Microenvironment Gene Signatures and Immune Checkpoint Inhibitor Effectiveness in Melanoma †
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Co-Expression Study Between the HDAC4 Gene and the T-Cell Inflamed TME Signature Genes
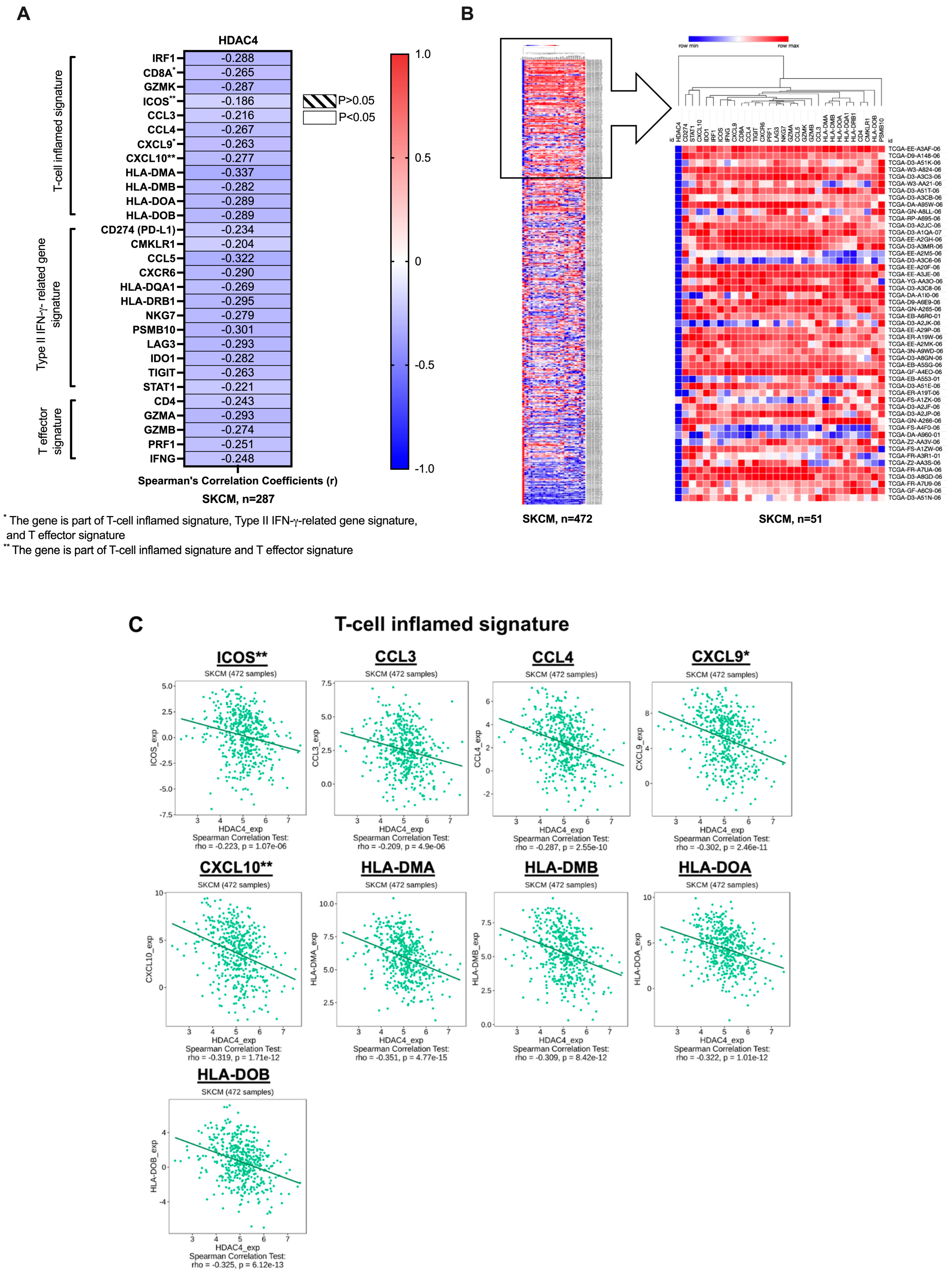
- T-cell inflamed signature: interferon regulatory factor 1 (IRF1), CD8A, granzyme K (GZMK), inducible T-cell co-stimulator (ICOS), C-C motif chemokine ligands (CCL3, CCL4), C-X-C motif chemokine ligands (CXCL9, CXCL10), and major histocompatibility complex, class II (HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB) [38,39].
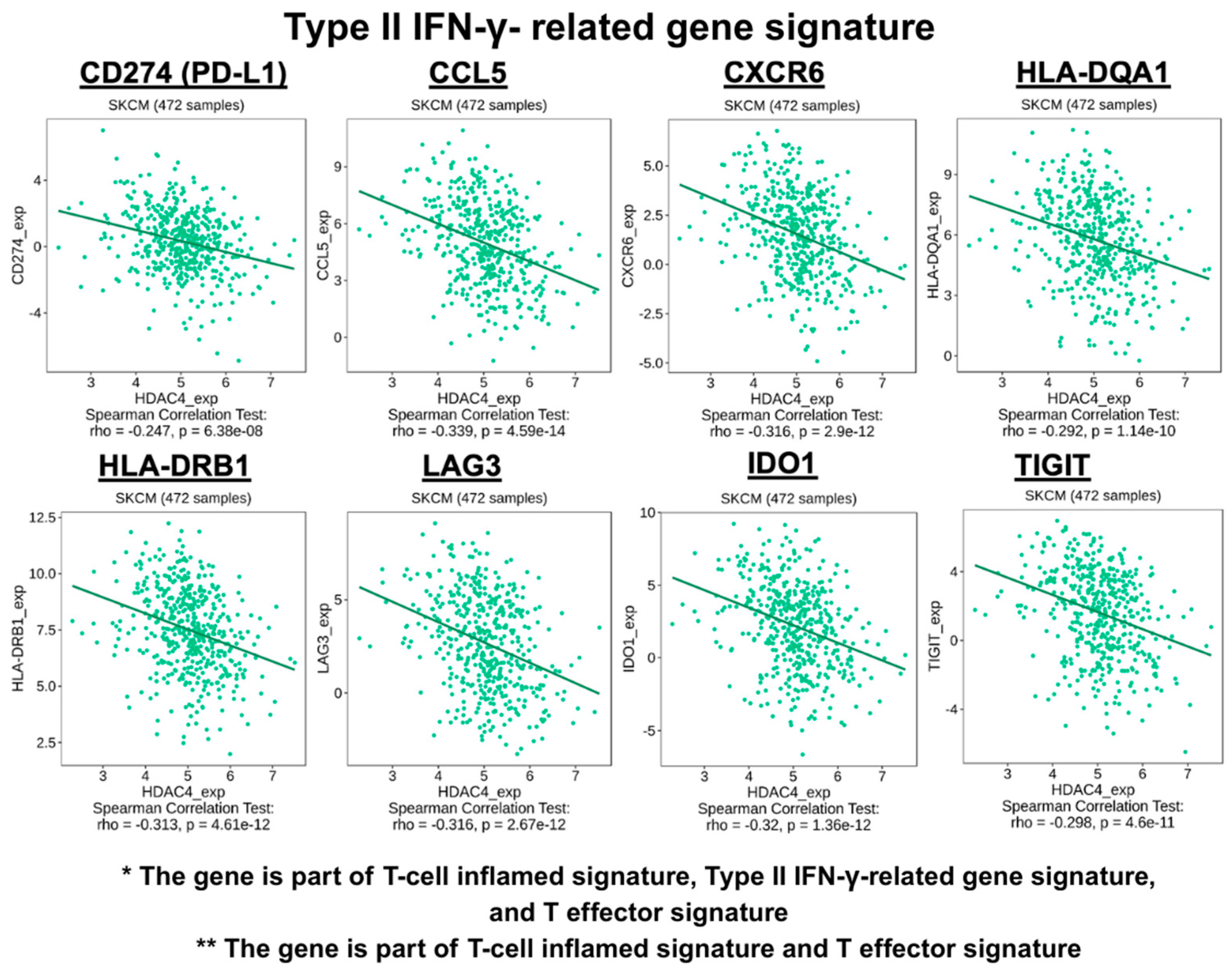
- Type II IFN-γ-related gene signature: CD8A, PD-L1 (also known as CD274), chemerin chemokine-like receptor 1 (CMKLR1), CCL5, CXCL9, C-X-C motif chemokine receptor 6 (CXCR6), HLA-DQA1, HLA-DRB1, natural killer cell granule protein 7 (NKG7), proteasome 20S subunit, beta type, 10 (PSMB10), lymphocyte activation gene 3 (LAG3), indoleamine 2,3-dioxygenase 1 (IDO1), T-cell immunoreceptor with Ig and ITIM domains (TIGIT), and signal transducer and activator of transcription 1 (STAT1) [38,40].
2.2. Immune Cell Association Study
2.3. The Transcription Analysis and the DNA Methylation Analysis of T-Cell Inflamed TME Signature Genes with Regard to HDAC4 Expression
2.4. Survival Prognosis Analysis
2.5. The Effect of HDAC4 Expression in ICI-Pretreated Melanoma Patients
2.6. Statistical Analysis
3. Results
3.1. The Co-Expression Profile of HDAC4 and T-Cell Inflamed TME Gene Signatures
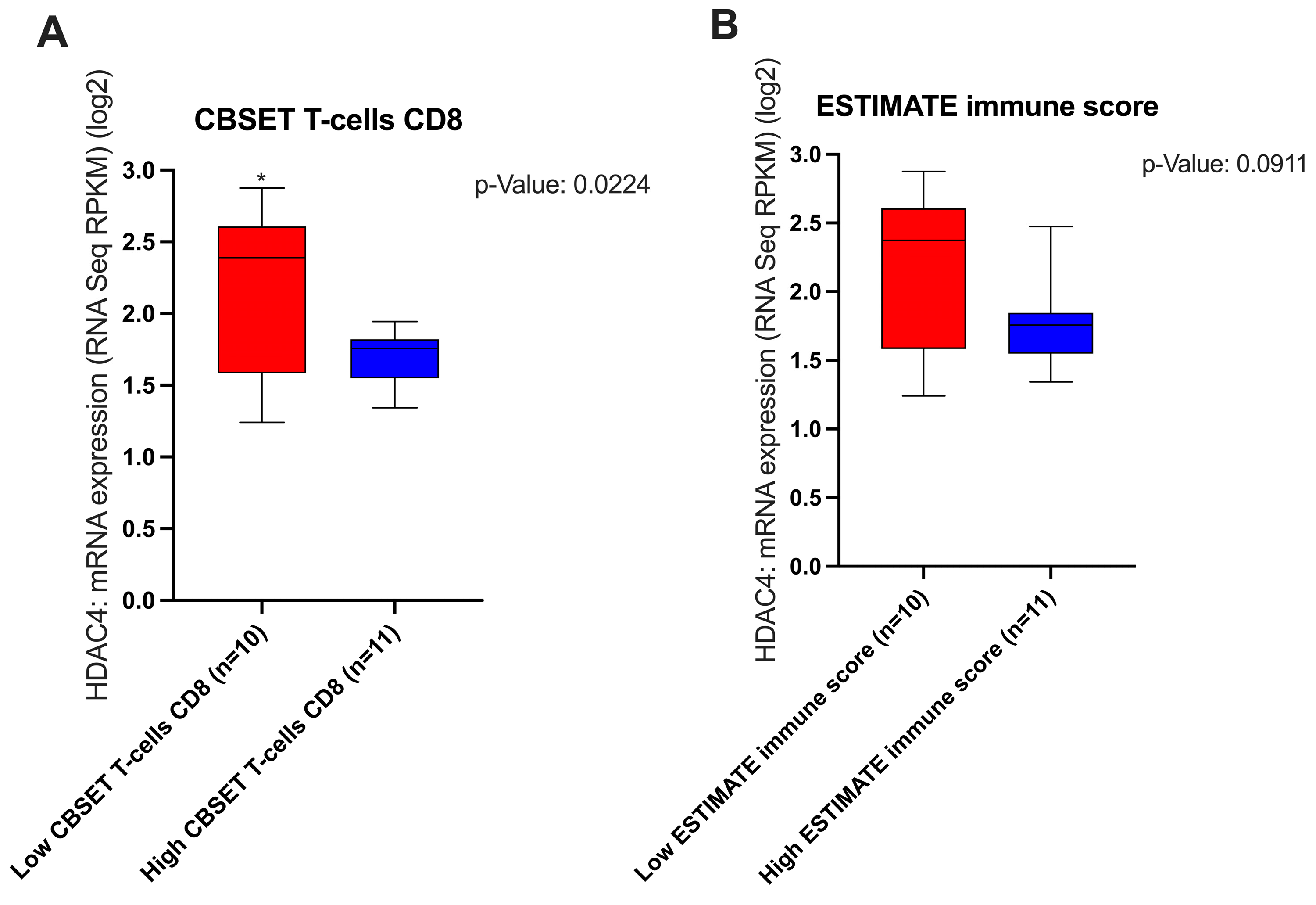
3.2. The Effect of HDAC4 Expression and T-Cell Inflamed TME Genes Signature Expression on the TME
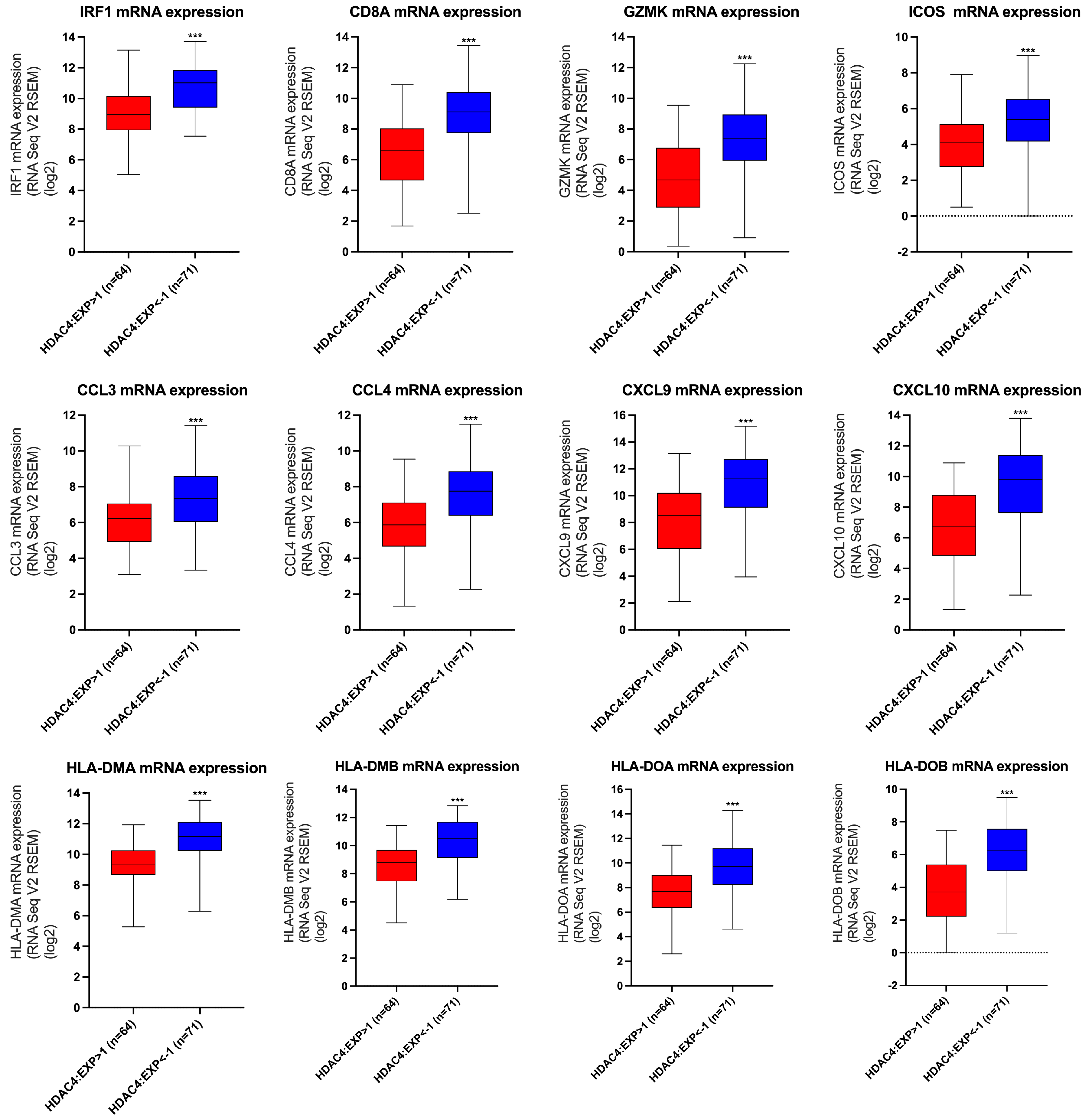
3.3. The Effect of HDAC4 Expression on the Transcription of T-Cell Inflamed TME Gene Signatures
3.4. The Effect of HDAC4 Expression on the DNA Methylation of T-Cell Inflamed TME Gene Signatures
3.5. Prognostic Values of HDAC4 and T-Cell Inflamed TME Gene Signatures
3.6. The Effect of HDAC4 in the ICI-Pretreated Melanoma Dataset
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CCL | C-C Motif Chemokine Ligand |
| CD | Cluster of Differentiation |
| CMKLR | Chemerin Chemokine-like Receptor 1 |
| CTLs | Cytotoxic T-cells |
| CTLA4 | Cytotoxic T-Lymphocyte-Associated Protein 4 |
| CXCL | C-X-C Motif Chemokine Ligand |
| CXCR | C-X-C Motif Chemokine Receptor |
| DCs | Dendritic Cells |
| DFS | Disease-Free Survival |
| GZM | Granzyme |
| HDACs | Histone Deacetylases |
| HLA | Major Histocompatibility Complex, class II |
| IC | Immune Checkpoint |
| ICIs | Immune Checkpoint Inhibitors |
| ICOS | Inducible T-Cell Co-stimulator |
| IDO1 | Indoleamine 2,3-dioxygenase |
| IFN | Interferon |
| IRF1 | IFN Regulatory Factor 1 |
| LAG3 | Lymphocyte Activation Gene 3 |
| mDCs | Myeloid Dendritic Cells |
| MDSCs | Myeloid-Derived Suppressor Cells |
| NK | Natural Killer Cells |
| NKG7 | Natural Killer Cell Granule Protein 7 |
| OS | Overall Survival |
| pDC | Plasmacytoid Dendritic Cells |
| PD-L1 | Programmed Cell Death Ligand 1 |
| PD-1 | Programmed Cell Death Protein 1 |
| PRF1 | Perforin 1 |
| PSMB10 | Proteasome 20S Subunit, Beta type, 10 |
| SKCM | Skin Cutaneous Melanoma |
| STAT1 | Signal Transducers and Activators of Transcription 1 |
| TCGA | The Cancer Genome Atlas |
| TH | T-helper Cells |
| Treg | Regulatory T-cells |
| TIGIT | T-Cell Immunoreceptor with Ig and ITIM Domains |
| TILs | Tumor-Infiltrating Lymphocytes |
| TIM-3 | T-cell Immunoglobulin and Mucin-domain containing-3 |
| TLTs | Tertiary Lymphoid Tissues |
| TMB | Tumor Mutational Burden |
| TME | Tumor Microenvironment |
References
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Carr, S.; Smith, C.; Wernberg, J. Epidemiology and Risk Factors of Melanoma. Surg. Clin. N. Am. 2019, 100, 1–12. [Google Scholar] [CrossRef]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current state of melanoma diagnosis and treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef]
- Tímár, J.; Ladányi, A. Molecular Pathology of Skin Melanoma: Epidemiology, Differential Diagnostics, Prognosis and Therapy Prediction. Int. J. Mol. Sci. 2022, 23, 5384. [Google Scholar] [CrossRef]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghì, A.; Spandidos, D.A.; Mccubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy. Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef]
- Sample, A.; He, Y.-Y. Mechanisms and prevention of UV-induced melanoma. Photodermatol. Photoimmunol. Photomed. 2017, 34, 13–24. [Google Scholar] [CrossRef]
- Watson, M.; Holman, D.M.; Maguire-Eisen, M. Ultraviolet Radiation Exposure and Its Impact on Skin Cancer Risk. Semin. Oncol. Nurs. 2016, 32, 241–254. [Google Scholar] [CrossRef]
- O’Neill, C.H.; Scoggins, C.R. Melanoma. J. Surg. Oncol. 2019, 120, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Donahue, L.R.; Choi, E.; Scumpia, P.O.; Lowry, W.E.; Grenier, J.K.; Zhu, J.; White, A.C. Melanocyte Stem Cell Activation and Translocation Initiate Cutaneous Melanoma in Response to UV Exposure. Cell Stem Cell 2017, 21, 665–678.e6. [Google Scholar] [CrossRef]
- Davis, E.J.; Johnson, D.B.; Sosman, J.A.; Chandra, S. Melanoma: What do all the mutations mean? Cancer 2018, 124, 3490–3499. [Google Scholar] [CrossRef]
- Tanda, E.T.; Vanni, I.; Boutros, A.; Andreotti, V.; Bruno, W.; Ghiorzo, P.; Spagnolo, F. Current State of Target Treatment in BRAF Mutated Melanoma. Front. Mol. Biosci. 2020, 7, 154. [Google Scholar] [CrossRef]
- Heppt, M.V.; Siepmann, T.; Engel, J.; Schubert-Fritschle, G.; Eckel, R.; Mirlach, L.; Kirchner, T.; Jung, A.; Gesierich, A.; Ruzicka, T.; et al. Prognostic significance of BRAF and NRAS mutations in melanoma: A German study from routine care. BMC Cancer 2017, 17, 536. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.Y.; Miller, D.; Tsao, H. Somatic driver mutations in melanoma. Cancer 2017, 123, 2104–2117. [Google Scholar] [CrossRef] [PubMed]
- Ponti, G.; Manfredini, M.; Greco, S.; Pellacani, G.; Depenni, R.; Tomasi, A.; Maccaferri, M.; Cascinu, S. BRAF, NRAS and C-KIT Advanced Melanoma: Clinico-pathological Features, Targeted-Therapy Strategies and Survival. Anticancer Res. 2017, 37, 7043–7048. [Google Scholar] [CrossRef]
- Richtig, G.; Hoeller, C.; Kashofer, K.; Aigelsreiter, A.; Heinemann, A.; Kwong, L.; Pichler, M. Beyond the BRAF V 600E hotspot: Biology and clinical implications of rare BRAF gene mutations in melanoma patients. Br. J. Dermatol. 2017, 177, 936–944. [Google Scholar] [CrossRef]
- Cheng, L.; Lopez-Beltran, A.; Massari, F.; Maclennan, G.T.; Montironi, R. Molecular testing for BRAF mutations to inform melanoma treatment decisions: A move toward precision medicine. Mod. Pathol. 2017, 31, 24–38. [Google Scholar] [CrossRef]
- Kozar, I.; Margue, C.; Rothengatter, S.; Haan, C.; Kreis, S. Many ways to resistance: How melanoma cells evade targeted therapies. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 313–322. [Google Scholar] [CrossRef]
- Mackiewicz, J.; Mackiewicz, A. BRAF and MEK inhibitors in the era of immunotherapy in melanoma patients. Contemp. Oncol. 2018, 22, 68–72. [Google Scholar] [CrossRef]
- Ribas, A.; Gonzalez, R.; Pavlick, A.; Hamid, O.; Gajewski, T.F.; Daud, A.; Flaherty, L.; Logan, T.; Chmielowski, B.; Lewis, K.; et al. Combination of vemurafenib and cobimetinib in patients with advanced BRAFV600-mutated melanoma: A phase 1b study. Lancet Oncol. 2014, 15, 954–965. [Google Scholar] [CrossRef]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Combined BRAF and MEK Inhibition versus BRAF Inhibition Alone in Melanoma. N. Engl. J. Med. 2014, 371, 1877–1888. [Google Scholar] [CrossRef]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Hiam-Galvez, K.J.; Allen, B.M.; Spitzer, M.H. Systemic immunity in cancer. Nat. Rev. Cancer 2021, 21, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lu, M.; Qin, Y.; Gao, W.; Tao, L.; Su, W.; Zhong, J. Neoantigen: A New Breakthrough in Tumor Immunotherapy. Front. Immunol. 2021, 12, 672356. [Google Scholar] [CrossRef]
- Demaria, O.; Cornen, S.; Daëron, M.; Morel, Y.; Medzhitov, R.; Vivier, E. Harnessing innate immunity in cancer therapy. Nature 2019, 574, 45–56. [Google Scholar] [CrossRef]
- Li, X.; Dai, H.; Wang, H.; Han, W. Exploring innate immunity in cancer immunotherapy: Opportunities and challenges. Cell. Mol. Immunol. 2021, 18, 1607–1609. [Google Scholar] [CrossRef]
- Jin, M.Z.; Jin, W.L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 2020, 5, 166. [Google Scholar] [CrossRef]
- Salemme, V.; Centonze, G.; Cavallo, F.; Defilippi, P.; Conti, L. The Crosstalk Between Tumor Cells and the Immune Microenvironment in Breast Cancer: Implications for Immunotherapy. Front. Oncol. 2021, 11, 610303. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, M.; Nie, H.; Yuan, Y. PD-1 and PD-L1 in cancer immunotherapy: Clinical implications and future considerations. Hum. Vaccines Immunother. 2019, 15, 1111–1122. [Google Scholar] [CrossRef]
- Ai, L.; Chen, J.; Yan, H.; He, Q.; Luo, P.; Xu, Z.; Yang, X. Research status and outlook of PD-1/PD-L1 inhibitors for cancer therapy. Drug Des. Dev. Ther. 2020, 14, 3625–3649. [Google Scholar] [CrossRef]
- Seth, R.; Messersmith, H.; Kaur, V.; Kirkwood, J.M.; Kudchadkar, R.; McQuade, J.L.; Provenzano, A.; Swami, U.; Weber, J.; Alluri, K.C.; et al. Systemic Therapy for Melanoma: ASCO Guideline. J. Clin. Oncol. 2020, 38, 3947–3970. [Google Scholar] [CrossRef] [PubMed]
- Pavlick, A.C.; Zhao, R.; Lee, C.-H.; Ritchings, C.; Rao, S. First-line immunotherapy versus targeted therapy in patients with BRAF-mutant advanced melanoma: A real-world analysis. Futur. Oncol. 2021, 17, 689–699. [Google Scholar] [CrossRef]
- van Breeschoten, J.; Wouters, M.W.J.M.; Hilarius, D.L.; Haanen, J.B.; Blank, C.U.; Aarts, M.J.B.; Berkmortel, F.W.P.J.V.D.; de Groot, J.-W.B.; Hospers, G.A.P.; Kapiteijn, E.; et al. First-line BRAF/MEK inhibitors versus anti-PD-1 monotherapy in BRAFV600-mutant advanced melanoma patients: A propensity-matched survival analysis. Br. J. Cancer 2021, 124, 1222–1230. [Google Scholar] [CrossRef]
- Kim, T.; Amaria, R.N.; Spencer, C.; Reuben, A.; Cooper, Z.A.; Wargo, J.A. Combining targeted therapy and immune checkpoint inhibitors in the treatment of metastatic melanoma. Cancer Biol. Med. 2014, 11, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Barrios, D.M.; Do, M.H.; Phillips, G.S.; Postow, M.A.; Akaike, T.; Nghiem, P.; Lacouture, M.E. Immune checkpoint inhibitors to treat cutaneous malignancies. J. Am. Acad. Dermatol. 2020, 83, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Harlin, H.; Meng, Y.; Peterson, A.C.; Zha, Y.; Tretiakova, M.; Slingluff, C.; McKee, M.; Gajewski, T.F. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer. Res. 2009, 69, 3077–3085. [Google Scholar] [CrossRef]
- Ulloa-Montoya, F.; Louahed, J.; Dizier, B.; Gruselle, O.; Spiessens, B.; Lehman, F.F.; Suciu, S.; Kruit, W.H.J.; Eggermont, A.M.M.; Vansteenkiste, J.; et al. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J. Clin. Oncol. 2013, 31, 2388–2395. [Google Scholar] [CrossRef]
- Trujillo, J.A.; Sweis, R.F.; Bao, R.; Luke, J.J. T Cell-Inflamed versus Non-T Cell-Inflamed Tumors: A Conceptual Framework for Cancer Immunotherapy Drug Development and Combination Therapy Selection. Cancer Immunol. Res. 2018, 6, 990–1000. [Google Scholar] [CrossRef]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015, 523, 231–235. [Google Scholar] [CrossRef]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef]
- Herbst, R.S.; Soria, J.C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Milazzo, G.; Mercatelli, D.; Muzio, G.D.; Triboli, L.; De Rosa, P.; Perini, G.; Giorgi, F.M. Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability. Genes 2020, 11, 556. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef]
- Que, Y.; Zhang, X.L.; Liu, Z.X.; Zhao, J.J.; Pan, Q.Z.; Wen, X.Z.; Xiao, W.; Xu, B.S.; Hong, D.C.; Guo, T.H.; et al. Frequent amplification of HDAC genes and efficacy of HDAC inhibitor chidamide and PD-1 blockade combination in soft tissue sarcoma. J. Immunother. Cancer 2021, 9, e001696. [Google Scholar] [CrossRef]
- Zheng, H.; Zhao, W.; Yan, C.; Watson, C.C.; Massengill, M.; Xie, M.; Massengill, C.; Noyes, D.R.; Martinez, G.V.; Afzal, R.; et al. HDAC Inhibitors Enhance T-Cell Chemokine Expression and Augment Response to PD-1 Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2016, 22, 4119–4132. [Google Scholar] [CrossRef]
- Yeon, M.; Kim, Y.; Jung, H.S.; Jeoung, D. Histone Deacetylase Inhibitors to Overcome Resistance to Targeted and Immuno Therapy in Metastatic Melanoma. Front. Cell Dev. Biol. 2020, 8, 486. [Google Scholar] [CrossRef]
- Deng, S.; Hu, Q.; Zhang, H.; Yang, F.; Peng, C.; Huang, C. HDAC3 Inhibition Upregulates PD-L1 Expression in B-Cell Lymphomas and Augments the Efficacy of Anti-PD-L1 Therapy. Mol. Cancer Ther. 2019, 18, 900–908. [Google Scholar] [CrossRef]
- Levidou, G.; Gajdzis, P.; Cassoux, N.; Donizy, P.; Masaoutis, C.; Gajdzis, M.; Gardrat, S.; Pergaris, A.; Danas, E.; Klijanienko, J.; et al. Histone Deacetylase (HDAC)-1, -2, -4, and -6 in Uveal Melanomas: Associations with Clinicopathological Parameters and Patients’ Survival. Cancers 2021, 13, 4763. [Google Scholar] [CrossRef]
- Shu, X.; Cheng, H.; Shu, S.; Tang, S.; Li, K.; Dong, Z. Correlation between HDAC4 enhancer DNA methylation and mRNA expression during palatal fusion induced by all-trans retinoic acid. J. Cell. Biochem. 2018, 119, 9967–9973. [Google Scholar] [CrossRef]
- Nada, S.; Kahaleh, B.; Altorok, N. Genome-wide DNA methylation pattern in systemic sclerosis microvascular endothelial cells: Identification of epigenetically affected key genes and pathways. J. Scleroderma Relat. Disord. 2022, 7, 71–81. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- De Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef] [PubMed]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Li, B.; Severson, E.; Pignon, J.-C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Rodriguez, I.P.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Booth, L.; Roberts, J.L.; Poklepovic, A.; Kirkwood, J.; Dent, P. HDAC inhibitors enhance the immunotherapy response of melanoma cells. Oncotarget 2017, 8, 83155–83170. [Google Scholar] [CrossRef] [PubMed]
- Banik, D.; Moufarrij, S.; Villagra, A. Immunoepigenetics Combination Therapies: An Overview of the Role of HDACs in Cancer Immunotherapy. Int. J. Mol. Sci. 2019, 20, 2241. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, S.; Redon, C.E.; Peer, C.J.; Bryla, C.; Lee, M.J.; Trepel, J.B.; Tomita, Y.; Rajan, A.; Giaccone, G.; Bonner, W.M.; et al. Phase I trial of belinostat with cisplatin and etoposide in advanced solid tumors, with a focus on neuroendocrine and small cell cancers of the lung. Anticancer Drugs 2018, 29, 457–465. [Google Scholar] [CrossRef]
- Ji, R.R.; Chasalow, S.D.; Wang, L.; Hamid, O.; Schmidt, H.; Cogswell, J.; Alaparthy, S.; Berman, D.; Jure-Kunkel, M.; Siemers, N.O.; et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol. Immunother. 2012, 61, 1019–1031. [Google Scholar] [CrossRef]
- Ugurel, S.; Schrama, D.; Keller, G.; Schadendorf, D.; Brocker, E.B.; Houben, R.; Zapatka, M.; Fink, W.; Kaufman, H.L.; Becker, J.C. Impact of the CCR5 gene polymorphism on the survival of metastatic melanoma patients receiving immunotherapy. Cancer Immunol. Immunother. 2008, 57, 685–691. [Google Scholar] [CrossRef]
- Uccellini, L.; De Giorgi, V.; Zhao, Y.; Tumaini, B.; Erdenebileg, N.; Dudley, M.E.; Tomei, S.; Bedognetti, D.; Ascierto, M.L.; Liu, Q.; et al. IRF5 gene polymorphisms in melanoma. J. Transl. Med. 2012, 10, 170. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Fessler, J.L.; Gajewski, T.F. The Microbiota: A New Variable Impacting Cancer Treatment Outcomes. Clin. Cancer Res. 2017, 23, 3229–3231. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Michalak, E.M.; Burr, M.L.; Bannister, A.J.; Dawson, M.A. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 573–589. [Google Scholar] [CrossRef]
- Goutas, D.; Theocharis, S.; Tsourouflis, G. Unraveling the Epigenetic Role and Clinical Impact of Histone Deacetylases in Neoplasia. Diagnostics 2021, 11, 1346. [Google Scholar] [CrossRef]
- Falkenberg, K.J.; Johnstone, R.W. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014, 13, 673–691. [Google Scholar] [CrossRef] [PubMed]
- Mann, B.S.; Johnson, J.R.; Cohen, M.H.; Justice, R.; Pazdur, R. FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist 2007, 12, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Coiffier, B.; Pro, B.; Prince, H.M.; Foss, F.; Sokol, L.; Grenwood, M.; Caballero, D.; Borchmann, P.; Morschhauser, F.; Wilhelm, M.; et al. Results from a pivotal, open-label, phase II study of romidepsin in relapsed or refractory peripheral T-cell lymphoma after prior systemic therapy. J. Clin. Oncol. 2012, 30, 631–636. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, O.A.; Horwitz, S.; Masszi, T.; Van Hoof, A.; Brown, P.; Doorduijn, J.; Hess, G.; Jurczak, W.; Knoblauch, P.; Chawla, S.; et al. Belinostat in Patients with Relapsed or Refractory Peripheral T-Cell Lymphoma: Results of the Pivotal Phase II BELIEF (CLN-19) Study. J. Clin. Oncol. 2015, 33, 2492–2499. [Google Scholar] [CrossRef]
- Borcoman, E.; Kamal, M.; Marret, G.; Dupain, C.; Castel-Ajgal, Z.; Tourneau, C.L. HDAC Inhibition to Prime Immune Checkpoint Inhibitors. Cancers 2021, 14, 66. [Google Scholar] [CrossRef]
- Baba, Y.; Huttenhower, C.; Nosho, K.; Tanaka, N.; Shima, K.; Hazra, A.; Schernhammer, E.S.; Hunter, D.J.; Giovannucci, E.L.; Fuchs, C.S.; et al. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol. Cancer 2010, 9, 125. [Google Scholar] [CrossRef]
- Pleyer, L.; Greil, R. Digging deep into “dirty” drugs-modulation of the methylation machinery. Drug Metab. Rev. 2015, 47, 252–279. [Google Scholar] [CrossRef]
- Bespalov, D.; Pino, D.; Vidal-Guirao, S.; Franquesa, J.; Lopez-Ramajo, D.; Filgaira, I.; Wan, L.; O’Sullivan, P.A.; Ley, S.C.; Forcales, S.V.; et al. Bioinformatic analysis of molecular characteristics and oncogenic features of CARD14 in human cancer. Sci. Rep. 2024, 14, 22972. [Google Scholar] [CrossRef]
- Wang, G.; Fu, J.; Liu, M.; Zheng, Q. CXC chemokines: Potential biomarker and immunotherapeutic target for uterine corpus endometrial carcinoma. PLoS ONE 2024, 19, e0277872. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Yang, Y. Identification of inhibitory immune checkpoints and relevant regulatory pathways in breast cancer stem cells. Cancer Med. 2021, 10, 3794–3807. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Guo, J.; Deng, S.; Chen, D.; Huang, M. A pan-cancer analysis of the role of USP5 in human cancers. Sci. Rep. 2023, 13, 8972. [Google Scholar] [CrossRef]
- Mak, I.W.Y.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114–118. [Google Scholar] [PubMed]
- Bracken, M.B. Why animal studies are often poor predictors of human reactions to exposure. J. R. Soc. Med. 2009, 102, 120–122. [Google Scholar] [CrossRef]
- Alamoudi, M.K.; Alsaleh, A.A.; Alkholifi, F.K.; Thyagarajan, A.; Sahu, R.P. A New Strategy to Boost the Efficacy of Melanoma Immune Checkpoint Inhibitor by Targeting HDAC4 to Modify the T-cell Inflamed Signature. J. Pharmacol. Exp. Ther. 2024, 389, 56. [Google Scholar] [CrossRef]
- Alamoudi, M.K.; Alsaleh, A.A.; Thyagarajan, A.; Sahu, R.P. A novel method to boost melanoma ICIs efficacy by targeting HDAC4 to modulate T-cell inflamed signature. In Proceedings of the 29th Dubai International Pharmaceutical & Technologies Conference & Exhibition—DUPHAT 2024, Dubai, United Arab Emirates, 9–11 January 2024. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alamoudi, M.K.; Alsaleh, A.A.; Thyagarajan, A.; Alkholifi, F.K.; Raza, M.L.; Sahu, R.P. Elevated HDAC4 Expression Is Associated with Reduced T-Cell Inflamed Tumor Microenvironment Gene Signatures and Immune Checkpoint Inhibitor Effectiveness in Melanoma. Cancers 2025, 17, 1518. https://doi.org/10.3390/cancers17091518
Alamoudi MK, Alsaleh AA, Thyagarajan A, Alkholifi FK, Raza ML, Sahu RP. Elevated HDAC4 Expression Is Associated with Reduced T-Cell Inflamed Tumor Microenvironment Gene Signatures and Immune Checkpoint Inhibitor Effectiveness in Melanoma. Cancers. 2025; 17(9):1518. https://doi.org/10.3390/cancers17091518
Chicago/Turabian StyleAlamoudi, Mariam K., Abdulmonem A. Alsaleh, Anita Thyagarajan, Faisal K. Alkholifi, Muhammad Liaquat Raza, and Ravi P. Sahu. 2025. "Elevated HDAC4 Expression Is Associated with Reduced T-Cell Inflamed Tumor Microenvironment Gene Signatures and Immune Checkpoint Inhibitor Effectiveness in Melanoma" Cancers 17, no. 9: 1518. https://doi.org/10.3390/cancers17091518
APA StyleAlamoudi, M. K., Alsaleh, A. A., Thyagarajan, A., Alkholifi, F. K., Raza, M. L., & Sahu, R. P. (2025). Elevated HDAC4 Expression Is Associated with Reduced T-Cell Inflamed Tumor Microenvironment Gene Signatures and Immune Checkpoint Inhibitor Effectiveness in Melanoma. Cancers, 17(9), 1518. https://doi.org/10.3390/cancers17091518





